Abstract
Approximately one third of children with autism spectrum disorder (ASD) reportedly lose skills within the first three years, yet a causal mechanism remains elusive. Considering evidence of strong genetic effects for ASD and findings that distinct phenotypes in ASD associate with specific genetic events, we examined rates of parent-reported regression in the Simons Simplex Collection with likely gene disrupting (LGD) mutations from five distinct classes: FMRP target genes, genes encoding chromatin modifiers, genes expressed preferentially in embryos, genes encoding postsynaptic density proteins, and essential genes. Children with ASD and mutations in postsynaptic density genes were more likely to experience regression, while a trend suggested that children with ASD and mutations in embryonic genes were less likely to have skill losses.
Keywords: autism, ASD, simplex, regression, genetics, mutation, exome
Developmental regression—the loss of previously acquired skills—in children with autism spectrum disorder (ASD) is reported by parents in approximately one third of affected children (Baird et al., 2008; Goldberg et al., 2003; Luyster et al., 2005; Werner, Dawson, Munson, & Osterling, 2005) at an average age of 21.4 months (Barger, Campbell, & McDonough, 2013). Several studies indicate a poorer prognosis for those with a history of regression (Giannotti et al., 2008; Matson, Wilkins, & Fodstad, 2010), with the bulk of this literature focused on cognitive and adaptive-functioning outcomes (Bernabei, Cerquiglini, Cortesi, & D’Ardia, 2007; Goin-Kochel, Esler, Kanne, & Hus, 2014; Lord, Shulman, & DiLavore, 2004; Richler et al., 2006; Shumway et al., 2011). Although regression has been reported as largely unique to ASD relative to other developmental disorders (Pickles et al., 2009), the cause of skill loss in children with ASD is unknown. Many parents vehemently believe that regression is triggered by vaccines (Goin-Kochel & Myers, 2005; Goin-Kochel, Mire, & Dempsey, 2015; Offit, 2008), although numerous epidemiological studies do not support this theory (e.g., DeStefano & Chen, 2001; Fombonne & Chakrabarti, 2001; Madsen et al., 2002; Parker et al., 2004; Taylor, Swerdfeger, & Eslick, 2014). Other investigators have examined risk factors for regression in ASD, demonstrating the regressive subtype to be associated with a family history of autoimmune thyroid disease (Molloy et al., 2006), abnormal brain enlargement among affected preschool-aged males (Nordahl et al., 2011), and increased rates of seizures and family history of neuropsychiatric disorders (Zhang et al., 2012), yet a clear biological mechanism or pathway remains elusive.
Given that ASD has strong genetic underpinnings (Bailey et al., 1995; Beaudet, 2007; Geschwind, 2011; Muhle et al., 2004; Ozonoff et al., 2011; Steffenburg et al., 1989; Szatmari et al., 1998), recent efforts have focused on identifying genetic contributions to regression in ASD. Molloy, Keddache, & Martin (2005) found evidence for linkage on 21q and 7q in a sample of children with ASD and regression, although Parr and colleagues (2006) found limited support for this association. Using data from full-biological siblings in the Autism Genetic Resource Exchange (AGRE), Goin-Kochel, Abbacchi, Duku, and Constantino (2010) used intraclass correlation coefficients (ICCs) to measure the degree to which there was variation in regression status between versus within sibships. Among non-twin siblings concordant for ASD, ICCs were small but significant for “social-engagment loss” or “any loss.” Among monozygotic (MZ) and dizygotic (DZ) twins that included concordant and discordant pairs, MZ-twin ICCs showed moderate correlations ranging from .27 to .34, whereas those for DZ twins were negligible, suggesting familiality for regression in ASD. However, researchers examining comparable data from the International Molecular Genetic Study of Autism Consortium (IMGSAC) found that concordance for regression was 18.9%, which was only modestly above the rate expected under independence (13.5%), leading them to conclude that there was not enough evidence to support familiality for regression aside from that already associated with ASD (Parr et al., 2011).
