Abstract
Limited knowledge of the molecular evolution of deep-sea fish proteomes so far suggests that a few widespread residue substitutions in cytosolic proteins binding hydrophilic ligands contribute to resistance to the effects of high hydrostatic pressure (HP). Structure-function studies with additional protein systems, including membrane bound proteins, are essential to provide a more general picture of adaptation in these extremophiles. We explored molecular features of HP adaptation in proteins binding hydrophobic ligands, either in lipid bilayers (cytochrome P450 1A - CYP1A) or in the cytosol (the aryl hydrocarbon receptor - AHR), and their partners P450 oxidoreductase (POR) and AHR nuclear translocator (ARNT), respectively. Cloning studies identified the full-length coding sequence of AHR, CYP1A and POR, and a partial sequence of ARNT from Coryphaenoides armatus, an abyssal gadiform fish thriving down to 5000 m depth. Inferred protein sequences were aligned with many non-deep-sea homologs to identify unique amino acid substitutions of possible relevance in HP adaptation. Positionally unique substitutions of various physicochemical properties were found in all four proteins, usually at sites of strong-to-absolute residue conservation. Some were in domains deemed important for protein-protein interaction or ligand binding. In addition, some involved removal or addition of beta-branched residues; local modifications of beta-branched residue patterns could be important to HP adaptation. In silico predictions further suggested that some unique substitutions might substantially modulate the flexibility of the polypeptide segment in which they are found. Repetitive motifs unique to the abyssal fish AHR were predicted to be rich in glycosylation sites, suggesting that post-translational changes could be involved in adaptation as well. Recombinant CYP1A and AHR showed functional properties (spectral characteristics, catalytic activity and ligand binding) that demonstrate proper folding at 1 atmosphere, indicating that they could be used as deep-sea fish protein models to further evaluate protein function under pressure.
1. Introduction
There are more than 32,000 extant fish species, with an extraordinary diversity and ecological range. The number of fully-sequenced fish genomes is expanding rapidly, and will continue in the coming years [1]. The diversity of cytochrome P450 (CYP) sequences in fishes, including the numbers and identity of gene families, subfamilies, and genes per species, is becoming fairly well understood [2, 3]. The diversity of nuclear and cytosolic xenobiotic receptors that participate in regulation of CYP genes also is becoming well known in fishes [4]. However, there are many unanswered questions about the functions of most of the CYPs, and the nuclear and cytosolic receptors responsible for their transcriptional regulation. One question has to do with how structures of these proteins may have changed during evolutionary adaptation to diverse environments on the planet. This study addresses CYP1A and the aryl hydrocarbon receptor (AHR), which regulates CYP1A transcription, and their partner proteins, cloned from fish from the abyssal ocean, and identifies structural features possibly linked to high hydrostatic pressure (HP).
HP increases by about 0.1 MPa for each ten-meter depth in the water column [5], and is thought to shape the vertical distribution of marine biota and drive speciation in the deep-sea [6–8]. Pressure effects on the static and dynamic behavior of biomolecules depend on the magnitude and direction of the volume change (ΔV) upon HP application. Reactions linked to a negative ΔV are promoted and to a positive ΔV are repressed [9]. Changes in cavity volume, bond length and hydration state all contribute to the ΔV term in biomolecular systems [10]. Invasion of the deep-sea by marine fishes seems tightly linked to an evolved minimization of ΔV of their enzyme arsenal. Achieving “life at low volume change” [11] was proposed to involve changes in amino acid residues at sites typically not affecting overall protein structure or basic function, and changes in composition of the intracellular milieu [12]. Amino acid side chains indeed govern the formation of non-covalent bonds and cavities and, in concert with the intracellular milieu, dictate a protein’s hydration state [9, 13].
There have been few structural studies on proteins of deep-sea fish. A few cytosolic enzymes have been shown to have HP-stable Km values for their hydrophilic substrates (e.g., [11, 14]). Lactate dehydrogenase (LDH) from a deep-sea fish has a few amino acid substitutions that appear to be responsible for resistance to HP [14, 15]. Similarly, HP tolerance of deep-sea fish actins seems to be due to the presence of a few residue substitutions [16–18]. Despite the importance of hydrophobic ligand binding in many cell functions, and a controversy regarding the stability of hydrophobic bonds with HP [19], no study has yet focused on the molecular adaptations of deep-sea fish proteins that bind hydrophobic ligands. As well, other than studies of Na+/K+-ATPase in gill membrane [20–22] and brain membrane G-proteins [23–25], there has been little analysis of molecular adaptation of membrane bound proteins in deep-sea fish, including the possible relevance of specific amino acid substitutions.
CYP1A and the AHR are good targets for assessing molecular adaptation in proteins relying on hydrophobic ligand binding, in a lipid (membrane) and an aqueous (cytosolic) environment, respectively. The AHR is a cytosolic basic-helix-loop-helix (bHLH) Per-Arnt-Sim (PAS) protein [26], activated by binding of planar hydrophobic ligands. Activated receptor dimerizes with the AHR nuclear translocator (ARNT; another bHLH-PAS protein), and then activates expression of the gene encoding the detoxification monooxygenase CYP1A and other genes. A flavin-containing P450 oxidoreductase (POR) transfers electrons to CYP1A necessary for catalytic function. While AHR is cytosolic, CYP1A and POR are membrane bound enzymes that function in a lipid environment.
Here we report on the cloning and sequence analysis of AHR, ARNT, CYP1A and POR, and the function of recombinant AHR and CYP1A of Coryphaenoides armatus. This rattail, also known as the abyssal grenadier, is a circumglobal gadiform that dominates megafaunal assemblages at 2000 – 5000 m depth [27, 28]. The results identify unique substitutions, based on alignment with many non-deep sea vertebrate homologs, which may be important in maintaining functionality at pressure.
2. Material and Methods
2.1. Animals
Individuals of C. armatus were retrieved from 3000 m depth in Monterey Bay Canyon during 2009. Within one hour of trawl recovery, specimens were sorted on ice, the liver excised and frozen in liquid nitrogen, and kept at −80°C until further processing.
2.2. Total RNA extraction
Total RNA was isolated from livers of two individuals using the Total Fatty and Fibrous Tissue kit from Bio-Rad (Hercules, USA). Two µg of total RNA from each liver were subjected to electrophoresis on 1% formaldehyde denaturing agarose gels to check for RNA integrity. Purity and concentration were determined with a Nanodrop ND-1000 (Wilmington DE, USA) with OD 260/280 and OD 260/230.
2.3. First-fragment PCR
As no C. armatus sequence information was available in databases, we first PCR amplified a cDNA fragment for each target sequence, using an ABI 2700 thermocycler (Grand Island, USA) and the Advantage 2 PCR kit (Clontech, Mountain View CA, USA). Primers were designed to consensus regions of non-deep-sea fish homologs (Table S1). Reverse transcription of 1-µg aliquots of total RNA from each sample was done with the iScript cDNA Synthesis kit (Bio-Rad, Hercules CA, USA). The 50-µl reactions contained 1 µM of primers and 1-µl aliquots of cDNA. The thermal profile was: 94°C for 1 min; 94°C for 15 s and 60°C for 30 s (35 cycles); 68°C for 5 min. The annealing-and-extension temperature was set at 55°C for both AHR and POR.
Aliquots of PCR products (15-µl) were resolved on 1.2% (w/v) agarose gels; for those samples yielding bands of the expected size, the remaining reaction volume was resolved on 1.2% (w/v) agarose gels and the material purified using the QIAquick Gel Extraction kit from Qiagen (Valencia CA, USA) following manufacturer’s guidelines. Gel-extracted amplicons (3 µl) were cloned into pGEM-T Easy (Promega, Madison WI, USA). Plasmid DNA was isolated with the QIAprep Spin Miniprep kit (Qiagen). Three clones of each amplicon were sent to Eurofins MWG Operon (Huntsville AL, USA) for forward and reverse sequencing with M13 primers.
