Abstract
Generation of intratumoral phenotypic and genetic heterogeneity has been attributed to clonal evolution and cancer stem cells that together give rise to a tumor with complex ecosystems. Each ecosystem contains various tumor cell subpopulations and stromal entities, which depending upon their composition can influence survival, therapy responses and global growth of the tumor. Despite recent advances in breast cancer management, the disease has not been completely eradicated as tumors recur despite initial response to treatment. In this review, using data from clinically relevant breast cancer models, we show that the fates of tumor stem cells/progenitors in the individual tumor ecosystems comprising a tumor are predetermined to follow a limited (unipotent) and/or unlimited (multipotent) path of differentiation which create conditions for active generation and maintenance of heterogeneity. The resultant dynamic systems respond differently to treatments, thus disrupting the delicate stability maintained in the heterogeneous tumor. This raises the question whether it is better then, to preserve stability by preventing take-over by otherwise dormant ecosystems in the tumor following therapy. The ultimate strategy for personalized therapy would require serial assessments of the patient’s tumor for biomarker validation during the entire course of treatment that is combined with their three dimensional mapping to the tumor architecture and landscape.
Keywords: Tumor ecosystem, heterogeneity, stem cells, clinical models, breast cancer
Introduction
Inter- and intratumoral heterogeneity refers to genetic diversity between and within patient tumors, and genomic instability is regarded as a major driving force for tumor heterogeneity. Cancer cells are intrinsically genomically unstable which predisposes them to increased mutation rates resulting in evolution of tumor subpopulations with notably distinct phenotypes. The presence of distinct subpopulations of cells within a tumor with distinguishable differences in tumorigenicity, metastatic potentials and therapy sensitivities was elegantly demonstrated several decades ago [1–3]. The relative abundance of the tumor subclones or subpopulations is dependent upon the selective pressures imposed by genetic- and epigenetic- (microenvironment) mediated constraints that allow tumor subclones to take different routes that enable survival and acquisition of malignant properties. Interestingly, despite the fact that tumor evolution is proposed to follow the laws of Darwinian evolution whereby tumor subclones accumulate new genetic alterations that confer growth, survival and metastatic advantages, it must be recognized that these “evolutionary changes” do not dramatically alter the major lesion morphologies or phenotypes within the tumor. Despite the genetic heterogeneity revealed by deep sequence analysis, breast tumors still preserve the major histotype architectures that pathologists use to classify them as hyperplastic, atypical hyperplasia, ductal carcinoma in situ (DCIS), lobular carcinoma in situ (LCIS), invasive carcinoma, etc., indicating a gap between genetic diversity and phenotypic stability. We propose that maintenance of phenotypic stability of the lesions despite their genetic variabilities is attributed to the presence of progenitor or precursor cells that carry defined sets of genes that “preordain” them to differentiate selectively into a specific histotype (e.g., atypia, any one of the many DCIS subtypes (comedo, cribriform, papillary, etc.), invasive ductal or lobular carcinoma).
Underlying causes of intratumoral heterogeneity
Two models have been proposed for generation of intratumoral heterogeneity – the clonal evolution and the cancer stem cell model, and studies show that these mechanisms are mutually inclusive [4]. In the clonal evolution model, cells acquire mutations that not only give rise to derivatives with different functionalities and behavior but also serve as a platform for further acquisition of genetic alterations. In the continuum of evolution, this process produces tumors with noticeably distinct and variant abilities for survival, malignancy and therapy tolerance at the regional and distant metastatic sites. This model predicts that cancers arise from a single cell [5], which over time can develop various combinations of mutations resulting in genetic drift and selection of the fittest [6–8]. According to the clonal evolution model, cancer progression is non-linear with clones branching out to produce diverse clones, which leads to heterogeneity [4,9]. One of the disadvantages of this model is that it ignores nongenetic variability and does not take into consideration the interactions among clones within the tumor ecosystem [10].
In contrast, the cancer stem cell (CSC) model proposes that only a small subpopulation of tumor cells with stem cell properties drive tumor initiation, progression, and recurrence because of their indefinite self-renewal capability [11,4,5], and eradication of this subpopulation is critical for tumor elimination. CSCs share fundamental properties of stem cells, but harbor tumor-initiating mutations which can be transferred to the progeny [12], and are recently referred as tumor initiating cells (TICs). Two theories have been proposed to explain the origin of CSC: they can arise through mutations in normal stem cells or through the acquisition of mutations in progenitor cells [13]. Heterogeneity in CSCs has been revealed by generation of a variety of differentiation states [14]. As discussed below, our studies suggest that distinct genetic alterations define CSC/TIC subsets which confer them with the ability to generate either unipotent (single phenotype) or multipotent (multiple phenotypes) derivatives (Fig. 1). Recent evidence shows that both of these models are mutually inclusive [4]. This is further amended by the recent hypothesis that differentiation of stem cells is not a unidirectional process as the plasticity of the cells can allow dedifferentiation of the differentiated cells into cells with stem-like properties [15–17]. Regardless of the mechanisms by which intratumoral heterogeneity is generated, the tumor ecosystem consists of variant cell populations that coexist and potentially influence each other’s behavior and survival.
Figure 1.