Recent work suggests that the behavioral heterogeneity observed in ASD may be due to the etiological heterogeneity, prompting a shift toward a genetics-first approach in understanding ASD (Stessman et al., 2014; Spiro et al, 2012). Indeed, recent efforts indicate distinct phenotypes within ASD are associated with specific genetic events. For example, increased rates of gastrointestinal disturbances and macrocephaly are associated with mutations to CHD8 (Bernier et al., 2014), differences in white matter and macrocephaly are linked with PTEN mutations (Frazier et al., 2014), and distinct facial features and feeding disturbances are tied to DYRK1A mutations (Deriziotis et al., 2014). Further, gene mutations falling within certain functional networks impact phenotypes, as well. For example, disruptions to genes affecting chromatin remodeling are linked with head size, with some mutations linked to microcephaly and others within the cluster linked to macrocephaly (O’Roak et al., 2014). The mixed findings observed in the literature regarding the genetic contributions to regression in ASD may similarly be confounded by the etiological heterogeneity in studied samples such that distinct genetic events may contribute differently to the presence or absence of regression in ASD.
In order to clarify and extend the current mixed findings in the literature regarding the potential genetic contribution to regression in ASD, we capitalized on recently reported genetic findings in a large cohort of children with ASD. The Simons Simplex Collection (SSC) is a sample of 2800 rigorously characterized and carefully phenotyped families with one child with ASD (Fischbach & Lord, 2010). The SSC has provided a valuable resource for gene discovery (O’Roak et al., 2010, 2012, 2012; Sanders et al., 2011, 2012; Iossifov et al., 2014). Most recently, whole exome sequencing of the SSC revealed that de novo mutations, including copy number variations, contribute to about 30% of all simplex cases of ASD and up to 45% of all simplex female cases and that the identified likely gene disrupting mutations primarily included chromatin modifiers, FMRP-associated genes, and embryonically expressed genes. The purpose of the current study was to examine the relationship between de novo genetic mutations and skill loss in ASD by comparing rates of parent-reported regression in the SSC with likely gene disrupting (LGD) mutations from five distinct classes identified by Iossifov and colleagues (2014).
Methods
Participants
Participants were 2508 children (86% male; M[SD] age = 8.9 years [3.6 years], range = 4 years to 17 years, 11 months; 78% White; 89% non-Hispanic) who had participated in the Simons Simplex Collection (N = 2477; SSC; Fischbach & Lord, 2010) or had participated in an ongoing study of individuals with ASD and likely gene disrupting mutations (N = 31) and who had available genotype data. The SSC is a repository of clinical and genetic data from families who have a single child diagnosed with ASD and no other first- through third-degree relatives with ASD or suspected ASD (see Fischbach and Lord [2010] for information about inclusion/exclusion criteria and recruitment tactics). All probands had confirmed ASD based on clinical judgment and research-reliable administrations of both the Autism Diagnostic Observation Schedule (ADOS; Lord, Rutter, DiLavore, & Risi; 1999) and the Autism Diagnostic Interview—Revised (ADI-R; Rutter et al. 2003).
Measure
Regression was operationalized using data from the ADI-R (Rutter et al., 2003). For the purposes of this study, we focused on the most salient losses that are likely the result or hallmark of ASD so as to enhance the potential for detecting a genotype-phenotype relationship. In this vein, regression-case status was defined as both language loss (#11 = 1) and social-skill loss (#25 > 0) at or before the age of 36 months, as our prior work showed these to be the most common types of skill losses in the SSC (Goin-Kochel, Esler, Kanne, & Hus, 2014). The reason for limiting the focus of regression to losses at/before 36 months is because we cannot be assured that regressions occurring after age three are truly associated with the child’s ASD. A small number of children with ASD reportedly experience skill loss after age three—approximately 2—3% in the SSC—but often these losses can be attributed to some known problem/event (e.g., regression following head injury, encopresis following bowel obstruction). Additionally, Goin-Kochel and colleagues (2014) observed that children with ASD in the SSC who experienced regression had significantly lower cognitive-functioning and adaptive-functioning scores than their counterparts without regression, which further validates the need to examine potential underlying mechanisms of skill loss.
