Abstract
The eukaryotic translational initiation factor 4G (eIF4G) interacts with the cap-binding protein eIF4E through a consensus binding motif, Y(X)4LΦ (where X is any amino acid and Φ is a hydrophobic residue). 4E binding proteins (4E-BPs), which also contain a Y(X)4LΦ motif, regulate the eIF4E/eIF4G interaction. The non- or minimally-phosphorylated form of 4E-BP1 binds eIF4E, preventing eIF4E from interacting with eIF4G, thus inhibiting translation initiation. 4EGI-1, a small molecule inhibitor of the eIF4E/eIF4G interaction that is under investigation as a novel anti-cancer drug, has a dual activity; it disrupts the eIF4E/eIF4G interaction and stabilizes the binding of 4E-BP1 to eIF4E. Here, we report the complete backbone NMR resonance assignment of an unliganded 4E-BP1 fragment (4E-BP144–87). We also report the near complete backbone assignment of the same fragment in complex to eIF4E/m7GTP (excluding the assignment of the last C-terminus residue, D87). The chemical shift data constitute a prerequisite to understanding the mechanism of action of translation initiation inhibitors, including 4EGI-1, that modulate the eIF4E/4E-BP1 interaction.
Keywords: translation initiation, 4E-BP1, eIF4E, NMR assignment
Biological context
Translation control of gene expression allows cells to respond quickly to external cues. In eukaryotic cells, this regulation occurs mostly at the translation initiation step, during which the ribosome is recruited to messenger RNAs (mRNAs) (Sonenberg and Hinnebusch 2009). Most eukaryotic mRNAs have a cap structure at their 5′ end, which is usually the modified nucleotide, 7-methylguanosine triphosphate (m7GTP). The translational initiation complex assembles at the m7GTP cap through the eIF4F complex (Gingras et al. 1999), which comprises a cap binding protein, eIF4E, a DEAD box RNA helicase, eIF4A, and a large scaffold protein, eIF4G. The scaffold protein eIF4G interacts with eIF4E through a consensus motif, Y(X)4LΦ, where X is any amino acid, and Φ is a hydrophobic residue (Mader et al. 1995; Altmann et al. 1997). eIF4G also interacts with eIF3, which in turn, binds the small ribosomal subunit. Thus, eIF4G, by binding to eIF3 and eIF4E, brings the small ribosomal subunit to the 5′ end of mRNAs, allowing translation initiation to occur. 4E-binding proteins (4E-BPs) regulate the ability of eIF4E to engage eIF4G. Among these, 4E-BP1 is the best characterized. 4E-BP1 contains a Y(X)4LΦ motif and competes with eIF4G for binding to eIF4E (Holz et al. 2005). Only non or hypo-phosphorylated 4E-BP1 binds to eIF4E (Beretta et al. 1996; Gingras et al. 2001).
Several translation initiation factors are overexpressed in cancer cells (Lazaris-Karatzas et al. 1990; Avdulov et al. 2004). Therefore, disrupting protein synthesis at the initiation step has emerged as a new anti-cancer strategy. 4EGI-1 is an eIF4E/eIF4G interaction inhibitor that has anti-cancer activity (Moerke et al. 2007; Chen et al. 2012; Yi et al. 2014). A crystal structure of 4EGI-1 bound to eIF4E revealed that 4EGI-1 induces eIF4G’s dissociation from eIF4E by causing a conformational change in eIF4E (Papadopoulos et al. 2014). The crystal structure of a 4E-BP1 fragment (4E-BP150–84) bound to eIF4E could not explain how 4EGI-1 stabilizes 4E-BP1’s binding to eIF4E (Sekiyama et al. 2015) (Peter et al. 2015). NMR experiments were used to show that 4EGI-1 and 4E-BP1 can indeed bind simultaneously to eIF4E and that they both contribute to inhibiting translation initiation. Here, we report the complete backbone NMR resonance assignment of an unliganded 4E-BP1 fragment (4E-BP144–87). We also report the near complete backbone assignment of the same fragment in complex to eIF4E/m7GTP (excluding the assignment of the last C-terminus residue, D87). We did not pursue the side-chain assignments, as we have previously determined a high resolution crystal structure of the eIF4E/4E-BP144–84 complex (2.1 Å) (Sekiyama et al. 2015), and because the free form of 4E-BP1 is unstructured. The backbone assignments reported here are crucial for understanding the dynamics and binding of small molecules to eIF4E/4E-BP1. Overall, the NMR backbone assignments provided in this study establish a general platform to study translation initiation inhibitors of the cap-binding complex.