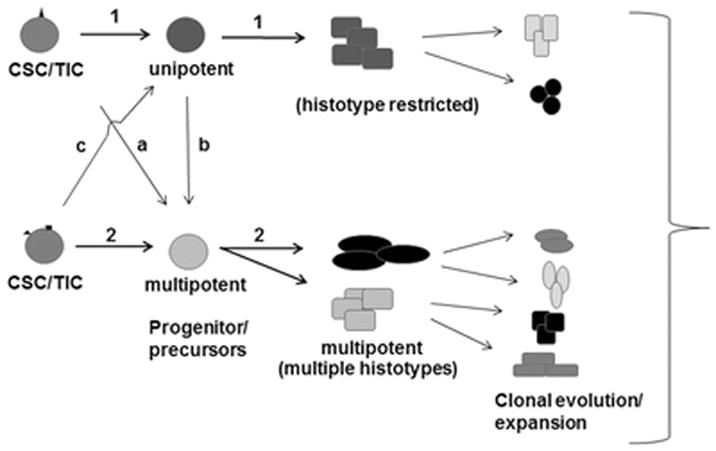
Model for origination of breast cancer heterogeneity. In route (1), CSC/TICs/progenitors differentiate into breast cancer histotypes of a specific lineage (e.g., hyperplasia, a specific DCIS subtype, etc.), signifying limited or restricted differentiation potential, whereas in route (2), multiple histotypes are generated from CSCs/TICs/progenitors suggesting multipotency. The unipotent and multipotent CSCs/progenitors may represent distinct subsets; alternatively, the CSCs/TICs may produce precursor cells that possess the ability to give rise to one or more histotypes (a, b, c). The histotype composition of a breast tumor or “heterogeneity” would depend upon the renewal and differentiation rates and routes taken by the CSCs/TICs/progenitors, and alterations impacted by clonal evolution and expansions of the differentiated derivatives.
Clinically relevant models for investigating the origin of intratumoral heterogeneity and therapy resistance
The MCF10AT xenograft model is a model of early human breast cancer as it faithfully recapitulates the key histogenetic pathways of premalignant breast cancer [18], and thus provides a unique model for studying human breast cancer heterogeneity. MCF10A cells from which MCF10AT cells were derived were established by spontaneous immortalization from benign fibrocystic breast disease [18]. MCF10A cells are non-transformed human breast epithelial cells with a stable pseudodiploid karyotype and possess normal stem cell properties. When orthotopically implanted, they produce normal ducts comprised of luminal and myoepithelial cells with a short life span. Stable transfection of MCF10A cells with mutant Ha-ras preserved the multipotent stem cell property of MCF10A cells as MCF10AT xenografts produce ductular structures with the myoepithelium properly oriented between a basement membrane and the luminal epithelium. When orthotopically implanted, MCF10AT xenografts produce lesions containing variable amounts of simple ducts, hyperplasia, atypia, DCIS and frank carcinoma [19]. In vivo implantation of single clones of MCF10AT cells showed that all clones produce simple and hyperplastic ducts surrounded by myoepithelium confirming that these ducts originate from stem cell/progenitors cells rather than from distinct populations of cells that either gives rise to myoepithelial or luminal subtypes. MCF10AT cells express functional estrogen receptor α (ERα) and respond in vivo to exogenous estrogen supplementation with increased frequency of index precursor lesions atypia and DCIS but with minimal impact on the frequency of invasive carcinomas [20,21]. While treatment with tamoxifen abolishes atypia and DCIS, tamoxifen treatment had no impact on invasive carcinoma despite being ER+ [22]. According to the clonal evolution model, a tumor cell gains malignant potential by acquiring new genetic alterations and resultant clonal expansion. This would require the presence of index precursor lesions for development of invasive carcinomas. However, since tamoxifen treated MCF10AT xenografts showed the presence of invasive carcinomas at a similar frequency as those exposed to estrogen despite the absence of atypia and DCIS [22], these data suggest that precursor (atypia and DCIS) and malignant (invasive carcinoma) components of a tumor can arise independently from a transformed stem cell/progenitor cell/tumor initiating cell (TIC), and that the proportion and frequency of specific histologic subtypes in a tumor would depend upon initiating alterations defining the “CSC or TIC subset” and their subsequent ability for clonal expansion. It is conceivable that hormonal therapies and other therapeutic agents could similarly exert specific effects upon the renewal and differentiation of CSC/TIC/progenitor cells and hence control the subsequent histogenic pathways of tumorigenesis and therapy sensitivity.
Our data also reveal a mechanism for emergence of drug resistance. The current thinking attributes drug resistance to the presence of CSC subpopulation that is elusive to therapy and their elimination is critical for complete therapy response. However, our data suggest that similar to the precursor lesions, malignant lesions can also arise from CSC/progenitor cells, albeit from a distinct “CSC” subset. Breast CSCs were first identified as a CD44+/CD24−/low population that has enhanced ability to initiate tumor growth when xenografted into immunocompromised mice [23]. CD44/CD24 expression analysis of MCF10AT xenografts showed CD24−/low and strong CD44 immunoreactivity in regions of DCIS and invasive carcinoma, and whereas CD44-expressing DCIS lesions were eliminated by tamoxifen therapy, tamoxifen had little impact on CD44+/ER+ invasive cancer cells. These data suggest that retention of CD44+ cells in the residual tumor is not responsible for the failure to achieve complete therapy response [24]. These data are consistent with a study by Liu et al. [25] who demonstrated that commonly used putative CSC markers CD24, CD44, ALDH, and SOX2 are not coexpressed in the same cells. The authors were unable to identify specific CSC subpopulations using these markers and found that the relative expression levels of these markers did not correlate with each other or with therapy resistance [25]. Further support for our hypothesis was provided by Miller et al. [26] who by single cell cloning of MCF10AT cells isolated MCF10DCIS.com cells, so named because of their ability to differentiate in vivo directly into pure DCIS lesions without going through lower grades of ductal differentiation. These data provide further support for the presence of distinct subsets of transformed stem cells/progenitors carrying specific genetic alterations that predetermine their differentiated progeny. Over a period of time the DCIS lesions progress to invasive carcinoma, potentially by clonal evolution and expansion. Similarly, Miller et al. have also isolated from MCF10AT xenografts, MCF10CA1A and MCF10CA1D cells that progress directly to invasive carcinomas providing additional support for this hypothesis [27]. These data suggest that differences in the rates of differentiation of different CSC/progenitor subpopulations influence the composition and relative amounts of the phenotypically distinguishable progeny histotypes or the heterogeneity that is characteristic of breast cancers. This raises an important question of whether despite phenotypic resemblances, are the invasive cancer cells derived from “CSC subsets” genetically similar to the invasive carcinomas arising by clonal evolution of DCIS? Depending on whether they represent related or distinct entities, this could significantly impact clinical responses of the tumors. Compounding these effects, differentiated mammary epithelial cells have been reported to undergo reprogramming to multipotent mammary stem cells by forced expression of stem cell transcription factors [28] illustrating the phenotypic plasticity of mammary cancer cells.