2.4. RACE-PCR
The SMARTer RACE cDNA Amplification kit from Clontech was used. For each sample, two ligand-adapted cDNA libraries were derived from 1 µg of total RNA (5'- and 3'-RACE ready). Gene-specific primers were designed using sequence information from Section 2.3. (Table S1). Touchdown PCR reactions were usually followed by nested PCR. Multiple 3'-RACE reactions, with additional gene-specific forward primers, were often required. The touchdown thermal profile was: 94°C for 30 s and 72°C for 3 min (5 cycles); 94°C for 30 s, 70°C for 30 s and 72°C for 3 min (5 cycles); 94°C for 30 s, 68°C for 30 s and 72°C for 3 min (25 cycles); 72°C for 7 min. The nested PCR profile was: 94°C for 30 s, 68°C for 30 and 72°C for 3 min (25 cycles); 72°C for 7 min. Electrophoresis, gel extraction and cloning-sequencing procedures followed protocols above, except that gene-specific primers were sometimes used for sequencing.
2.5. Full-length cDNA
Conventional PCR was performed using additional primers (Table S1), to ensure that the 5' and 3' cDNA fragments amplified in Section 2.4. corresponded to the same RNA, and to obtain full-length coding sequences for heterologous expression. The thermal profile followed that of a nested RACE-PCR, except that the extension step was increased to 4 min for AHR. The cDNA libraries from Section 2.3 were used, and primers were tested at 1 µM. Electrophoresis, gel extraction and subsequent cloning-sequencing procedures were as in Section 2.3., except that gene-specific primers were sometimes used for sequencing. The associated Genbank accession numbers are: KY829467 (AHR), KY829468 (ARNT), KY829469 (CYP1A) and KY829470 (POR).
2.6. Inference of primary and secondary structure, and in silico predictions
The protein sequences of full-length AHR, CYP1A and POR, and a partial sequence for ARNT were inferred from coding sequence, and secondary structure predicted using the consensus information obtained with NetSurfP1.1 [29] and PsiPred 3.3 [30] at a cutoff of 80%, as previously performed [31]. Additional in silico analyses addressed the location and identity of functional domains with the NCBI Conserved Domain Database [32], InterPro [33], hydrophobicity (i.e., grand average of hydropathicity, or GRAVY) and instability indexes with ProtParam [34], and glycosylation and myristoylation sites with PredictProtein [35]. MacVector 12.0.2 (Oxford Molecular Group, Madison WI, USA) was used to predict protein aliphatic and flexibility indices at atmospheric pressure (0.1 MPa). The latter index was used to estimate possible variations of local rigidity between aligned regions of surface and deep-sea fish homologs. A threshold of 0.025 (absolute variation of index) was applied arbitrarily for relative comparisons. To add stringency, only segments with 3 or more consecutive residues showing variation above this threshold were considered.
Well-curated homologous sequences available in Ensembl or GenBank were aligned with the 4 inferred sequences of C. armatus, using MacVector. The accession numbers for the non-deep-sea proteins and the associated BlastP data are in Tables S2 and S3, respectively. Sequence analyses of residue “burial” were conducted with NetsurfP1.1 and PsiPred 3.3. Estimation of the volume change associated with a particular residue substitution was based on the information available in Harpaz et al. [36], and determination of the change in residue hydrophobicity was obtained from [37].
2.7. Phylogenetic analyses
Phylogenetic relationships of AHR and ARNT sequences were inferred by using the Maximum Likelihood method based on the JTT matrix-based model [38]. Initial tree(s) for a heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. All positions containing gaps and missing data were eliminated. Residues corresponding to 1–408 (N-terminal half) of the mouse AHR were used in the analysis, because the substantial divergence in the C-terminal half of the proteins precluded the construction of a well-supported multiple alignment in this region. There were a total of 282 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [39].
Phylogenetic relationships of POR and CYP1A were inferred using the maximum likelihood method in RAxML (v8.2.8) [40, 41], using the LG matrix [42] and the PROTCAT model of amino acid substitution. All positions of uncertain alignment were eliminated, based on the internal alignment scoring function of Muscle (v3.8.31) [43].
2.8. In vitro protein expression and functional analyses
Expression and Isolation of CYP1A and reconstitution of catalytic activity
The C. armatus CYP1A protein was heterologously expressed in Escherichia coli employing methods described for the successful expression of many eukaryotic P450s [44]. Briefly, the first 8 amino acids were changed to MALLLAVF and a tag encoding four histidines was inserted immediately before the stop codon. A NdeI restriction site spanning the start codon was introduced at the 5'-end sequence, and the 3'-flanking sequence contained a HindIII restriction site. This modified gene was cloned into the NdeI/HindIII digested pCWori+ expression plasmid and transformed into competent E. coli DH5α cells. Gene integrity was confirmed by DNA sequencing. Preparation of enzyme samples and spectrophotometric analysis of the reduced, CO-ligated protein was carried out as described previously [45]. 7-Ethoxyresorufin O-deethylase (EROD) activity of recombinant C. armatus CYP1A was assayed as described previously [46], by fluorometric detection of resorufin. Rat cytochrome P450 reductase (POR) was used to drive CYP1A activity and all measurements were performed in triplicate.
AHR Ligand binding assay
For protein expression analysis, C. armatus AHR2 construct was expressed in vitro using the T7 TNT-Quick Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine, and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by fluorography. AHR ligand-binding was determined using unlabeled protein by velocity sedimentation (sucrose density gradient centrifugation) analysis in a vertical tube rotor as described previously [47]. The TNT reactions were incubated overnight at 4°C with 2 nM [3H]TCDD; nonspecific binding was determined using reactions containing an empty vector (unprogrammed lysate, UPL).
2.9 Homology modeling and location of residues
Homology models were built based on the crystal structures for human CYP1A1 [PDB:4IV8] [48] or the bHLH-PAS domains of mammalian EPAS1 [PDB:1P97], BMAL1 [PDB:4F3L], and HIF1A [PDB:4H6J] for AHR. Sequences were initially aligned using ClustalW and refined using the salign_2d function of Modeller (v 9.16; [49, 50]). Default parameters in Modeller were applied, excluding water molecules and ions that were part of any of the templates, with the exception of the heme and heme iron. Homology modeling was carried out by satisfaction of spatial restraints using the automodel function of Modeller, with very thorough VTFM, thorough MD, and two repeated cycles of minimization. Fifty randomly seeded models were generated for each protein. Both Procheck [51, 52] and the Modeller zDOPE score [53] were used to assess model quality and pick one best homology model for further work. The APBS plugin [54, 55] for Pymol (Schrodinger LLC) was used to evaluate surface charge distribution. Evaluation of potential substrate access channels was performed using CAVER 3.0 [56].
3. Results
3.1. CYP1A and POR
3.1.1. Phylogeny and sequence analysis
CYP1A
The cloned abyssal fish CYP1A encodes a protein of 521 aa that shares 60–87% identity with CYP1A orthologs of surface fish, with the highest identity being to CYP1A of the shallow water cod Gadus morhua (Figure S1). A slightly higher content of the basic residue Lys and the acidic residue Glu are seen in the abyssal fish enzyme compared to G. morhua CYP1A. The content of Pro and Ile was also higher in the abyssal CYP1A, and the content of non-polar residues (especially Val, Leu and Met) was slightly lower (Table S4). Modest decreases of the hydrophobicity and instability indices were associated with such changes (Table S4). No additional loops or helices are evident in the abyssal monooxygenase, and the P450 signature motifs are also typical of vertebrate CYP1s.
Alignment of known CYP1A and POR sequences reveal unique residue substitutions in C. armatus enzymes (Table 1). The abyssal fish CYP1A has four unique substitutions in the primary structure (Figure 1A). Ile 91 is located within a beta strand, about 20 aa upstream of SRS 1, at a site of absolute residue conservation in all other vertebrate CYP1As. This Leu-to-Ile transition (non-polar for non-polar beta branched) is associated with an increased residue hydrophobicity. Tyr 225 occurs within an alpha helix in SRS 2 at the surface of the protein, adjacent to a substrate access channel. This unique substitution, from a Leu-to-Tyr transition (non-polar for polar aromatic), results in an increased side chain volume and reduced residue hydrophobicity. Cys 376 occurs at a solvent-accessible site within the K helix, at the interface to helix J, and is located a few residues upstream of the K-helix signature motif and SRS 5. The Phe-to-Cys transition (non-polar aromatic for thiol-containing polar) leads to a strongly reduced side chain volume and a slightly reduced hydrophobicity. The unique substitution Lys 499 is located at a site of moderate residue conservation in vertebrates (though a much higher level of conservation is seen when only fish sequences are considered; Table 1). Lys 499 is located adjacent to a substrate access channel, and this substitution can be regarded as either a Glu-to-Lys (negatively charged polar for positively charged polar) or an Ile-to-Lys transition (non-polar, beta branched for positively charged polar). The former transition is associated with an increased side chain volume, a slightly reduced hydrophobicity and charge reversal. The latter is associated with a strongly reduced hydrophobicity, the removal of beta branching and appearance of a positive charge.