Procedures and Data Analysis
Data for version 15 of the SSC were downloaded from the SFARI website and imported into SAS® 9.3 for cleaning and merging files to create the final dataset. Descriptive statistics were used to characterize the sample demographically, as well as to calculate the frequencies of skill loss. LGD mutation data were extracted from published exome sequencing experiments on 2508 individuals with ASD from the SSC collection (Iossifov et al., 2014) or who had participated in an ongoing study of individuals with ASD with LGD mutations falling into one of six functional clusters. For our analysis, a dichotomous variable was used to indicate the presence of any identified LGD mutation for each participant. Iossifov and colleagues (2014) further classified mutations into 6 functional clusters: FMRP target genes, genes encoding chromatin modifiers, genes expressed preferentially in embryos, genes encoding postsynaptic density (PSD) proteins, essential genes, and genes identified as Mendelian disease genes. Because many genes fall into more than one category, a given participant could have a mutation or mutations falling into multiple functional pathways; however, for the current analysis, only individuals with LGD mutations falling into one functional cluster were included for nonparametric analyses examining the relationship between functional clusters and regression. This restricted the sample, but provided a cleaner signal for genotype-phenotype comparisons.
Analysis first included nonparametric assessment of presence of LGD mutation and gender on rate of regression. Subsequent nonparametric analysis examined the relationship between LGD mutations within functional clusters on rate of regression across genders. Given the restricted sample size, both genders were combined for functional mutation analyses. To account for multiple comparisons within functional mutation clusters, a stricter p value of .01 was utilized to establish statistical significance.
Results
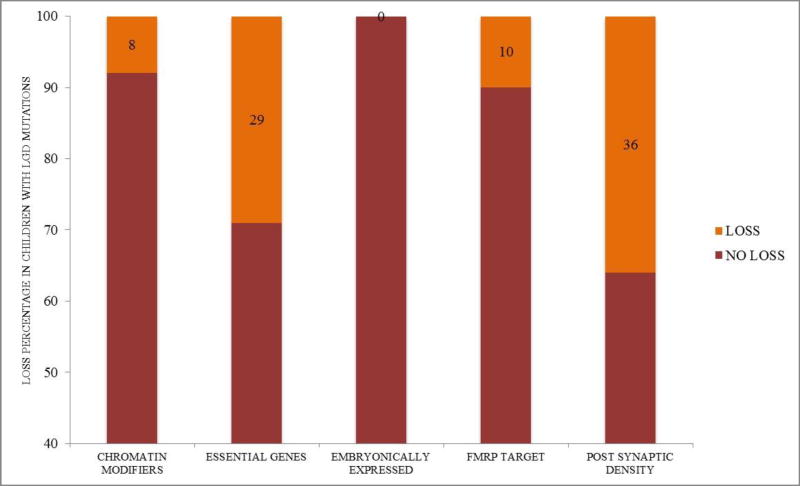
In the sample of 2508 children overall, 768 (31%) children reportedly had either language or social loss and 295 (12%) children had a reported loss in both domains. A trend toward overall decreased rates of regression in girls was observed (χ2(1) = 3.6, p = .057, Φ = .038), with 8.7% of girls and 12.3% of boys having a language + social loss. Chi square (χ2(1) = 3.3, p = .07) indicated no differences in regression rates between individuals with any type of LGD mutation (N = 35 of 388 with regression) and those without (N = 260 of 2120 without regression; see descriptive information in Table 1). However, significantly higher rates of regression (2-sided Fisher’s Exact p = .01, Φ = .175, small effect size [Fisher’s computed given sample size]) were observed in individuals with mutations to post-synaptic density genes (N = 4 of 11 with mutations to PSD genes) relative to children with mutations to other genes (N = 27 of 342 with any LGD mutation). A trend (2-sided Fisher’s Exact p = .09, Φ = .101, small effect size [Fisher’s computed given sample size]) was observed toward decreased rates of loss in individuals with mutations to genes expressed preferentially in embryos (N = 0 of 31 with mutations to embryonically expressed genes) compared to children with mutations to other genes (N = 32 of 312 with any LGD mutation). Similarly, a trend toward increased rates of loss (2-sided Fisher’s Exact p = .02, Φ = .150, small effect size [Fisher’s computed given sample size]) was observed in individuals with mutations to essential genes (N = 4 of 14 with mutations to essential genes) relative to children with mutations to other genes (N = 24 of 306 with any LGD mutation). As shown in Figure 1, no children with LGD mutations to embryonic genes had a loss, while 36% of children with post-synaptic density gene mutations experienced regression and 29% of children with LGD mutations to essential genes experienced regression. Mutations to FMRP target genes and genes encoding chromatin modifiers were not related to regression rates. Gene descriptions for those who exhibited regression can be found in Appendix 1, while additional clinical characteristics of those who exhibited regression can be found in Appendix 2.