Unipotent or multipotent differentiation of CSC/progenitors
Our findings support the emergence of precursor or malignant lesions from separate putative CSC/progenitor cells, which could either, have restricted potential for differentiation (unipotent lineage) or have the ability to give rise to multiple lineages (multipotent). Molecular analysis of comedo-DCIS derived from MCF10DCIS.com cells showed that the majority of the comedo-DCIS are Her2/neu-negative with the basal marker p63/cytokeratin 5-expressing cells restricted to the myoepithelial layer. However, interestingly the tumors also contain small areas of comedo-DCIS that coexpress basal (p63) and luminal (Her2/neu) markers [29]. Progression of comedo-DCIS in these tumors results in invasive carcinomas that are p63+/Her2− as well as p63+/Her2+. Clinical comedo-DCIS similarly show the presence of p63/Her2-colabeled and p63+/Her2- cells, providing clinical support for the MCF10DCIS.com data and validating a novel link between comedo-DCIS and basal-like breast cancer [29]. Several studies have reported that a subset of in situ ductal carcinomas as defined by genomic [30,31] or immunohistochemical [32–36] definitions are basal. In most cases, basal DCIS were associated with high nuclear grade, central necrosis (resembling comedo DCIS.com lesions) and high proliferative indices [34]. Basal DCIS was often found to be admixed with invasive basal breast cancers suggesting that basal DCIS could serve as precursor lesions for invasive cancers [34]; however, interestingly, earlier precursor lesions such as atypical ductal hyperplasia for basal DCIS have not been identified [37].
The emergence of DCIS with distinct molecular subtypes (Her2+ and Her2−) from MCF10DCIS.com clone suggests the presence of multipotent CSC/progenitor cells, and that their differentiation rates may ultimately determine the relative amounts of Her2/neu-expressing or Her2/neu-nonexpressing DCIS, and their subsequent invasive potential. Based on our data that p63 and Her2/neu are coexpressed in clinical comedo-DCIS and the MCF10DCIS.com comedo-DCIS model, we posit that the p63 and Her2/neu expressors share a common precursor and that the p63+/Her2+ cells represent an intermediate progeny of stem cell differentiation. Since the p63+/Her2+ coexpressing cells are detected both in the myoepithelial and luminal compartments of comedo-DCIS, we suggest that these transitional precursors probably experience a block in differentiation into discrete p63+/Her2/neu− (basal cells of myoepithelial lineage) and Her2+/p63− (Her2-overexpressing) lineages. It is conceivable that p63+/Her2 coexpression could potentially direct novel or modified gene expression programs and depending upon their relative amounts in the tumor they could potentially alter their growth potentials and therapy sensitivities. Thus while patients with p63+/Her2 coexpressing DCIS may benefit from Her2-targeted therapy, this opens up the clinical dilemma whether targeting Her2/neu would allow for expansion of p63+/Her2− progeny and consequently promotes transition to typical basal-like breast cancer.
Stromal contribution to heterogeneity
Molecular profiling studies have revealed that heterogeneity is not limited to cancer cells but also to components of the tumor microenvironment. The rate and frequency of occurrence of a specific or general pathway(s) of differentiation is not only determined by genetic features intrinsic to tumor subpopulations but is also determined by extraneous elements such as dietary factors, environmental agents, therapy, or diagnosis-induced stress (e.g., biopsy collection) on the tumor cells and the stromal microenvironment. Genetic and microenvironment-mediated epigenetic events can trigger activation and/or prevent the return to quiescence of activated stem/progenitor cells, thus trapping the activated cells in a state of continuous renewal. The importance of growth regulatory role of breast stroma in normal development and cancer is well documented [38–44]. Studies from our laboratory using three dimensional cocultures of nontransformed or premalignant human breast epithelial cells with normal or tumor derived fibroblasts and/or endothelial cells have revealed distinct functional roles for these stromal elements in reconstitution of an ecosystem that is more favorable towards either a benign or transformed phenotype [45]. When placed in a microenvironment containing normal breast fibroblasts, the growth and aberrant ductal branching morphogenesis of both nontransformed and transformed breast epithelial cells are inhibited. However, growth and aberrant ductal branching morphogenesis of both normal and transformed breast cells are stimulated by tumor-derived fibroblast microenvironment. Interestingly, the growth inhibitory effects of normal fibroblasts are not relieved by addition of endothelial cells to the microenvironment, whereas endothelial cells augment the growth stimulatory effects of tumor derived fibroblasts [45,46]. These data not only reveal the dominant epigenetic regulatory roles of the stromal microenvironment in activation or maintenance of quiescence of progenitor cells but also demonstrate that stroma-mediated epigenetic forces not only override the genetic constraints of breast epithelial cells but also take advantage of tumor cell plasticity.
Histologic analysis of breast tumors provides evidence for the reciprocal/symbiotic relationships between the epithelium and its stromal microenvironment. Consistent with the varying proportions of precursor index and malignant lesions in a tumor, the composition and proportions of the stroma surrounding individual lesions are also variable, suggesting a reciprocal and active relationship between the epithelial cells and the stroma (Fig. 2). The assembly of a rich stromal microenvironment comprising of fibroblasts, endothelial cells, immune cells and/or inflammatory cells would not only provide a rich soil and matrix for renewal, differentiation and clonal expansion and evolution of CSC/progenitor cells that are “marked for a particular histotype” but would also provide a barrier or shield against attack by therapy or immune surveillance.
Figure 2.