Table 1.
Positionally-unique residues identified in Coryphaenoides armatus CYP1A and POR protein sequences. Primary sequences were inferred from the full-length coding sequences amplified by polymerase chain reactions, and were aligned with the highest number of homologous fish sequences available in databases (as of September 2016) using MacVector software. The location and identity of the unique substitutions identified this way are shown, along with corresponding residues in shallower-living fish homologs and residue depth predictions (E for exposed, B for buried), obtained with two different open-access software programs (see Material and Methods for details). (N) Refers to the number of species from which orthologous sequences were compared in alignments.
| Protein | Site | Unique residue |
Residues in amphibians birds reptiles and mammals (N) |
Residues in shallower-living fish (N) |
Depth |
|---|---|---|---|---|---|
| CYP1A | 91 | I | L(132) | L(67) | B/B |
| 225 | Y | L(122), M(12), P(2), V(2), D(1), I(1) | L(61), M(8), P(2), V(1) | B/B | |
| 376 | C | F(143), Y(3), V(1) | F(74), Y(3) | B/B | |
| 499 | K | E(59), I(45), Q(15), V(3), T(2), L(2), N(1), A(1) | E(59), Q(2), N(1) | E/B | |
|
| |||||
| POR | 194 | Q | E(71), A(3), N(1) | E(16), A(3), N(1) | E/B |
| 230 | Q | E(77) | E(20) | E/B | |
| 333 | L | N(69), S(6), T(2), I(1) | N(19), I(1) | E/I | |
| 584 | G | D(44), E(11), A(5), T(5), N(5), K(3), S(2), M(1) | T(5), N(5), A(4), K(2), S(2), D(1), M(1) | E/I | |
| 622 | N | G(44), D(21), A(5), E(4), Q(1), R(1) | D(16), E(3), Q(1) | E/E | |
| 654 | P | E(43), T(18), S(9), D(3), A(2), N(1) | T(16), S(3) | E/E | |
|
| |||||
| 560* | L | V(17), I(3) | B/B | ||
| 588* | S | T(20) | B/I | ||
| 644* | C | Y(15), S(4) | E/E | ||
| 656* | A | T(15), P(2), Y(2) | E/I | ||
Figure 1.
Homology models of C. armatus CYP1A (A) and POR (B) showing location of residues unique to these deep-sea proteins. The models were generated using Modeller. The CYP1A structure was based on human crystal structures for CYP1A, and the access channels were calculated using Caver (see Methods). The POR model was based on both human and rat POR crystal structures (see Methods). The CYP1A model shows unique amino acids in blue, the locations of several calculated substrate access channels, in red, and the heme in green. Note that most of the unique residues are on the surface of the CYP1A, and that two are immediately adjacent to the largest substrate access channels. The POR model shows that most of the unique residues in POR also are on the surface of the protein (residues in orange; residue labeling is C. armatus residue number). The POR cofactors extracted from crystal structures are shown in black.
POR
The cloned abyssal fish POR codes for a protein of 677 aa that shares 76–92% identity with surface fish homologs, and 65–73% with mammalian homologs. As with CYP1A, the highest percentage of identity is found with POR of G. morhua (Fig. S2). The cloned abyssal fish POR contains the typical POR binding motifs, the flavin-binding domains (flavin mononucleotide or FMN, and flavin adenine dinucleotide or FAD) and the nicotinamide adenine dinucleotide phosphate (NADPH) binding domain [57]. Similarly to CYP1A, few changes of amino acid composition are apparent in the abyssal fish reductase, compared to shallower-living species (Table S5). Thr residues were less abundant (−8), while there was an increased content of Pro (+4), Ser (+3) and Leu (+3). The changes result in a slightly higher aliphatic index in C. armatus POR, along with a reduced instability index and a modest increase of the hydrophobicity index (Table S5).
Six unique substitutions were found in C. armatus POR. All are on the surface of the protein (Fig. 1B). Gln 194 is within an alpha helix in the FMN binding domain. The Glu-to-Gln transition (negatively charged polar for uncharged polar) results in an increased side chain volume. A second Glu-to-Gln transition occurs at position 230 within the FMN binding domain. Leu 333 is found within a predicted alpha helix of the connecting domain. It corresponds to an Asn-to-Leu transition that comes with a strong increase of side chain volume and residue hydrophobicity. The unique substitution at position 584 within the NADPH binding domain has high sequence variability in surface vertebrate homologs, especially in fish, but is uniquely a Gly in C. armatus. Asn 622 is also solvent-exposed within the NADPH binding domain. It can either be viewed as a Gly-to-Asn (non-polar for polar) or an Asp-to-Asn (negatively charged polar for uncharged polar) transition. The former comes with a strong increase of side chain volume and a strong reduction of residue hydrophobicity. The latter predominates in fish, and is associated with a slight increase of side chain volume and the removal of a negative charge. The unique substitution at position 654 (i.e., within the NADPH binding domain) can be viewed as either a Glu-to-Pro (negatively charged polar for pyrrolidine-containing non-polar) or a Thr-to-Pro (beta branched, polar for pyrrolidine-containing non-polar) at a solvent-exposed site in a loop. The former comes with a strong increase of residue hydrophobicity. The latter situation predominates in fish, and is associated with a slightly increased side chain volume and increased residue hydrophobicity.
Additional substitutions unique relative only to fish (rather than other vertebrates, where variability masks these residues) were identified (Table 1). All of these are located within the NADPH binding domain. The fish-specific substitution at position 560 is a Leu-to-Val transition (non-polar for non-polar, beta branched) at a buried site within a predicted beta strand. It comes with an increased side chain volume and a slight reduction of residue hydrophobicity. Thr 588 Ser, Tyr 644 Cys, and Thr 656 Ala replace conserved residues and all result in reduced side chain volume, and typically increased residue hydrophobicity (except T588S).
3.1.2. Local flexibility at atmospheric pressure
Local flexibility was predicted for both the surface and deep-sea CYP1A, and sites where flexibility significantly diverged are reported in Figure 2. Significant changes of local flexibility were predicted in the abyssal CYP1A compared to the most identical surface fish homolog. Thus, a region of increased flexibility was found in SRS 3, as well as immediately downstream of SRS 3, 5 and 6. Also, a short region of increased flexibility was found associated with Ile 91, a substitution unique to C. armatus CYP1A.
Figure 2.
Local flexibility analysis for Coryphaenoides armatus CYP1A and POR. Local flexibility values and associated graphs for both CYP1A (A) and POR (B) were predicted through the protein analysis toolbox of MacVector software. Above each plot is a scheme of the linearized primary structure (grey lines) with orange regions corresponding to substrate recognition sites (SRS) for CYP1A, and flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide phosphate (NADPH) binding regions for PORb. Also shown are positionally unique residue substitutions (within vertebrates: red asterisks, within fish: yellow asterisks). Red and green vertical bars respectively reflect regions with increased or reduced basal flexibility in C. armatus when compared to the surface cod Gadus morhua (CYP1A, accession: ENSGMOP00000000312; POR, accession: ENSGMOP00000018048).
Local flexibility was also predicted for POR sequences of surface and deep-sea gadids (Figure 2). Relative increases of local flexibility in C. armatus reductase were found upstream of the FMN binding domain, and within the FAD- and NADPH- binding domains. None of these predicted changes were found associated with unique residue substitutions. Reductions of local flexibility were also identified in the abyssal fish POR (i.e., within the FMN and NADPH binding domains)relative to G. morhua. When only homologous fish sequences were considered to identify “unique” substitutions, two of these were found associated with reduced local flexibility values compared to G. morhua (i.e., Cys 644 and Ala 656, within the NADPH binding domain; see below).