Table 1.
Rates of regression with presence of de novo LGD mutations
| Overall | Presence of LGD mutation |
Regression | Number of individuals with: | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Any LGD Mutation |
Chromatin Modifiers |
Essential Genes |
Embryonically Expressed |
FMRP Targets |
Post Synaptic Density Genes |
||||
| Full | 2508 | 388 (13.4% of total) | 295 (11.8% of total) | 388 Loss=35 No Loss=353 | 52 Loss=4 No Loss=48 | 82 Loss=11 No Loss=71 | 76 Loss=3 No Loss=73 | 98 Loss=10 No Loss=88 | 46 Loss=8 No Loss=38 |
| Male | 2166 | 317 (14.6% of males) | 266 (12.3% of males) | With mutations to genes only falling in one functional cluster: | 13 Loss=1 No Loss=12 | 14** Loss=4 No Loss=10 | 31*** Loss=0 No Loss=31 | 20 Loss=2 No Loss=18 | 11* Loss=4 No Loss=7 |
| Female | 342 | 71 (20.8% of females) | 29 (8.5% of females) | ||||||
Significantly higher rate of regression with de novo LGD mutations to post synaptic density genes (Fisher’s Exact p=.01)
Trend toward higher rate of regression with de novo LGD mutations to essential genes (Fisher’s Exact p=.02)
Trend toward lower rate of regression with de novo LGD mutations to embryonically expressed genes (Fisher’s Exact p=.09)
Figure 1.
Percentage of regression rates within gene mutation clusters. **=Significantly greater rate of regression (p = .01), *greater rate of regression (trend, p = .09); ^= lower rate of regression (trend, p = .02). Loss percentages are listed in each functional cluster.
Discussion
The current study aimed to clarify the discrepant findings in the literature regarding genetic risk for loss in ASD by examining whether distinct genetic events contribute differently to the presence or absence of regression. These findings highlight the potential contribution of specific LGD mutations (e.g., post synaptic density proteins) to regression in ASD and underscore the relevance of understanding genotypic variability to the regression phenotype.
Given the rarity of individual LGD mutations, the examination of de novo mutations falling into distinct functional clusters increases power to detect relationships. However, even with this approach, sample sizes remain small, limiting power. This is compounded by the observation that the relationship between LGD mutations and regression is complex, given some children have more than one LGD mutation and that some LGD mutations fall into multiple functional pathways.