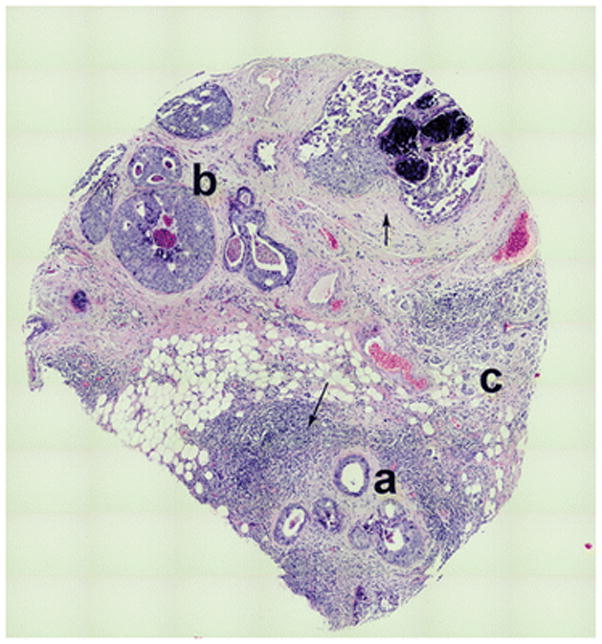
Tile map of a breast cancer section showing histologic homogeneity within a heterogeneous tumor milieu. Note the preservation of orderly ecosystems as defined by areas composed of individual histologic subtypes, (a) hyperplasia, (b) DCIS, and (c) invasive cancer within a complex and heterogeneous tumor milieu, implicating their origination from separate progenitors. Also note the heterogeneity in the stromal microenvironments surrounding each ecosystem (denoted by arrow) that implicate their roles in generation/maintenance of tumor heterogeneity.
Impact of heterogeneity on clinical management and outcome
Broad range chemotherapeutic regimens utilize the maximum tolerated dose to eradicate tumors by inducing lethal toxicity to the bulk of the tumor cells. However, these regimens induce systemic toxicity. More recently, combination targeted therapies have been implemented in clinical practice to overcome systemic toxicity and to simultaneously target multiple cell subpopulations within the tumor ecosystem to eliminate tumor burden. Targeted therapies represent a class of agents that have been designed to interact with specific molecules involved in cancer development and progression [47–51]. Current FDA-approved targeted therapies in breast cancer include Her2 targeted therapies (trastuzumab, pertuzumab, everolimus, lapatinib, and ado-trastuzumab emtansine), estrogen modulators (tamoxifen, toremifene, fulvestrant, anastrozole, exemestane, and letrozole) and cyclin-dependent kinase (CDK) inhibitors (palbociclib) [52–56]. Combination therapy utilizes these targeted therapies in conjugation with broad range chemotherapeutic agents and/or other targeted therapies. Using combination therapy, clinicians can target the same molecular target (i.e., the use of trastuzumab and pertuzumab to target HER2), compensatory molecular pathways (i.e., the use of platinum-based compounds and PARP1 inhibitors to target DNA damage response pathways), or multiple nodes within a single pathway (i.e., using lapatinib to target both EGFR and HER2) [57–59], and clinical trials have shown improved efficacy and/or drug resistance reversal with combination therapy [53,57,59–63]. Indeed, the results of the phase III PALOMA3 trail examining ER-positive, HER2-negative patients with drug resistance found that combination therapy with palbociclib and fulvestrant resulted in significant improvement in progression free survival (9.5 months versus 4.6 months) compared to the fulvestrant alone arm [60]. Additionally, it was found from the CLEOPATRA study using patients with Her2+ metastatic breast cancer that the addition of pertuzumab to trastuzumab and docetaxel significantly increased progression-free survival from 40.8 months to 56.5 months when compared with placebo, trastuzumab, and docetaxel [61], further showing evidence for the use of combination therapy to enhance drug efficacy. While these chemotherapeutic regimens have been found to enhance drug efficacy and revert drug resistance, new resistance develops [4,64–70]. Emergence of therapy resistance has been attributed to the failure of eliminating “drug resistant” CSC subpopulations and the resulting therapy-induced alterations in the tumor ecosystem [71–73]. Accordingly, while therapy-sensitive clones are eliminated, systemic therapies could alter the tumor composition by providing a mutagenic stimulus that promotes selection of resistant clones and consequent alteration of the proportions of tumor histotypes [74,75; Fig. 3]. Since therapies target the most vulnerable lesion phenotypes in the tumor ecosystem, questions to consider include (i) whether the susceptible cells are required for maintaining a stable ecosystem that suppresses the outgrowth of resistant cells, and (ii) whether the disruption of the tumor ecosystem enables reconstitution of a more homogeneous yet resistant tumor.
Figure 3.
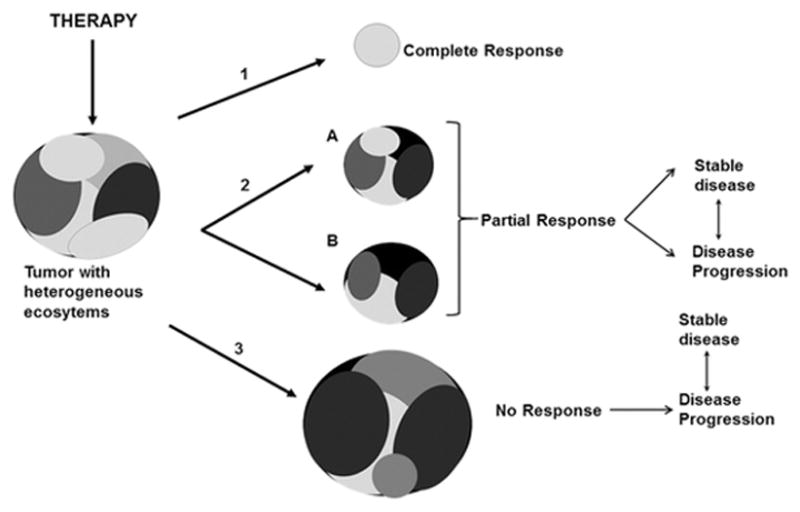
Tumors are comprised of heterogeneous ecosystems that have variable therapy sensitivities and the potential to influence growth, survival and therapy responses of neighboring tumor cells through cell-microenvironment mediated interactions. (1) Pathologic complete response, a surrogate endpoint that is predictive of long term disease-free survival, is associated with complete or near complete resolution of the lesion and potentially its heterogeneous landscape. (2) Partial response defined as a ≥30% decrease in tumor size could either result in the residual tumor remaining dormant or stable, or eventually progressing depending upon the compositions and activities of the residual tumor. (3) An increase or no change in tumor size is defined as a “no response” outcome where the most vulnerable tumor subpopulations are eliminated with potential enrichment of the tumor with ecosystems that are more or less heterogeneous and containing therapy resistant variants. In scenarios (2) and (3), the tumors could either attain a state of tumor homeostasis (stable disease) or imbalance (disease progression) depending upon the nature of reestablished tumor ecosystems.