3.2. AHR and ARNT
3.2.1. Phylogeny and sequence analysis
AHR
The cloned AHR encodes a protein of 1239 aa that shares 65–66% identity with the AHR2s from tomcod (Microgadus tomcod) and cod (G. morhua), shallow-living gadiforms closely related to C. armatus. AHRs have highly variable C-terminal transactivation regions, contributing to low identities with surface fish AHRs [47]. When only the first 400 aa of the C. armatus AHR (a region encompassing the bHLH-PAS domains; Figure 3) are considered, shared identities are increased to 87% with AHRs of M. tomcod and G. morhua. Phylogenetic analysis of the C. armatus AHR together with other vertebrate AHRs indicates that it belongs to the AHR2A subfamily (Figure S3).
Figure 3.
Alignment of Coryphaenoides armatus AHR protein with surface cod homologs. The abyssal fish sequence was aligned with homologs of two surface cods (Microgadus tomcod AHR2, Genbank accession: ACI25083.1; Gadus morhua AHR2, Ensembl accession: ENSGMOP00000005051) using MacVector software. Three functional domains--the basic helix-loop-helix (bHLH) and two Per-Arnt-Sim domains (PAS A and B)--are labeled with blue lines above the sequences. Additional sequence motifs are labeled with lines below the sequences; predicted nuclear localization signals (green line), the LxxLL protein-protein interaction motif (black line) and the xLxxLxxLxLx nuclear export signal (orange line). Red brackets indicate the boundaries of the ligand-binding domain (corresponding to mouse aa 230–397 [58]). The region with the repeated motifs is shown with vertical red lines and the NQ repeats are highlighted in red.
The region downstream of the predicted ligand-binding domain (LBD; corresponding to residues 230–397 of the mouse AHR [58]) is quite unusual in the abyssal fish AHR. Two stretches of short repetitive motifs were observed, adjacent to similar stretches also present in M. tomcod (Figure 3). In silico analysis of putative functional domains in these peculiar protein segments suggests the presence of an ARG80 domain (regulator of arginine and related MADS box-containing transcription factors) or a pentapeptide-like motif involved in protein ubiquitination and negative regulation of cell growth (SSF141571 domain). Many putative glycosylation sites are also detected in this region. Glycosylation is known to increase the stability of proteins and limit risks of aggregation [59]. Note that 3'-RACE experiments with cDNA libraries from the two individuals identified transcript variants in that region (not shown). Several additional motifs of possible functional relevance were also identified in the C-terminal region. These are a polyglycine sequence extending from residue 430 to 438, a polyglutamine motif from residue 1021 to 1027 that is close to a similar motif shared with surface living cod homologs (residue 1013 to 1016), and a polyglutamine motif from residue 1138 to 1140.
To the best of our knowledge, the AHR we cloned is the longest so far identified in fish. The inferred C. armatus AHR2 is longer than the M. tomcod AHR variants, by about 130 residues. Slightly lower percentages of non-polar, acidic and basic residues were found in the abyssal fish AHR (Table S6). However, there were a high number of polar residues in the C. armatus AHR, with 20 Gln, 20 Asn, 12 Ser and 8 Thr residues in the repetitive motifs identified (see below). These motifs also are enriched with the non-polar Pro, Val and Met (9, 8 and 7 residues, respectively). Both the aliphatic and hydrophobicity indices of C. armatus AHR2 were lower than those of M. tomcod AHRs, while the instability index was similar (Table S3).
Seven substitutions were identified within the first 400 aa of the abyssal AHR2 (Fig. 3; Table 2), including in the ligand binding domain (LBD; aa 230–397), although none fall within the ligand-binding pocket (residues corresponding to mouse aa 278–384 [60]) within the LBD. Val 5 is found at a site of low residue conservation in fish homologs, and is expected to be solvent exposed. Val 115 and Thr 125 both are located within the PAS A domain. Val 115 is an Ala-to-Val transition (non-polar for non-polar beta branched) calculated to occur at a buried (i.e., non-solvent exposed) position neighboring a beta strand. Such a substitution is associated with a strong increase of hydrophobicity and side chain volume. The latter (Thr 125) is an Ala-to-Thr transition (non-polar for polar, beta-branched) associated with a reduced residue hydrophobicity and a strong increase of side-chain volume, although some surface fish homologs have a Ser residue at position 125. A transition from Ser to Thr (polar for polar, beta-branched) is associated with increases of side chain volume but no major change in hydrophobicity. Thr 199, located a few residues downstream of PAS A, is a Pro-to-Thr transition (pyrrolidine-containing non-polar for polar, beta branched) that is associated with a reduced hydrophobicity and few changes of side chain volume. Ala 255 and Ala 268 are located on both edges of a putative beta strand upstream of PAS B and the ligand-binding pocket [60] but within the LBD. Interestingly, both substitutions are Pro-to-Ala transitions (pyrrolidine-containing non-polar for non-polar). Such substitutions are associated with mild increases of hydrophobicity and reductions of side chain volume at both positions. Cys 390 is located downstream of the ligand binding pocket [60] (but within the LBD) at a solvent-exposed position, and is an Arg-to-Cys transition (positively-charged polar for thiol-containing polar) associated with a strong reduction of side chain volume and a strong increase of residue hydrophobicity.
Table 2.
Positionally-unique residues identified in Coryphaenoides armatus AHR and ARNT protein sequences. Primary sequences were inferred from the full-length (AHR) and partial (ARNT) coding sequences amplified by polymerase chain reactions. These were aligned with the highest number of homologous fish sequences available in databases (as of September 2016) using MacVector software. The location and identity of the unique substitutions identified this way are shown, along with corresponding residues in surface fish homologs and residue depth predictions (E for exposed, B for buried) obtained with two different open-access online software (see Material and Methods for details). (N) Refers to the number of species from which orthologous sequences were compared in alignments.
| Protein | Site | Unique residue |
Residues in Shallower-living fish (N) |
Depth | ||
|---|---|---|---|---|---|---|
| AHR2 | 5 | V | P(8), T(8), A(6), G(4), I(2), N(1) | E/E | ||
| 115 | V | A(32), S(1) | B/B | |||
| 125 | T | A(21), S(13) | B/B | |||
| 199 | T | P(34), L(2), F(1) | E/B | |||
| 255 | A | P(23), S(7), V(3), L(1), I(1) | B/E | |||
| 268 | A | P(17), T(9), Q(5), S(3), N(2), R(1) | E/B | |||
| 390 | C | R(31), H(1) | E/E | |||
|
| ||||||
| ARNT1 (partial) | 16 | H | D(12), N(2), E(2) | E/E | ||
| 25 | S | G(16), I(1) | E/E | |||
| 28 | E | A(12), G(2), N(2), T(1) | E/E | |||
| 29 | A | S(12), T(2), N(1) | B/E | |||
| 33 | L | V(13), G(3), C(1) | B/E | |||
| 36 | Q | K(18) | E/E | |||
| 44 | E | A(15), T(2), S(1) | E/E | |||
| 46 | A | D(18) | E/E | |||
| 55 | D | No residue | E/E | |||
| 134 | S | G(17), A(2) | E/E | |||
| 136 | A | No residue | E/B | |||
| 191 | A | S(19) | E/B | |||
| 245 | L | M(19) | E/B | |||
| 308 | A | V(18), M(1) | B/B | |||
| 309 | T | S(17), N(2) | E/E | |||
| 310 | A | L(19) | B/B | |||
| 347 | M | V(19) | E/E | |||
ARNT
It was not possible to get the full-length coding region for the C. armatus ARNT despite several attempts with various primers and RACE-ready libraries coming from different fish. Amplicons from such reactions always had intronic sequences downstream of residue 453. Attempts to circumvent the problem by designing primers for full-length cDNA amplification by conventional PCR (i.e., using a reverse primer that was designed based on consensus region of surface fish homologs) proved unsuccessful (not shown). Thus, characterization is based on the partial sequence, containing the bHLH and the two PAS domains (Figure S4).
The partial C. armatus ARNT is 90% identical to an ARNT isoform of M. tomcod and 89% to an ARNT1 isoform of G. morhua. Phylogenetic analysis supports the inclusion of the abyssal sequence in the ARNT1 subfamily (Supplemental Figure S5). A slightly lower content of the non-polar Ile (−4 residues) and Leu (+3 residues) was found in the abyssal ARNT sequence, along with a slightly higher content of the polar Ser (+4 residues), and of the non-polar Ala (+3 residues) and Met (+3 residues) (Table S7). The aliphatic and instability indices of C. armatus ARNT are lower than those in M. tomcod, and there also was a slightly decreased hydrophobicity index (Table S7).