Methods and Experiments
Sample preparation
We amplified the cDNA encoding human 4E-BP144–87 (4E-BP144–87) and sub-cloned it into a previously described pH-GB1 Escherichia coli expression vector (Sekiyama et al. 2015). Briefly, we inserted 4E-BP144–87 such that it is expressed as a fusion protein with a hexahistidine (His6) tag, a protein G B1 domain tag (GB1-tag), and a TEV protease cleavage site at its N terminus (pH-TEV-4E-BP144–87). For eIF4E, we used an E. coli expression plasmid containing the full-length cDNA of mouse eIF4E, inserted such that it contains a GB1-tag followed by a TEV cleavage site at its N terminus (pET-GB1-TEV-eIF4E). We used 4E-BP144–87 and eIF4E constructs to transform E. coli (BL21) cells individually. We grew cells expressing 4E-BP144–87 overnight at 20 °C in M9 minimal media containing 95 % 2H2O and appropriate isotopes (15NH4Cl, 13C6-deuterated glucose, as sole nitrogen and carbon sources, respectively). We harvested the cells and lysed these by sonication. After centrifugation, we loaded the supernatant on a metal-affinity column (Ni-NTA; Qiagen). After elution, we concentrated the fusion proteins and incubated overnight with TEV protease to cleave the tags, followed by size-exclusion chromatography using a Superdex 75 16/60 preparative column (GE Healthcare Bio-Sciences). For 15N13C4E-BP1/eIF4E/m7GTP complex preparation, we grew cells expressing 4E-BP144–87 in M9 minimal media as described above and grew cells expressing eIF4E in Luria Broth (LB) overnight at 25 °C. We harvested both groups of cells and combined both pellets before lysing cells by sonication. We then centrifuged the cells and loaded the supernatant containing the 15N13C4E-BP144–87/eIF4E complex on an adipic-agarose-m7GDP column (Edery et al. 1988). For the complex, we also incubated overnight with TEV protease to cleave the tags. The TEV reaction was followed by size-exclusion chromatography using a Superdex 75 16/60 preparative column.
NMR experiments
We recorded all NMR spectra at 298 K on a Varian Inova 600 MHz spectrometer or a Bruker 750 MHz spectrometer, both equipped with cryogenic probes. We processed and analyzed the data using NMRPipe (Delaglio et al. 1995) and CARA (Keller 2004), respectively. We assigned the backbone chemical shifts of 15N13C4E-BP144–87 and 15N13C4E-BP144–87/eIF4E/m7GTP using TROSY versions of the traditional triple-resonance experiments: HNCA, HNCOCA, HNCO, HNCACO, HNCACB, and HNCOCACB. We deuterated all NMR observable samples except for 4E-BP1 alone. We used Non Uniform Sampling (NUS) in the two indirect dimensions to collect triple resonance data and used Poisson Gap Sampling to sample 12–15% of the indirect grid (Hyberts et al. 2010). We used the hmsIST program to reconstruct and process the data (Hyberts et al. 2012).