Using a mixture of two sister subclones 168FAR and 4T07 derived from a single mouse mammary tumor and with varying metastatic propensities, Miller et al. injected orthotopically different mixture ratios of the cells into mice [76]. The relative proportions of the two sublines in the tumors were analyzed by colony forming assays using medium selective for 168FAR or 4T07 cells. Regardless of the initial injection ratios of the two sublines, the resulting tumor primarily consisted of 4T07 cells [76]. Additionally, the growth inhibition elicited by 4T07 cells was seen in monolayer cocultures, which was diminished when 4T07 cells were subjected to lethal irradiation prior to mixing with 168FAR cells. These data highlight the impact of tumor variants on the survival and growth potentials of tumor subpopulations in a tumor ecosystem. Crespi et al. recently described the tumor ecosystem to exist in a state of dynamic equilibrium between tumor cells that function either as cheaters or helpers, wherein the helpers provide the nurturing factors which the cheaters usurp to gain selective growth and survival advantages [77]. Utilizing a human Rad6B promoter-driven Zs Green reporter construct, Gerard et al [78] isolated Rad6B-overexpressing and underexpressing subpopulations of MDA-MB-231 breast cancer cells. Rad6B is an ubiquitinating enzyme that upregulates stability and oncogenic transcriptional activity of β-catenin [78,79]. Although the Rad6B-overexpressing subpopulations produced smaller tumors compared to the control polyclonal parental cells, the tumors produced by Rad6B–overexpressing subpopulations were composed exclusively of cancer cells with a homogeneous EMT phenotype consistent with activated Wnt/β-catenin signaling, and displayed high propensity for lymph node and lung metastasis whereas loss of Rad6B impaired their tumor growth potential [78]. These data suggest that while Rad6B is required for tumor growth and aggressiveness, the presence of “low Rad6B” expressing subpopulation is necessary for development of large tumors as seen with the control polyclonal parental cells [78]. Similarly, coculturing parental MDA-MB-468 triple negative breast cancer cells with MDA-MB-468 clones engineered to overexpress IL-11 enhanced the tumor growth of MDA-MB-468 parental cells, while loss of the IL-11 subclone reduced the tumor growth of polyclonal parental cells [80]. Addition of IL-11 and FGF overexpressing subclones were needed to recapitulate the metastatic phenotype of the polyclonal parental tumor [80]. These results indicate that it is the interaction between tumor subclones that create cancerous phenotypes. These data lend support to the idea that the cells within the tumor ecosystem depend upon their biochemical interactions with the neighboring subpopulations for survival and expansion. However, these tumor cells can also exert inhibitory effects that prevent outgrowth of more resistant and aggressive subclones. Thus, chemotherapy aimed to disrupt this tumor ecosystem and induce apoptosis in responsive cells may ultimately eliminate this inherent inhibition, resulting in resistant disease. Changes in tumor heterogeneity following neoadjuvant chemotherapy at mid and post-treatment phases were assessed by T2-weighted MRI changes in entropy (a measure of heterogeneity) and uniformity (a measure of homogeneity) by MRI imaging. Reduction in entropy with increase in uniformity was found to correlate better at mid-treatment than after completion of therapy [81]. While this study suggests that treatment response may correlate with breast tumor homogeneity, since the analysis was limited to the maximum axial diameter it may not be representative of the entire heterogeneous tumor and scoring systems incorporating degrees of partial response may be required to validate this observation.
Conclusion
Intratumoral heterogeneity has been viewed as a clinical challenge to be combated. Although the obvious treatment option for breast cancer is surgical removal of the tumor, therapeutic options such as chemotherapy and radiation therapy are also followed in cases where surgery is not the first line of treatment. The end goal of therapeutic regimens is to induce apoptosis in the bulk of the tumor and eradicate/shrink the tumor. The advent of therapies targeted to critical molecules required for tumor growth have improved drug efficacy and clinical response. While combining targeted therapies with a broad range of chemotherapeutics and/or other targeted therapies aimed to disrupt multiple oncogenic pathways have shown clinical benefits, development of drug resistance is inevitable in most cases. We posit that the elimination of certain vulnerable tumor subpopulations could disrupt an otherwise stable or dormant tumor ecosystem, and inadvertently create new opportunities for activation and outgrowth of “quiescent” therapy resistant or aggressive histotypes by generating permissive microenvironmental conditions. The goal so far has been to reduce tumor heterogeneity so that the resulting tumor can be rendered more suitable for elimination by therapy.