However, in children with ASD and with disruptions to genes involved in postsynaptic density proteins, we observed significantly higher rates of regression. Proteins localized to the postsynaptic density (PSD), a region of specialization of the postsynaptic neuron’s cytoskeleton at the synaptic junction, play a role in the regulation of synaptic function in human neocortex, as well as cell adhesion and signaling (Bayés et al., 2011). In the case of regression, it is possible that foundational synaptic functions are established, allowing for intact neural systems underlying social and behavioral functioning and the development of early foundational skills. However, given the key role in activity dependent synaptic plasticity of these proteins (Meyer et al., 2014), these foundational skills are then lost as necessary synaptic pruning and fine-tuning of these circuits fail. Relevantly, the structure and composition of postsynaptic densities have been shown to vary with developmental timing, with different proteins preferentially expressed during synaptic formation (e.g., SAP102 and NR2B) and synaptic remodeling and differentiation (e.g., PSD-95 and NR2A; Swulius, Kubota, Forest, & Waxham, 2010). Disruptions in genes coding for PSD proteins primarily involved later in developmental timing would disrupt synaptic remodeling and differentiation (Elias et al., 2006) but would leave intact the process of synaptic formation. This developmental timing-dependent involvement of PSD proteins may underlie the behavioral regression seen following apparently typical development observed with some PSD gene disruptions. Interestingly, in the absence of certain PSD proteins, expression of functionally similar PSD proteins may increase as a compensatory mechanism (e.g., SAP102 and PSD-95; Murata & Constantine-Paton, 2013).
In addition to loss-of-function effects, which reduce or abolish functional levels of coded proteins at the post-synaptic density, gain-of-function effects may occur when PSD mutations lead to the retention of misfolded protein products in the endoplasmic reticulum (ER; Comoletti et al., 2004; Zhang et al., 2009). ER retention triggers the Unfolded Protein Response (UPR), a stress-induced signaling cascade that enhances protein folding, increases protein degradation, and decreases translation of the misfolded protein (Chakrabarti, Chen, & Varner, 2011). Prolonged UPR may result in altered neuronal function and even cell death (Matus, Glimcher, & Hetz, 2011). This secondary gain-of-function effect has been linked to several ASD-associated PSD genes (e.g., Fujita et al., 2010; Falivelli et al., 2012) and could also contribute to behavioral regression reported in individuals with ASD. Additionally, a gain-of-function mutation causing misfolded NLGN3 cell-adhesion proteins produces downstream upregulation of a long term potentiation inhibitor protein CREB-2, ultimately disrupting synaptic remodeling and differentiation (Momoi, Fujita, Senoo & Momoi, 2010). Anatomically, decreased pruning has been associated with increased head size (Courchesne et al., 2011; Pierce & Eyler, 2011), and increased rates of regression have been associated with increased head size (Nordahl et al., 2011), lending support for a complex relationship between risk factors and ASD.
In contrast to the above patterns, we observed a trend suggesting that children with mutations to embryonically expressed genes may have a lower rate of regression. This hints at the possibility that disruption to functioning of these genes results in faulty neural architecture at early stages, thereby disrupting underlying social and behavioral functioning at the outset of development. As such, the neural systems are unable to function even at foundational levels such that early social communicative skills are never able to develop and, subsequently, regression never occurs. We also observed a trend suggesting that children with disruptive mutations to essential genes may have a higher rate of regression. This finding of a trend toward increased regression is observed in published reports of genetics-first studies of ASD. For example, Bernier et al. (2014) reported on 15 children with CHD8 loss of function mutations, a gene falling into the essential gene cluster, for which 47% had regression. The rate for this particular gene exceeds our observed rate of 29%, which spans multiple genes. Essential genes are characterized by an enrichment of functional categories related to gene expression, cell growth, cell death, and cell proliferation (Georgi, Voight, & Bućan, 2013). Similar to PSD genes, essential genes have been found to be differentially expressed throughout various stages of development. For example, some essential genes involved in transcriptional regulation are preferentially expressed in early prenatal development (e.g., HDAC1; Li et al., 2015) while others play an important role during postnatal development (e.g., FOXP1; Bacon et al., 2015). As with PSD genes, this differential expression among essential genes may explain the variability in observed behavioral regression seen in individuals with essential gene mutations.