The partial ARNT1 sequence identified contained the bHLH and PAS domains typical of this family of proteins [61] (Figure S4). Two additional residues are present in the C. armatus ARNT1 compared to cod homologs (positions 55 and 136). The former corresponds to an additional Asp residue in a polyaspartate motif extending from residue 46 to 56. The ARNT from C. armatus is suggested to contain one fewer glycosylation site than the M. tomcod ARNT1 (Table S7).
Substitutions identified in the ARNT1 of C. armatus were based on alignment with a smaller set (<20) of surface fish homologs. Seventeen substitutions were identified, nine of which are located in the first few dozen residues (Table 2). A substantial number could represent false positives due to the limited amount of aligned shallow fish homologs. It seems reasonable, nevertheless, to discuss the possible relevance to HP adaptation of some likely candidates (i.e., those that occur at sites of absolute residue conservation in the few surface fish homologs).
Gln 36 and Ala 46 are located upstream of the bHLH domain, at predicted solvent-exposed positions. Both result in the substitution of charged for uncharged polar residues (i.e., Lys-to-Gln and Asp-to-Ala, respectively). The former change is also associated with a slight reduction of side chain volume, and the latter with a reduction of side chain volume and a strong increase of hydrophobicity. Note that the Asp-to-Ala transition occurs at the beginning of the polyaspartate motif identified above. The Ser-to-Ala transition at position 191 (polar for non-polar), located within PAS A, is associated with a strong increase of hydrophobicity. The Met-to-Leu transition at position 245 within PAS A (non-polar for non-polar) is associated with a strong increase of residue hydrophobicity as well. The Leu-to-Ala transition at position 310 (non-polar for non-polar) comes with a reduced side chain volume and hydrophobicity. The Val-to-Met transition at position 347 within PAS B (beta-branched non-polar for non-polar; solvent-exposed residue) is in the vicinity of putative beta strands. This substitution is associated with an increased side chain volume and a strong reduction of residue hydrophobicity.
3.2.2 Local flexibility at atmospheric pressure
The flexibility of both AHR and ARNT proteins of C. armatus and M. tomcod (i.e., homologous sequences with the highest % identity) at atmospheric pressure (0.1 MPa) was calculated with the alignment software toolbox. Sites where flexibility significantly diverged between the surface and deep-sea fish homologs (i.e., based on an arbitrarily-fixed threshold) are shown in Figure 4. The flexibility values were used to estimate the possible conformational requirements for HP adaptation of the deep-sea receptor system (see Section 2.6.).
Figure 4.
Local flexibility analysis for Coryphaenoides armatus AHR and ARNT. Local flexibility values and associated graphs for both AHR2 (A) and ARNT1 (B) were predicted through the protein analysis toolbox of MacVector software. Above each plot is a scheme of the linearized primary structure (grey lines) with regions corresponding to basic helix-loop-helix (bHLH) and Per-Arnt-Sim (PAS) domains highlighted in orange. Also shown are positionally unique residue substitutions (yellow asterisks), including the most promising ones in the case of ARNT (blue asterisks), and regions that are only found in C. armatus proteins and not Microgadus tomcod homologs (black line; AHR, accession: ACI25083.1; ARNT, accession: ACX53265.1). Red and green vertical bars respectively reflect regions with increased or reduced local flexibility in C. armatus when compared to M. tomcod homologs.
The bHLH domains of the deep-sea and surface fish AHR proteins exhibit similar flexibility values. An increased local flexibility was however noticed in the PAS A domain of C. armatus AHR2, within a segment containing the unique residue substitution Thr 125 (see below). Two regions of increased local flexibility were also identified in PAS B of C. armatus AHR2, but these did not contain unique substitutions. Many regions that are not in the bHLH and PAS domains exhibit increased or decreased flexibility at 0.1 MPa in the abyssal fish AHR. Interestingly, three segments with predicted rigidified conformation at 0.1 MPa in the abyssal fish receptor contain unique substitutions (i.e., Ala 255, Ala 268 and Cys 390; see below). The search for unique residues in C. armatus AHR2 was restricted to the first 400 aa, due to downstream sequence divergence with cod relatives and other surface fishes. It is therefore not possible to determine whether other substitutions unique to the abyssal fish AHR are associated with changes of local flexibility (or other parameters) in the C-terminal region. Note that the repetitive motifs unique to the abyssal fish AHR are moderately to highly flexible. The polyglycine segment extending from residue 430 to 438 and the polyglutamine segments (residue 1022 to 1026 and 1138 to 1140) show high local flexibility.
The comparison of local flexibility (at 0.1 MPa) in the ARNT partial sequence of C. armatus and the corresponding region of M. tomcod ARNT suggested no difference between these homologs in the bHLH domain. However, upstream of bHLH in the abyssal fish ARNT are several “unique” substitutions (i.e., as judged from a limited set of surface fish homologs) that are associated with altered local flexibility at 0.1 MPa. Briefly, Gln 36 is found in a segment of increased local flexibility, while His 16 and Ala 46 are located at sites of reduced local flexibility (compared to M. tomcod). Two PAS A segments exhibit reduced local flexibility in the abyssal fish ARNT. One of these is associated with the “unique” substitution Ala 190 (see below). Also, upstream of the PAS B domain is a short segment of increased flexibility in the abyssal fish ARNT that contains two consecutive “unique” substitutions (i.e., Ala 307 and Thr 308; see below). The “unique” substitution Met 347 (see below) is located in a segment of reduced flexibility.
3.3 Observed trends across proteins
Features of the substitutions in each C. armatus protein were analyzed jointly to decipher commonalities potentially relevant to HP adaptation. Substitutions involving various physicochemical residue types were found widespread in the primary structures, at both buried and exposed positions, and sometimes in functional domains (Table S8). There was a clear enrichment with Ala (non-polar, non-voluminous side chain) among substituting residues, followed by Gln (polar, voluminous side chain), Leu (non-polar, voluminous side chain) and Cys (thiol-containing polar, non-voluminous side chain). Substitutions in the AHR-ARNT pair seem to predominantly involve non-polar residues. The opposite is seen with CYP1A-POR. Several substitutions are linked to the appearance or removal of a beta-branched residue. Changes of residue hydrophobicity (most often, increases are noted) and side chain volume are frequently observed, and overall the four abyssal fish proteins exhibit higher values for GRAVY index.
3.4 Recombinant proteins
Analysis by sucrose density gradient centrifugation at atmospheric pressure showed that the TNT-expressed C. armatus AHR2 protein showed specific binding of [3H]TCDD similar in sedimentation properties and binding capacity to the binding of [3H]TCDD displayed by AHRs from Fundulus heteroclitus (Figure 5).
Figure 5.
Ligand binding assay with Coryphaenoides armatus AHR2. Ligand ([3H]TCDD) specific binding to C. armatus (Ca) AHR2 along with AHR1a and AHR2a from F. heteroclitus (Fh) was assessed using sucrose density gradient centrifugation as described in Methods. A. Confirmation of in vitro expression of C. armatus AHR2 by incorporation of [35S]methionine. Numbers indicate sizes of molecular weight markers. B. Binding of 2 nM [3H]TCDD to the in vitro-expressed AHR proteins. Peaks of specific binding appear in fractions 10–15. Nonspecific binding was determined using reactions containing an empty vector (unprogrammed lysate, UPL).