In this era of personalized medicine, much effort is focused on taking advantage of single cell-based deep sequence analysis and robust bioinformatics approaches to identify genetic alterations that define intratumoral heterogeneity or have predictive biomarker power. Single cell-based sequence analysis elegantly reveals genetic diversity; however, the success of treatment strategies based on outcomes of such analyses will be complicated by the dynamic nature of cell-cell, cell-stromal and cell-matrix interactions and the ensuing heterogeneity within the complex tumor architecture. As the predictive value of biomarkers are confounded and compromised by intratumoral heterogeneity, identification of biomarkers with strong predictive power and accuracy will require simultaneous tracking of intratumoral heterogeneity during the course of clinical management. Heterogeneity trials such as the Breast Cancer Proteomics and Molecular Heterogeneity trial NCT01840293 is focused on analyzing the associations between proteomic/molecular heterogeneity and the characteristics of primary and recurrent/metastatic breast tumors. Elimination of vulnerable cell populations while reducing the heterogeneity could revive otherwise dormant or minor subpopulations that restore heterogeneity and transform the tumor into a more resistant and aggressive type. This raises the question if it would be better to maintain stable disease or preserve heterogeneity by not disturbing the tumor ecosystem (preserving homeostasis). The ultimate strategy for personalized therapy would require sequential assessments of the patient’s tumor for identified/predicted vulnerabilities or intended targets during the entire course of treatment combined with their three dimensional mapping to the tumor architecture and landscape.
Acknowledgments
Funding Work related to the analysis of histologic origins and stromal contributions to therapy response was funded by grants DAMD-17-02-1-0618 and W81XWH-09-1-0608 from the Department of Defense. BH was supported by T32-CA009531 and a fellowship from the DeRoy Testamentary Foundation.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Research. 1978;38:2651–2660. [PubMed] [Google Scholar]
- 2.Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Reviews. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- 3.Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Research. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 4.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nature Reviews Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 6.Heppner GH, Miller FR. The cellular basis of tumor progression. International Reviews in Cytology. 1998;177:1–56. doi: 10.1016/s0074-7696(08)62230-5. [DOI] [PubMed] [Google Scholar]
- 7.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nature Reviews Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 8.Wolman SR, Heppner GH. Genetic heterogeneity in breast cancer. Journal of National Cancer Institute. 1992;84:469–470. doi: 10.1093/jnci/84.7.469. [DOI] [PubMed] [Google Scholar]
- 9.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica Biophysica Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janiszewska M, Polyak K. Clonal evolution in cancer: a tale of twisted twines. Cell Stem Cell. 2015;16:11–12. doi: 10.1016/j.stem.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Bapat SA. Evolution of cancer stem cells. Seminars in Cancer Biology. 2007;17:204–213. doi: 10.1016/j.semcancer.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nature Reviews Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- 13.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 14.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clinical Chemistry. 2013;59:168–179. doi: 10.1373/clinchem.2012.184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshamy WM, Duhe RJ. Overview: cellular plasticity, cancer stem cells and metastasis. Cancer Letters. 2013;341:2–8. doi: 10.1016/j.canlet.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Rhiannon F, Richard C. The Complex Nature of Breast Cancer Stem-Like Cells: Heterogeneity and Plasticity. Journal of Stem Cells Research and Therapy. 2012;S7(009) [Google Scholar]
- 18.Miller FR, Soule HD, Tait L, Pauley RJ, Wolman SR, Dawson PJ, Heppner GH. Xenograft model of progressive human proliferative breast disease. Journal of National Cancer Institute. 1993;85:1725–1732. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- 19.Miller FR. Xenograft models of premalignant breast disease. Journal of Mammary Gland Biology and Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 20.Shekhar MP, Nangia-Makker P, Wolman SR, Tait L, Heppner GH, Visscher DW. Direct action of estrogen on sequence of progression of human preneoplastic breast disease. American Journal of Pathology. 1998;152:1129–1132. [PMC free article] [PubMed] [Google Scholar]
- 21.Shekhar PV, Chen ML, Werdell J, Heppner GH, Miller FR, Christman JK. Transcriptional activation of functional endogenous estrogen receptor gene expression in MCF10AT cells: a model for early breast cancer. International Journal of Oncology. 1998;13:907–915. doi: 10.3892/ijo.13.5.907. [DOI] [PubMed] [Google Scholar]
- 22.Visscher DW, Nanjia-Makker P, Heppner G, Shekhar PV. Tamoxifen suppresses histologic progression to atypia and DCIS in MCFIOAT xenografts, a model of early human breast cancer. Breast Cancer Research & Treatment. 2001;65:41–47. doi: 10.1023/a:1006490000659. [DOI] [PubMed] [Google Scholar]
- 23.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of National Academy of Sciences U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shekhar MP, Tait L. Breast cancer stem cell paradigm. In: Parsons DW, editor. Stem Cells and Cancer. New York: Nova Science Publishers; pp. 47–64. [Google Scholar]
- 25.Liu Y, Nenutil R, Appleyard MV, Murray K, Boylan M, Thompson AM, Coates PJ. Lack of correlation of stem cell markers in breast cancer stem cells. British Journal of Cancer. 2014;110:2063–2071. doi: 10.1038/bjc.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. Journal of National Cancer Institute. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 27.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Research & Treatment. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 28.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shekhar MP, Kato I, Nangia-Makker P, Tait L. Comedo-DCIS is a precursor lesion for basal-like breast carcinoma: identification of a novel p63/Her2/neu expressing subgroup. Oncotarget. 2013;4:231–241. doi: 10.18632/oncotarget.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Research. 2006;8:R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu K, Lee CH, Tan PH, Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clinical Cancer Research. 2004;10:5508–5517. doi: 10.1158/1078-0432.CCR-04-0085. [DOI] [PubMed] [Google Scholar]