Assignments and data deposition
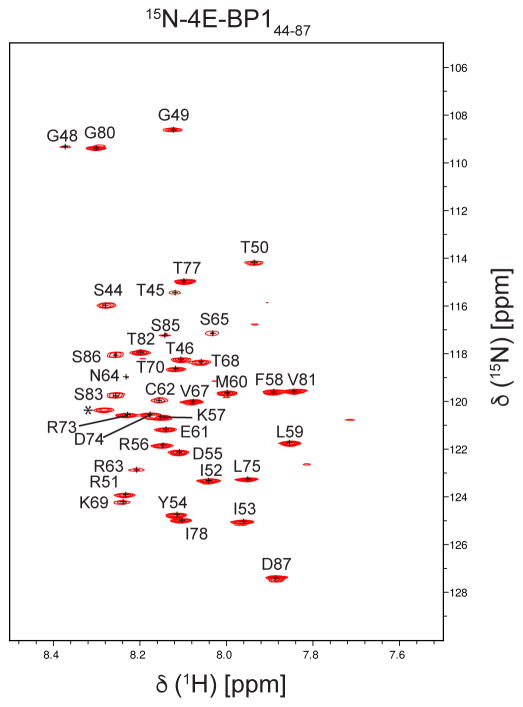
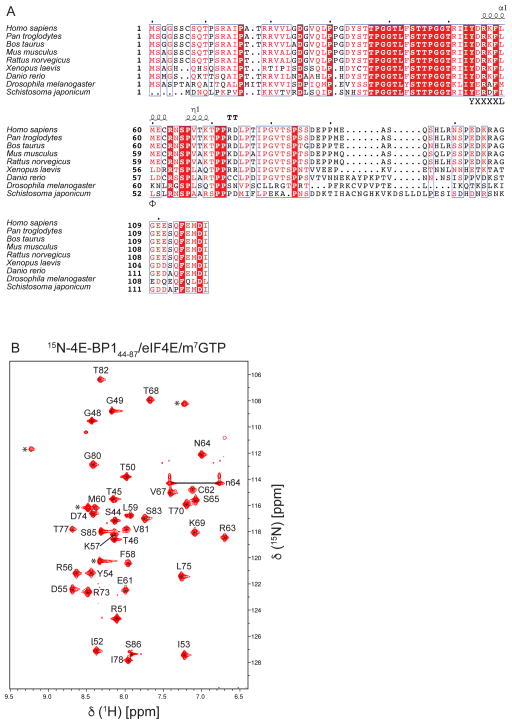
We obtained the complete backbone assignment of a fragment of 4E-BP1 (free form, residues 44–87), as shown in the 15N TROSY-HSQC spectrum (Fig. 1). However, we do lack the assignment of the last residue of 4E-BP144–87 (D87) when it is bound to eIF4E/m7GTP. The lack of assignment for this residue is consistent with the NMR relaxation data showing that when 4E-BP144–87 is bound to eIF4E/m7GTP, the residues S85 and S86 are dynamic, with S86 being even more mobile than S85 (Sekiyama et al. 2015). Moreover, in the spectra shown in Figure 2b, the intensity of the peak corresponding to S86 is a lot weaker and broader than of the peak corresponding to S85. These data agree with the suggestion that the peak corresponding to D87 is also very dynamic and hence, is most likely broadened beyond detection, preventing its assignment. All chemical shifts were deposited in the BioMagResBank (www.bmrb.wisc.edu) under accession number 27003 and 26997 for the free and bound form, respectively.
Fig. 1.
1H–15N TROSY-HSQC of the unbound form of 4E-BP1 (residues 44–87). Peaks indicated by a star may correspond to additional residues coming from the fusion tag after cleavage (not belonging to the native sequence of 4E-BP1), or from the C terminus.
Fig. 2.
A. Sequence alignment of 4E-BPs. Secondary structure prediction of 4E-BP1s per 4E-BP144–87/eIF4E/m7GTP crystal structure (PDB 5BXV). The consensus binding motif of 4E-BP1, YXXXXLΦ, is indicated. The figure was generated using ESPrit3 (Robert and Gouet 2014). B. 1H–15N TROSY-HSQC of 4E-BP1 (residues 44–87) bound to mouse eIF4E/m7GTP. Peaks indicated by a star may correspond to additional residues coming from the fusion tag after cleavage (not belonging to the native sequence of 4E-BP1). Deduced N64 sidechain resonances are indicated by “n64”.
Acknowledgments
This research was supported by NIH Grants GM047467, CA200913, AI108718, and EB002026.
References
- Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdulov S, Li S, Michalek V, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, et al. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Chen L, Aktas BH, Wang Y, et al. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget. 2012;3:869–881. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Edery I, Altmann M, Sonenberg N. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene. 1988;74:517–525. doi: 10.1016/0378-1119(88)90184-9. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Hyberts SG, Milbradt AG, Wagner AB, et al. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J Biomol NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial 2004 [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Papadopoulos E, Jenni S, Kabha E, et al. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc Natl Acad Sci USA. 2014;111:E3187–95. doi: 10.1073/pnas.1410250111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter D, Igreja C, Weber R, et al. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell. 2015;57:1074–1087. doi: 10.1016/j.molcel.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Arthanari H, Papadopoulos E, et al. Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc Natl Acad Sci USA. 2015;112:E4036–45. doi: 10.1073/pnas.1512118112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Kabha E, Papadopoulos E, Wagner G. 4EGI-1 targets breast cancer stem cells by selective inhibition of translation that persists in CSC maintenance, proliferation and metastasis. Oncotarget. 2014;5:6028–6037. doi: 10.18632/oncotarget.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]