In order to increase the likelihood of identifying key relationships between genetic risk factors and a regression phenotype, we focused on a strict definition of regression, operationalized as having both language and social-skill loss as retrospectively reported by parents. As expected, while overall rates of regression in our sample are similar to previous findings, our restricted definition decreased the number of identified individuals. This limits the generalizability of our results but starts to illuminate a relationship between disruptions in functional pathways and significant regression. It is possible that by focusing on specific components of loss (e.g., focusing on only loss of language), additional or different patterns may emerge. Prospective study of regression has revealed very subtle and gradual declines in social skills among a much larger proportion of children with ASD than has typically been reported by parents (Ozonoff et al., 2010). While this may also be an important type of loss to examine, our initial goal was to focus on clear, comprehensive losses in the functional areas most notably affected by ASD (i.e., social-communication skills). Further work examining distinct aspects of skill loss is warranted.
We also focused only on de novo LGD mutations in an effort to elucidate mechanisms in regression. These functional disruptions tend to lead to phenotypes characterized by more severe intellectual, social, and behavioral impairments and, as such, may not represent the full genetic contribution to regression across ASD. Additionally, we employed an existing genetic-classification system (i.e., Iossifov et al., 2014), and it is important to note that not all disrupting variants cause a disorder/condition. Some of the identified mutations may have little to no bearing on skill loss or ASD (e.g., OR4C11, MUC5B), and genes that conclusively explain regression in ASD were not identified. Although some chromatin regulators (e.g., MeCP2) have been found to be relevant to developmental regression in the literature (e.g., Lyst & Bird, 2015), no association was observed between regression and mutations in genes encoding chromatin modifiers in this study, likely due to heterogeneity in the role of chromatin modifiers in developmental processes.
In sum, these preliminary findings indicating trends for further workup with larger samples have important etiological and clinical implications. By clarifying key phenotypic outcomes for distinct genetic events in ASD, molecular pathways of ASD can be illuminated, offering insight into mechanisms and treatment. Further, if genetic mechanisms are associated with regression, then it may be possible to link specific types of mutations with functional outcomes. This has the potential to improve clinical care by providing predictive power for clinicians and families and enabling truly personalized health intervention.
Supplementary Material
Acknowledgments
Funding for this study was provided by the National Institutes for Mental Health (#R01MH100047 to R.B.) and by the Simons Foundation (SSC-15 to R.G.K. and SFARI #89368 to R.B.). We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). We appreciate obtaining access to phenotypic data on SFARI Base. Approved researchers can obtain the SSC population dataset described in this study by applying at https://base.sfari.org.
Footnotes
Conflict of Interest: Robin P. Goin-Kochel declares that she has no conflict of interest. Sandy Trinh declares that she has no conflict of interest. Shelley Barber declares that she has no conflict of interest. Raphael Bernier declares that he has no conflict of interest.
Compliance with Ethical Standards
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Informed consent was obtained from all individual participants included in the current study through their initial participation in the Simons Simplex Collection.
Literature Cited
- Bacon C, Schneider M, Le Magueresse C, Froehlich H, Sticht C, Gluch C, Rappold GA. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Molecular Psychiatry. 2015;20(5):632–639. doi: 10.1038/mp.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, LeCouteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychological Medicine. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Baird G, Charman T, Pickles A, Chandler S, Loucas T, Meldrum D, Carcani-Rathwell I, Serkana D, Simonoff E. Regression, developmental trajectory and associated problems in disorders in the autism spectrum: The SNAP study. Journal of Autism and Developmental Disorders. 2008;38(10):1827–1836. doi: 10.1007/s10803-008-0571-9. [DOI] [PubMed] [Google Scholar]