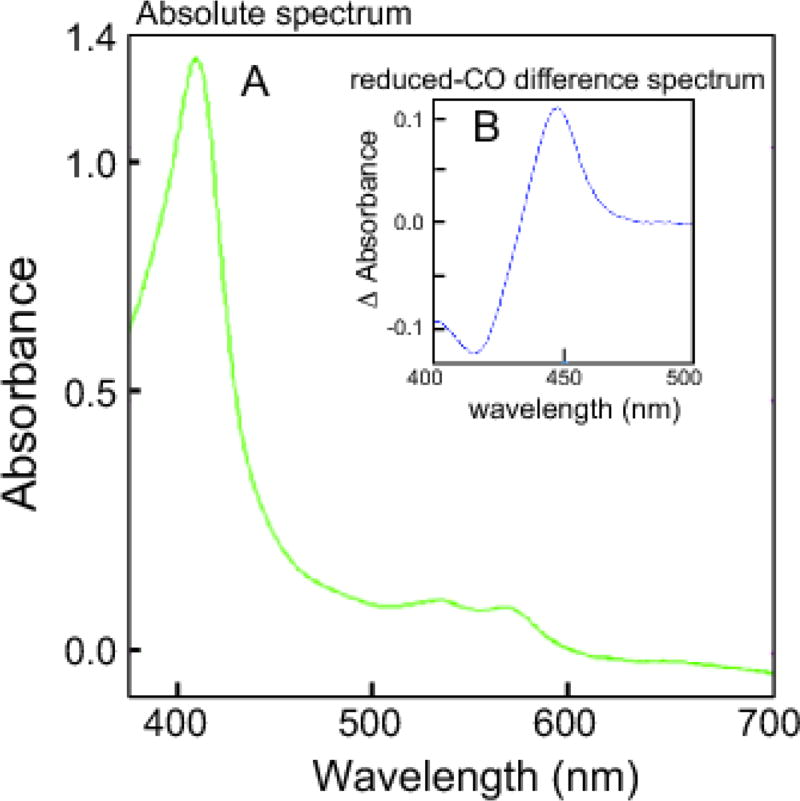
Heterologous expression of C. armatus CYP1A in E. coli yielded 100–200 nmol P450 per liter culture as determined by reduced CO difference spectra. Following purification on Ni2+ -NTA agarose, spectral analysis revealed the oxidized form of the recombinant CYP1A was typical for a low-spin ferric cytochrome P450 enzyme with α, β and γ bands appearing at 565, 535 and 419 nm respectively (Figure 6A). The reduced CO-difference spectrum Soret peak at 450 nm (Figure 6B) indicates a properly folded P450 protein with a cysteine thiolate trans to the CO ligand to the heme iron. Reconstitution of EROD activity with CYP1A and wild type rat P450 reductase gave a turnover rate of 2 nmol product min−1 (nmol P450)−1 at atmospheric pressure, revealing a functionally active P450 enzyme. The EROD turnover activity for recombinant C. armatus CYP1A was comparable to that determined for recombinant human CYP1A1, which was 2.54 nmol product min−1 (nmol P450)−1 [62].
Figure 6.
Spectral Characterization of Coryphaenoides armatus CYP1A. C. armatus CYP1A was heterologously expressed in E. coli cells after N-terminal amino acid modification. A. The absolute oxidized spectrum was typical for a low-spin ferric cytochrome P450 enzyme. B. The reduced CO-difference spectrum Soret peak was located at about 450 nm, indicating a native P450 protein with a cysteine thiolate trans to the CO ligand to the heme iron.
4. Discussion
The ligand activated AHR – CYP1A target gene pathway is a major mechanism by which many xenobiotic chemicals, including PAHs and planar halogenated compounds (some PCB and dioxin congeners) initiate effects in vertebrate animals. The functional properties of the AHR-ARNT and CYP-POR systems have implications for understanding the effects of chemicals that are globally distributed and tend to accumulate in the deep ocean [63]. This is an essential concern in that the deep ocean including the mesopelagic zone are places where chemical effects ultimately may indicate the status of the environmental health of the planet [63–65], The studies here address molecular features of the AHR, ARNT, CYP1A, and POR in a cosmopolitan rattail from the deep ocean. We identify sequence differences between the C. armatus proteins and those of species living at shallow depths, or at 0.1 MPa (atmospheric pressure). Some of these potentially contribute to molecular adaptation to function at high HP.
The rattail C. armatus was chosen for the studies, due to its global distribution and abundance [27]. Also, analysis of liver microsomes of several deep-sea fish species showed C. armatus to have native microsomal P450 (spectrally determined) and higher rates of EROD and benzo[a]pyrene metabolism, both catalyzed prominently by CYP1A, than other species tested [66]. In 1986 Stegeman et al., reported a correlation between levels of hepatic CYP1A (aka P450E) and EROD activity, and the levels of polychlorinated biphenyls in C. armatus, consistent with induction of CYP1A in this species in situ in the deep ocean [63]. Studies also suggested chemical induction of CYP1A in a congeneric though less deep-living species C. rupestris (200 – 2600 m depth) [67]. However, the extent of CYP1A increase in liver slices of C. rupestris exposed to an AHR agonist suggests that the induction response could be diminished under pressure [68]. Analysis of sequences and assays with recombinant systems will help to address the questions of CYP function and AHR responsiveness in fishes in the deep ocean.
Sequence Analysis
A substantial proportion of the substitutions identified are located in or near functional domains deemed important for protein-protein interaction or ligand binding (i.e., PAS A and the PAS B-containing LBD for AHR2, PAS A and B for ARNT1, SRS 2 and 6 for CYP1A, and FMN- and NADPH-binding regions for POR), and some are predicted to occur within secondary structure motifs. Interestingly, 9 out of 27 substitutions are suggested to participate, at least at 0.1 MPa, in changes of the flexibility of the local polypeptide segment in which they are found. Both positive and negative changes are predicted.
CYP1A
Four amino acid substitutions that are unique among the orthologous CYP1As known or inferred from more than 130 species, including more than 60 fishes, were detected in C. armatus CYP1A. That these unique amino acid substitutions occur more on the surface of the protein than in the protein’s core suggests that substrate binding within the active site is not a prime target for adaptation. While some substitutions are within or close to substrate recognition sites [69] and might influence substrate access, it is likely that changes in the hydration of amino acid side chains will affect the volume changes and hence the flexibility and stability of the enzyme associated with function under pressure. Two of the substitutions appear most interesting in relation to hydrophobic ligand binding. First, a leucine-to-isoleucine transition was observed at a site of absolute residue conservation. This unique substitution is suggested to occur within beta sheet 1–2, a region involved in substrate recognition and possibly interacting with the lipid bilayer [70]. Second, the leucine-to-tyrosine transition within SRS 2 suggests a possible role of the aromatic residue in orienting the ligand by means of pressure-stabilized stacking interactions [71]. It could also be that the hydroxyl group acquisition increases interactions with water, thereby reducing system volume.
AHR
The pattern of unique substitutions identified in the abyssal receptor shows that none are located within HLH domain. A similar observation is made with ARNT. Interestingly, 4 substitutions out of 7 concern beta-branched residues. Overall, the sequence modifications appear more complex than for CYP1A. It is perhaps particularly important that the transactivation region of the abyssal receptor contains unique repetitive motifs of possible functional relevance (as suggested by in silico domain predictions) that are notably enriched with serine, proline and threonine. Compared to surface cod homologs, the transactivation region also is enriched with polyglutamine and polyasparagine stretches. The literature suggests that, at least in mammals, proline/serine/threonine-rich regions, along with the latter types of stretches, actively participate in transactivation [72]. This could mean that additional contact points with co-regulators (and perhaps additional co-regulators as well) are needed in the AHR protein for optimal transactivation capacity of the system in the abyss.
POR
Among the substitutions identified in C. armatus POR, there are two glutamate-to-glutamine transition (i.e., from negatively charged to uncharged) in the vicinity of acidic clusters deemed important for CYP-POR interaction - at least in mammals [73]. A similar observation was made for lactate dehydrogenase of the deep-sea Sebastolobus altivelis when compared to the shallower living congeneric S. alascanus [11]. In the former, an asparagine-to-histidine transition is observed, which is suggested to limit volume fluctuation resulting from residue exposure during loop movement [11]. Future studies with the abyssal fish POR will test the hypothesis that charge removal in that region is necessary for protein-protein interaction and efficient electron transfer at abyssal pressures.
The present data also provide further insights into a possible general feature of HP adaptation in fishes. It appears that the set of unique substitutions identified here are associated with the appearance or removal of beta-branched residues (i.e., Val, Ile, Thr). Change in such residues is seen also in deep-sea fish lactate dehydrogenases [14, 74], and actin and myosin [16, 75]. Furthermore, significant changes in the transcriptome-wide content of beta-branched residues are noticed also in bathyal bacteria when compared to an intertidal related species [76]. Beta branching results in bulkier and more rigid side chains. Thus, substitutions promoting or repressing rigidity through local changes of the pattern of beta branched residues could be crucial for HP tolerance. Second, the possible presence of high numbers of glycosylation sites in the repetitive motifs unique to C. armatus AHR2 is clue to a possible implication of post-translational modifications (especially sugar addition) in HP adaptation of deep-sea fish proteins. To our knowledge, this possibility has not yet been studied. It is perhaps relevant that Saccharomyces cerevisiae cultivated in the presence of increasing concentrations of various sugars exhibit increased HP tolerance, presumably due to increased ordering of water molecules [77]. In the same line, glycosylation is considered as a valid bioengineering strategy to improve the stability of protein pharmaceuticals to various physicochemical conditions [59].