- 32.Bryan BB, Schnitt SJ, Collins LC. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Modern Pathology. 2006;19:617–621. doi: 10.1038/modpathol.3800570. [DOI] [PubMed] [Google Scholar]
- 33.Dabbs DJ, Chivukula M, Carter G, Bhargava R. Basal phenotype of ductal carcinoma in situ: recognition and immunohistologic profile. Modern Pathology. 2006;19:1506–1511. doi: 10.1038/modpathol.3800678. [DOI] [PubMed] [Google Scholar]
- 34.Livasy CA, Perou CM, Karaca G, Cowan DW, Maia D, Jackson S, Tse CK, Nyante S, Millikan RC. Identification of a basal-like subtype of breast ductal carcinoma in situ. Human Pathology. 2007;38:197–204. doi: 10.1016/j.humpath.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Paredes J, Lopes N, Milanezi F, Schmitt FC. P-cadherin and cytokeratin 5: useful adjunct markers to distinguish basal-like ductal carcinomas in situ. Virchows Archives. 2007;450:73–80. doi: 10.1007/s00428-006-0334-y. [DOI] [PubMed] [Google Scholar]
- 36.Tang P, Wang X, Schiffhauer L, Wang J, Bourne P, Yang Q, Quinn A, Hajdu SI. Relationship between nuclear grade of ductal carcinoma in situ and cell origin markers. Annals of Clinical Laboratory Science. 2006;36:16–22. [PubMed] [Google Scholar]
- 37.Bertucci F, Finetti P, Birnbaum D. Basal breast cancer: a complex and deadly molecular subtype. Current Molecular Medicine. 2012;12:96–110. doi: 10.2174/156652412798376134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 39.Hanley CJ, Noble F, Ward M, Bullock M, Drifka C, Mellone M, Manousopoulou A, Johnston HE, Hayden A, Thirdborough S, Liu Y, Smith DM, Mellows T, Kao WJ, Garbis SD, Mirnezami A, Underwood TJ, Eliceiri KW, Thomas GJ. A subset of myofibroblastic cancer-associated fibroblasts regulate collagen fiber elongation, which is prognostic in multiple cancers. Oncotarget. 2016;7:6159–6174. doi: 10.18632/oncotarget.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He K, Lv W, Zheng D, Cheng F, Zhou T, Ye S, Ban Q, Ying Q, Huang B, Chen L, Wu G, Liu D. The stromal genome heterogeneity between breast and prostate tumors revealed by a comparative transcriptomic analysis. Oncotarget. 2015;6:8687–8697. doi: 10.18632/oncotarget.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junk DJ, Cipriano R, Bryson BL, Gilmore HL, Jackson MW. Tumor microenvironmental signaling elicits epithelial-mesenchymal plasticity through cooperation with transforming genetic events. Neoplasia. 2013;15:1100–1109. doi: 10.1593/neo.131114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natrajan R, Sailem H, Mardakheh FK, Arias Garcia M, Tape CJ, Dowsett M, Bakal C, Yuan Y. Microenvironmental Heterogeneity Parallels Breast Cancer Progression: A Histology-Genomic Integration Analysis. PLoS Medicine. 2016;13:e1001961. doi: 10.1371/journal.pmed.1001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roman-Perez E, Casbas-Hernandez P, Pirone JR, Rein J, Carey LA, Lubet RA, Mani SA, Amos KD, Troester MA. Gene expression in extratumoral microenvironment predicts clinical outcome in breast cancer patients. Breast Cancer Research. 2012;14:R51. doi: 10.1186/bcr3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biology & Therapy. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 45.Shekhar MP, Werdell J, Santner SJ, Pauley RJ, Tait L. Breast stroma plays a dominant regulatory role in breast epithelial growth and differentiation: implications for tumor development and progression. Cancer Research. 2001;61:1320–1326. [PubMed] [Google Scholar]
- 46.Shekhar MP, Santner S, Carolin KA, Tait L. Direct involvement of breast tumor fibroblasts in the modulation of tamoxifen sensitivity. American Journal Pathology. 2007;170:1546–1560. doi: 10.2353/ajpath.2007.061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arif Harahap W, Ramadhan, Khambri D, Haryono S, Nindrea RD. Outcomes of trastuzumab therapy for 6 and 12 months in Indonesian national health insurance system clients with operable HER2-positive breast cancer. Asian Pacific Journal of Cancer Prevention. 2017;18:1151–1156. doi: 10.22034/APJCP.2017.18.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng YC, Shi Y, Zhang MJ, Brazauskas R, Hemmer MT, Bishop MR, Nieto Y, Stadtmauer E, Ayash L, Gale RP, Lazarus H, Holmberg L, Lill M, Olsson RF, Wirk BM, Arora M, Hari P, Ueno N. Long-term outcome of inflammatory breast cancer compared to non-inflammatory breast cancer in the setting of high-dose chemotherapy with autologous hematopoietic cell transplantation. Journal of Cancer. 2017;8:1009–1017. doi: 10.7150/jca.16870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasir A, Holzer TR, Chen M, Man MZ, Schade AE. Differential expression of VEGFR2 protein in HER2 positive primary human breast cancer: potential relevance to anti-angiogenic therapies. Cancer Cell International. 2017;17:56. doi: 10.1186/s12935-017-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rier HN, Levin MD, van Rosmalen J, Bos M, Drooger JC, de Jong P, Portielje JEA, Elsten EMP, Ten Tije AJ, Sleijfer S, Jager A. First-line palliative HER2-targeted therapy in HER2-positive metastatic breast cancer is less effective after previous adjuvant trastuzumab-based therapy. Oncologist. 2017 doi: 10.1634/theoncologist.2016-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, Wang L, Shen Y, Wang C, Zhang Y, Meng Y, Yang Y, Liang B, Zhou B, Wang H, Wei H, Lei C, Hu S, Li B. Targeting EGFR/HER2 heterodimerization with a novel anti-HER2 domain II/III antibody. Molecular Immunology. 2017;87:300–307. doi: 10.1016/j.molimm.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 52.Drakaki A, Hurvitz SA. Her2-positive breast cancer: Update on new and emerging agents. The American Journal of Hematology/Oncology. 2015;11:17–23. [Google Scholar]