- Barger B, Campbell J, McDonough J. Prevalence and onset of regression within autism spectrum disorders: A meta-analytic review. Journal of Autism & Developmental Disorders. 2013;43(4):817–828. doi: 10.1007/s10803-012-1621-x. [DOI] [PubMed] [Google Scholar]
- Bayés A, van de Lagemaat L, Collins M, Croning M, Whittle I, Choudhary J, Grant S. Characterisation of the proteome, diseases and evolution of the human postsynaptic density. Nature Neuroscience. 2011;14(1):19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaud AL, et al. Autism: highly heritable but not inherited. Nature Medicine. 2007;13(5):534–536. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- Bernabei P, Cerquiglini A, Cortesi F, D’Ardia C. Regression versus no regression in the autistic disorder: Developmental trajectories. Journal of Autism and Developmental Disorders. 2007;37(3):580–588. doi: 10.1007/s10803-006-0201-3. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman H, Coe B, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout A, Schuurs-Hoeijmakers J, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers L, Francescatto L, Mefford H, Rosenfeld J, Bakken T, O’Roak B, Pawlus M, Moon R, Shendure J, Amaral D, Lein E, Rankin J, Romano C, de Vries B, Katsanis N, Eichler E. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Chen AW, Varner JD. A review of the mammalian unfolded protein response. Biotechnology and Bioengineering. 2011;108(12):2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, De Jaco A, Jennings LI, Flynn RE, Gaietta G, Tsigelny I, Ellisman MH, Taylor P. The Arg451 Cys-Neuroligin-3 mutation associated with autism reveals a defect in protein processing. The Journal of Neuroscience. 2004;24(20):4889–4893. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, Barnes CC, Pierce K. Neuron number and size in prefrontal cortex of children with autism. The Journal of the American Medical Association. 2011;306(18):2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- Darnell J, Van Driesche S, Zhang C, Ying K, Hung S, Mele A, Fraser C, Stone E, Chen C, Fak S, Chi S, Licatalosi1 D, Richter J, Darnell R. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriziotis P, O’Roak B, Graham S, Estruch S, Dimitropoulou D, Bernier R, Gerdts J, Shendure J, Eichler E, Fisher S. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nature Communications. 2014 Sep 18;5:4954. doi: 10.1038/ncomms5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano F, Chen RT. Autism and measles-mumps-rubella vaccination: Controversy laid to rest? CNS Drugs. 2001;15(11):831–837. doi: 10.2165/00023210-200115110-00002. [DOI] [PubMed] [Google Scholar]
- Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52(2):307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Falivelli G, De Jaco A, Favaloro FL, Kim H, Wilson J, Dubi N, Ellisman MH, Abrahams BS, Taylor P, Comoletti D. Inherited genetic variants in autism-related CNTNAP2 show perturbed trafficking and ATF6 activation. Human Molecular Genetics. 2012;21(21):4761–4773. doi: 10.1093/hmg/dds320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: A resource for identification of autism genetic risk factors. Neuron. 2010;68(2):192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Fombonne E, Chakrabarti S. No evidence for a new variant of measles-mumps-rubella-induced autism. Pediatrics. 2001;8(4):991–998. doi: 10.1542/peds.108.4.e58. [DOI] [PubMed] [Google Scholar]
- Frazier T, Embacher R, Tilot A, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Molecular Psychiatry. 2014 doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita E, Dai H, Tanabe Y, Zhiling Y, Yamagata T, Miyakawa T, Tanokura M, Momoi MY, Momoi T. Autism spectrum disorder is related to endoplasmic reticulum stress induced by mutations iin the synaptic cell adhesioin molecule, CADM1. Cell Death and Disease. 2010;1(e47):1–7. doi: 10.1038/cddis.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi B, Voight BF, Bućan M. From mouse to human: Evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9(5):e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH. Genetics of autism spectrum disorders. Trends in Cognitive Sciences. 2011;15(9):409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannotti F, Cortesi F, Cerquiglini A, Miraglia D, Vagnoni C, Sebastiani T, et al. An investigation of sleep characteristics, EEG abnormalities and epilepsy in developmentally regressed and non-regressed children with autism. Journal of Autism and Developmental Disorders. 2008;38(10):1888–1897. doi: 10.1007/s10803-008-0584-4. [DOI] [PubMed] [Google Scholar]