Functional assays
The analysis of functional properties of the AHR and the CYP1A are necessary if these recombinant proteins are to be used in assessing HP effects. At 0.1 MPa, the binding capacity of C. armatus AHR2 for TCDD is similar to that seen with other fish AHR2s expressed by in vitro transcription and translation and assayed in the same way. Likewise, the EROD turnover number (2.0 nmol/min/nmol CYP1A) of the C. armatus CYP1A at 0.1 MPa is in the same order of magnitude as that observed with recombinant CYP1A orthologs from other species expressed and assayed the same way, including human CYP1A1 (2.54 nmol/min/nmol CYP1A1) [62]. The turnover of ER by recombinant C. armatus CYP1A also is similar to the turnover of ER by CYP1A estimated in liver microsomes of C. armatus (previously referred to as P450E) [63, 66]. The POR used in reconstitution was recombinant rat POR. Recombinant CYP1A of another fish functioned equally well when reconstituted with rat or fish POR [46], however it is plausible that the deep-sea fish CYP1A may function better with the conspecific reductase, especially under pressure.
The recombinant C. armatus CYP1A spectral properties and activity and the AHR ligand binding data indicate that synthesis at atmospheric pressure results in proteins that are folded properly for function. This sets the stage for direct analysis of the influence of pressure on the functions of the proteins - crucial experiments to address issues of function in situ in the deep sea. It also points to studies with directed alteration of the C. armatus sequences, and the reciprocal mutagenesis of shallow-living or even human CYP orthologs, to determine empirically the role of specific residues in pressure adaptation. Such studies are ongoing in our laboratories.
Supplementary Material
Highlights.
Cytochrome P450 and regulatory components were cloned and sequenced from a deep sea fish Coryphaenoides armatus.
CYP1A, P450 reductase, AH Receptor and ARNT all had unique sequence features compared to homologs in other vertebrates.
Amino acid substitutions in CYP1A and POR were on the surface of the proteins rather than in the active sites.
The changes are predicted to render the enzymes functional at habitat depth, with up to 500 atmospheres pressure.
Recombinant CYP1A and AHR were functional, allowing for studies to confirm the molecular adaptation to pressure in these enzymes.
Acknowledgments
We thank Diana Franks for assistance with the binding assays. This research was supported by a Belgian American Educational Foundation grant to BL, by the Boston University Superfund Research Program NIH 5P42ES007381 (JJS and MEH), by NIH 5U41HG003345 (JVG), by NIH R01ES006272 (MEH), by NSF OCE#0727135 (JCD), and by a Fulbright Scholarship to DCL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koepfli KP, Paten B, Genome KCoS, O’Brien SJ. The Genome 10K Project: a way forward. Annu Rev Anim Biosci. 2015;3:57–111. doi: 10.1146/annurev-animal-090414-014900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jonsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn ME, Karchner SI, Merson RR. Diversity as Opportunity: Insights from 600 Million Years of AHR Evolution. Curr Opin Toxicol. 2017;2:58–71. doi: 10.1016/j.cotox.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders PM, Fofonoff NP. Conversion of pressure to depth in the ocean. Deep-Sea Research. 1976;23 [Google Scholar]
- 6.Morita T. Molecular phylogenetic relationships of the deep-sea fish genus Coryphaenoides (Gadiformes: Macrouridae) based on mitochondrial DNA. Mol Phylogenet Evol. 1999;13:447–454. doi: 10.1006/mpev.1999.0661. [DOI] [PubMed] [Google Scholar]
- 7.Yancey PH, Gerringer ME, Drazen JC, Rowden AA, Jamieson A. Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc Natl Acad Sci U S A. 2014;111:4461–4465. doi: 10.1073/pnas.1322003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaither MR, Violi B, Gray HW, Neat F, Drazen JC, Grubbs RD, Roa-Varon A, Sutton T, Hoelzel AR. Depth as a driver of evolution in the deep sea: Insights from grenadiers (Gadiformes: Macrouridae) of the genus Coryphaenoides. Mol Phylogenet Evol. 2016;104:73–82. doi: 10.1016/j.ympev.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 9.Pradillon F, Gaill F. Pressure and life: some biological strategies. Reviews in Environmental Science and Biotechnology. 2007;6 [Google Scholar]
- 10.Heremans K, Smeller L. Protein structure and dynamics at high pressure. Biochim Biophys Acta. 1998;1386:353–370. doi: 10.1016/s0167-4838(98)00102-2. [DOI] [PubMed] [Google Scholar]
- 11.Somero GN. Life at Low-Volume Change - Hydrostatic-Pressure as a Selective Factor in the Aquatic Environment. American Zoologist. 1990;30:123–135. [Google Scholar]
- 12.Somero GN. Protein adaptations to temperature and pressure: complementary roles of adaptive changes in amino acid sequence and internal milieu. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2003;136:577–591. doi: 10.1016/s1096-4959(03)00215-x. [DOI] [PubMed] [Google Scholar]
- 13.Ohmae E, Gekko K, Kato C. Environmental Adaptation of Dihydrofolate Reductase from Deep-Sea Bacteria. Subcell Biochem. 2015;72:423–442. doi: 10.1007/978-94-017-9918-8_21. [DOI] [PubMed] [Google Scholar]
- 14.Brindley AA, Pickersgill RW, Partridge JC, Dunstan DJ, Hunt DM, Warren MJ. Enzyme sequence and its relationship to hyperbaric stability of artificial and natural fish lactate dehydrogenases. PLoS One. 2008;3:e2042. doi: 10.1371/journal.pone.0002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiguchi Y, Ito N, Okada M. Structure and function of lactate dehydrogenase from hagfish. Marine Drugs. 2010;8 doi: 10.3390/md8030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita T. Structure-based analysis of high pressure adaptation of alpha-actin. J Biol Chem. 2003;278:28060–28066. doi: 10.1074/jbc.M302328200. [DOI] [PubMed] [Google Scholar]
- 17.Morita T. High-pressure adaptation of muscle proteins from deep-sea fishes, Coryphaenoides yaquinae and C. armatus. Ann N Y Acad Sci. 2010;1189:91–94. doi: 10.1111/j.1749-6632.2009.05181.x. [DOI] [PubMed] [Google Scholar]
- 18.Wakai N, Takemura K, Morita T, Kitao A. Mechanism of deep-sea fish alpha-actin pressure tolerance investigated by molecular dynamics simulations. PLoS One. 2014;9:e85852. doi: 10.1371/journal.pone.0085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boonyaratanakornkit BB, Park CB, Clark DS. Pressure effects on intra-and intermolecular interactions within proteins. Biochim Biophys Acta. 2002;1595:235–249. doi: 10.1016/s0167-4838(01)00347-8. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs A, Somero GN. Pressure adaptation of Na+/K+-ATPase in gills of marine teleosts. J Exp Biol. 1989;143:475–492. doi: 10.1242/jeb.143.1.475. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs A, Somero GN. Na+-K+-Adenosine Triphosphatase Activities in Gills of Marine Teleost Fishes - Changes with Depth Size and Locomotory Activity Level. Marine Biology. 1990;106:315–321. [Google Scholar]
- 22.Gibbs A, Somero GN. Pressure Adaptation of Teleost Gill Na+/K+-Adenosine Triphosphatase - Role of the Lipid and Protein Moieties. Journal of Comparative Physiology B-Biochemical Systemic and Environmental Physiology. 1990;160:431–439. [Google Scholar]
- 23.Murray TF, Siebenaller JF. Comparison of the binding properties of A1 adenosine receptors in brain membranes of two congeneric marine fishes living at different depths. Journal of Comparative Physiology Part B. 1987;157 doi: 10.1007/BF00693353. [DOI] [PubMed] [Google Scholar]
- 24.Siebenaller JF. Pressure effects on the GTPase activity of brain membrane G proteins of deep-living marine fishes. Comp Biochem Physiol B Biochem Mol Biol. 2003;135:697–705. doi: 10.1016/s1096-4959(03)00165-9. [DOI] [PubMed] [Google Scholar]