- 53.Dickler MN, Tolaney S, Rugo HS, Cortes J, Dieras V, Patt DA, Wildiers H, Hudis CA, O’Shaughnessy JA, Zamora E, Yardley D, Frenzel M, Koustenis AG, Baselga J. MONARCH 1, a phase 2 study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR+/HER2- metastatic breast cancer. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-17-0754. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dean L. Pertuzumab therapy and ERBB2 (HER2) genotype. In: Pratt V, McLeod H, Dean L, Malheiro A, Rubinstein W, editors. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information; 2012. [PubMed] [Google Scholar]
- 55.Dean L. Trastuzumab (herceptin) therapy and ERBB2 (HER2) genotype. In: Pratt V, McLeod H, Dean L, Malheiro A, Rubinstein W, editors. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information; 2012. [Google Scholar]
- 56.Dean L. Tamoxifen therapy and CYP2D6 genotype. In: Pratt V, McLeod H, Dean L, Malheiro A, Rubinstein W, editors. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information; 2012. [PubMed] [Google Scholar]
- 57.Gu G, Dustin D, Fuqua SA. Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Current Opinion in Pharmacology. 2016;31:97–103. doi: 10.1016/j.coph.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 58.Yap TA, Omlin A, de Bono JS. Development of therapeutic combinations targeting major cancer signaling pathways. Journal of Clinical Oncology. 2013;31:1592–1605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- 59.Tryfonidis K, Senkus E, Cardoso MJ, Cardoso F. Management of locally advanced breast cancer-perspectives and future directions. Nature Reviews Clinical Oncology. 2015;12:147–162. doi: 10.1038/nrclinonc.2015.13. [DOI] [PubMed] [Google Scholar]
- 60.Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, Colleoni M, DeMichele A, Loi S, Verma S, Iwata H, Harbeck N, Zhang K, Theall KP, Jiang Y, Bartlett CH, Koehler M, Slamon D. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncology. 2016;17:425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 61.Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, Ciruelos E, Ferrero JM, Schneeweiss A, Heeson S, Clark E, Ross G, Benyunes MC, Cortés J CLEOPATRA Study Group. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. New England Journal of Medicine. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ates O, Sunar V, Aslan A, Karatas F, Sahin S, Altundag K. The short-term safety of adjuvant paclitaxel plus trastuzumab - A single centre experience. J Balkan Union of Oncology. 2017;22:320–324. [PubMed] [Google Scholar]
- 63.Liu Z, He K, Ma Q, Yu Q, Liu C, Ndege I, Wang X, Yu Z. Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PLoS One. 2017;12(5):e0177694. doi: 10.1371/journal.pone.0177694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Di Nicolantonio F, Mercer SJ, Knight LA, Gabriel FG, Whitehouse PA, Sharma S, Fernando A, Glaysher S, Di Palma S, Johnson P, Somers SS, Toh S, Higgins B, Lamont A, Gulliford T, Hurren J, Yiangou C, Cree IA. Cancer cell adaptation to chemotherapy. BMC Cancer. 2005;5:78. doi: 10.1186/1471-2407-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan SH, Sapari NS, Miao H, Hartman M, Loh M, Chng WJ, Iau P, Buhari SA, Soong R, Lee SC. High-throughput mutation profiling changes before and 3 weeks after chemotherapy in newly diagnosed breast cancer patients. PLoS One. 2015;10:e0142466. doi: 10.1371/journal.pone.0142466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy C, Dickler M. Endocrine resistance in hormone responsive breast cancer: mechanisms and therapeutic strategies. Endocrine Related Cancer. 2016 doi: 10.1530/ERC-16-0121. ERC-16-0121. [DOI] [PubMed] [Google Scholar]
- 67.Jeselsohn R, Brown M. How drug resistance takes shape. Elife. 2016;5 doi: 10.7554/eLife.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ter Brugge P, Kristel P, van der Burg E, Boon U, de Maaker M, Lips E, Mulder L, de Ruiter J, Moutinho C, Gevensleben H, Marangoni E, Majewski I, Józwiak K, Kloosterman W, van Roosmalen M, Duran K, Hogervorst F, Turner N, Esteller M, Cuppen E, Wesseling J, Jonkers J. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. Journal of National Cancer Institute. 2016;108 doi: 10.1093/jnci/djw148. [DOI] [PubMed] [Google Scholar]
- 69.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 70.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, Fan C, Zhang X, He X, Pavlick A, Gutierrez MC, Renshaw L, Larionov AA, Faratian D, Hilsenbeck SG, Perou CM, Lewis MT, Rosen JM, Chang JC. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proceedings of National Academy of Sciences U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leder K, Holland EC, Michor F. The therapeutic implications of plasticity of the cancer stem cell phenotype. PLoS One. 2010;5:e14366. doi: 10.1371/journal.pone.0014366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. Journal of National Cancer Institute. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 74.Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;174:2557–2566. doi: 10.1534/genetics.105.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou X, Zhang J, Yun H, Shi R, Wang Y, Wang W, Lagercrantz SB, Mu K. Alterations of biomarker profiles after neoadjuvant chemotherapy in breast cancer: tumor heterogeneity should be taken into consideration. Oncotarget. 2015;6:36894–36902. doi: 10.18632/oncotarget.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller BE, Miller FR, Wilburn D, Heppner GH. Dominance of a tumor subpopulation line in mixed heterogeneous mouse mammary tumors. Cancer Research. 1988;48:5747–5753. [PubMed] [Google Scholar]
- 77.Crespi B, Foster K, Ubeda F. First principles of Hamiltonian medicine. Philosophical Transactions of the Royal Society of London Series B Biological Sciences. 2014;369:20130366. doi: 10.1098/rstb.2013.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerard B, Tait L, Nangia-Makker P, Shekhar MP. Rad6B acts downstream of Wnt signaling to stabilize beta-catenin: Implications for a novel Wnt/beta-catenin target. Journal of Molecular Signaling. 2011;6:6. doi: 10.1186/1750-2187-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Research. 2008;68:1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- 80.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parikh J, Selmi M, Charles-Edwards G, Glendenning J, Ganeshan B, Verma H, Mansi J, Harries M, Tutt A, Goh V. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 2014;272:100–112. doi: 10.1148/radiol.14130569. [DOI] [PubMed] [Google Scholar]