- 25.Siebenaller JF, Hagar AF, Murray TF. The effect of hydrostatic pressure on A1 adenosine receptor signal transduction in brain membranes of two congeneric marine fishes. The Journal of Experimental Biology. 1991;159 [Google Scholar]
- 26.Hahn ME. Aryl hydrocarbon receptors: diversity and evolution. Chem Biol Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 27.King NJ, Bagley PM, Priede IG. Depth zonation and latitudinal distribution of deep-sea scavenging demersal fishes of the Mid-Atlantic Ridge 42 to 53 degrees N. Marine Ecology Progress Series. 2006;319:263–274. [Google Scholar]
- 28.Priede IG, Godbold JA, Niedzielski T, Collins MA, Bailey DM, Gordon JDM, Zuur AF. A review of the spatial extent of fishery effects species vulnerability of the deep-sea demersal fish assemblage of the Porcupine Seabight Northeast Atlantic Ocean (ICES Subarea VII) ICES Journal of Marine Science. 2010;68:281–289. [Google Scholar]
- 29.Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire B, Kubota A, O’Meara CM, Lamb DC, Tanguay RL, Goldstone JV, Stegeman JJ. Cytochrome P450 20A1 in zebrafish: Cloning regulation and potential involvement in hyperactivity disorders. Toxicol Appl Pharmacol. 2016;296:73–84. doi: 10.1016/j.taap.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor M, Lamb DC, Cannell R, Dawson M, Kelly SL. Cytochrome P450105D1(CYP105D1) from Streptomyces griseus: heterologous expression activity and activation effects of multiple xenobiotics. Biochem Biophys Res Commun. 1999;263:838–842. doi: 10.1006/bbrc.1999.1427. [DOI] [PubMed] [Google Scholar]
- 46.Klotz AV, Stegeman JJ, Woodin BR, Snowberger EA, Thomas PE, Walsh C. Cytochrome P-450 isozymes from the marine teleost Stenotomus chrysops: their roles in steroid hydroxylation and the influence of cytochrome b5. Arch Biochem Biophys. 1986;249:326–338. doi: 10.1016/0003-9861(86)90009-3. [DOI] [PubMed] [Google Scholar]
- 47.Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus. Evidence for a novel subfamily of ligand-binding basic helix loop helix-Per-ARNT-Sim (bHLH-PAS) factors. J Biol Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- 48.Walsh AA, Szklarz GD, Scott EE. Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem. 2013;288:12932–12943. doi: 10.1074/jbc.M113.452953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5 Unit 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:561–5632. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 52.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 53.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unni S, Huang Y, Hanson RM, Tobias M, Krishnan S, Li WW, Nielsen JE, Baker NA. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J Comput Chem. 2011;32:1488–1491. doi: 10.1002/jcc.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukunaga BN, Probst MR, Reiszporszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. Journal of Biological Chemistry. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 59.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraccalvieri D, Soshilov AA, Karchner SI, Franks DG, Pandini A, Bonati L, Hahn ME, Denison MS. Comparative analysis of homology models of the AH receptor ligand binding domain: verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochemistry. 2013;52:714–725. doi: 10.1021/bi301457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Powell WH, Karchner SI, Bright R, Hahn ME. Functional diversity of vertebrate ARNT proteins: identification of ARNT2 as the predominant form of ARNT in the marine teleost Fundulus heteroclitus. Arch Biochem Biophys. 1999;361:156–163. doi: 10.1006/abbi.1998.0992. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Ericksen SS, Sivaneri M, Besspiata D, Fisher CW, Szklarz GD. The effect of reciprocal active site mutations in human cytochromes P450 1A1 and 1A2 on alkoxyresorufin metabolism. Arch Biochem Biophys. 2004;424:33–43. doi: 10.1016/j.abb.2003.12.040. [DOI] [PubMed] [Google Scholar]
- 63.Stegeman JJ, Kloepper-Sams PJ, Farrington JW. Monooxygenase Induction and Chlorobiphenyls in the Deep-Sea Fish Coryphaenoides armatus. Science. 1986;231:1287–1289. doi: 10.1126/science.231.4743.1287. [DOI] [PubMed] [Google Scholar]
- 64.Stegeman JJ, Schlezinger JJ, Craddock JE, Tillitt DE. Cytochrome P450 1A expression in midwater fishes: potential effects of chemical contaminants in remote oceanic zones. Environ Sci Technol. 2001;35:54–62. doi: 10.1021/es0012265. [DOI] [PubMed] [Google Scholar]
- 65.Jamieson AJ, Malkocs T, Piertney SB, Fujii T, Zhang Z. Bioaccumulation of persistent organic pollutants in the deepest ocean fauna. Nature Ecology & Evolution. 2017;1:0051. doi: 10.1038/s41559-016-0051. [DOI] [PubMed] [Google Scholar]
- 66.Stegeman JJ. Hepatic Microsomal Monooxygenase Activity and the Biotransformation of Hydrocarbons in Deep Benthic Fish from the Western North Atlantic. Can. J. Fish. Aquat. Sci. 1983;40:78–85. [Google Scholar]
- 67.Lemaire B, Priede IG, Collins MA, Bailey DM, Schtickzelle N, Thomé JP, Rees JF. Effects of organochlorines on cytochrome P450 activity and antioxidant enzymes in liver of roundnose grenadier Coryphaenoides rupestris. Aquatic Biology. 2010;8:161–168. [Google Scholar]
- 68.Lemaire B, Debier C, Calderon PB, Thome JP, Stegeman J, Mork J, Rees JF. Precision-cut liver slices to investigate responsiveness of deep-sea fish to contaminants at high pressure. Environ Sci Technol. 2012;46:10310–10316. doi: 10.1021/es301850e. [DOI] [PubMed] [Google Scholar]
- 69.Gotoh O. Substrate Recognition Sites in Cytochrome P450 Family 2 (CYP2) Proteins Inferred from Comparative Analyses of Amino Acid and Coding Nucleotide Sequence. J. Biol. Chem. 1992;267:83–92. [PubMed] [Google Scholar]
- 70.Graham SE, Peterson JA. How similar are P450s and what can their differences teach us? Arch Biochem Biophys. 1999;369:24–29. doi: 10.1006/abbi.1999.1350. [DOI] [PubMed] [Google Scholar]
- 71.Balny C, Masson P, Heremans K. High pressure effects on biological macromolecules: from structural changes to alteration of cellular processes. Biochim Biophys Acta. 2002;1595:3–10. doi: 10.1016/s0167-4838(01)00331-4. [DOI] [PubMed] [Google Scholar]
- 72.Rowlands JC, McEwan IJ, Gustafsson JA. Trans-activation by the human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins: direct interactions with basal transcription factors. Mol Pharmacol. 1996;50:538–548. [PubMed] [Google Scholar]
- 73.Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- 74.Nishiguchi Y, Miwa T, Abe F. Pressure-adaptive differences in lactate dehydrogenases of three hagfishes: Eptatretus burgeri, Paramyxine atami and Eptatretus okinoseanus. Extremophiles. 2008;12:477–480. doi: 10.1007/s00792-008-0140-3. [DOI] [PubMed] [Google Scholar]
- 75.Morita T. Comparative sequence analysis of myosin heavy chain proteins from congeneric shallow- and deep-living rattail fish (genus Coryphaenoides) J Exp Biol. 2008;211:1362–1367. doi: 10.1242/jeb.017137. [DOI] [PubMed] [Google Scholar]
- 76.Van Campenhout J, Vanreusel A, Van Belleghem S, Derycke S. Transcription Signaling Receptor Activity Oxidative Phosphorylation and Fatty Acid Metabolism Mediate the Presence of Closely Related Species in Distinct Intertidal and Cold-Seep Habitats. Genome Biol Evol. 2015;8:51–69. doi: 10.1093/gbe/evv242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujii S, Obuchi K, Iwahashi H, Fujii T, Komatsu Y. Saccharides that protect yeast against hydrostatic pressure stress correlated to the mean number of equatorial OH groups. Bioscience Biotechnology and Biochemistry. 1996;60:476–478. doi: 10.1271/bbb.60.476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.