- Goin-Kochel RP, Abbacchi A, Duku E, Constantino JN. Familial aggregation of regression status and ADOS parameters among individuals with ASD from the AGRE collection. Oral session presented at the 9th Annual International Meeting for Autism Research; Philadelphia, PA. 2010. May, [Google Scholar]
- Goin-Kochel RP, Esler AN, Kanne SM, Hus V. Developmental regression among children with autism spectrum disorders: Onset, duration, and effects on functional outcomes. Research on Autism Spectrum Disorders. 2014;8(2):890–898. [Google Scholar]
- Goin-Kochel RP, Mire SS, Dempsey AG. Emergence of autism spectrum disorder in children from simplex families: Relations to parental perceptions of etiology. Journal of Autism and Developmental Disorders. 2015;45(5):1451–1463. doi: 10.1007/s10803-014-2310-8. [DOI] [PubMed] [Google Scholar]
- Goin-Kochel RP, Myers BJ. Congenital versus regressive onset of autism spectrum disorders: Parents' beliefs about causes. Focus on Autism and Other Developmental Disabilities. 2005;20(3):169–179. [Google Scholar]
- Goldberg WA, Osann K, Filipek PA, Laulhere T, Jarvis K, Modahl C, Flodman P, Spence MA. Language and other regression: Assessment and timing. Journal of Autism and Developmental Disorders. 2003;33(6):607–616. doi: 10.1023/b:jadd.0000005998.47370.ef. [DOI] [PubMed] [Google Scholar]
- Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. Journal of Autism and Developmental Disorders. 2009;39(5):693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham K, Pickles A, Lord C. Trajectories of autism severity in children using standardized ADOS scores. Pediatrics. 2012;130(5):e1278–e1284. doi: 10.1542/peds.2011-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak B, Sanders S, Ronemus M, Krumm N, Levy D, Stessman H, Witherspoon K, Vives L, Patterson K, Smith J, Paeper B, Nickerson D, Dea J, Dong S, Gonzalez L, Mandell J, Mane S, Murtha M, Sullivan C, Walker M, Waqar Z, Wei L, Willsey A, Yamrom B, Lee Y, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz M, Ye K, McCombie W, Shendure J, Eichler E, State M, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Kreienkamp H. The role of the postsynaptic density in the pathology of the fragile X syndrome. Results Probl Cell Differ. 2012;54:61–80. doi: 10.1007/978-3-642-21649-7_5. [DOI] [PubMed] [Google Scholar]
- Li J, Ma Z, Shi M, Malty RH, Aoki H, Minic Z, Urban AE. Identification of human neuronal protein complexes reveals biochemical activities and convergent mechanisms of action in autism spectrum disorders. Cell Systems. 2015;1(5):361–374. doi: 10.1016/j.cels.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore PC, Risi S. Autism Diagnostic Observation Schedule Manual. Los Angeles, CA: Western Psychological Services; 1999. [Google Scholar]
- Lord C, Shulman C, DiLavore P. Regression and word loss in autistic spectrum disorders. Journal of Child Psychology and Psychiatry. 2004;45(5):936–955. doi: 10.1111/j.1469-7610.2004.t01-1-00287.x. [DOI] [PubMed] [Google Scholar]
- Luyster R, Richler J, Risi S, Hsu W, Dawson G, Bernier R, Dunn M, Hepburn S, Hyman SL, McMahon WM, Goudie-Nice J, Minshew N, Rogers S, Sigman M, Spence MA, Goldberg WA, Tager-Flusberg H, Volkmar FR, Lord C. Early regression in social communication in autism spectrum disorders: A CPEA study. Developmental Neuropsychology. 2005;27(3):311–336. doi: 10.1207/s15326942dn2703_2. [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nature Review Genetics. 2015;16(5):261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- Madsen KM, Hviid A, Vestergaard M, Schendel D, Wohlfahrt J, Thorsen P, Olsen J, Melbye M. A population-based study of measles, mumps, and rubella vaccination and autism. The New England Journal of Medicine. 2002;347(19):1477–1482. doi: 10.1056/NEJMoa021134. [DOI] [PubMed] [Google Scholar]
- Matson JL, Wilkins J, Fodstad JC. Children with autism spectrum disorders: A comparison of those who regress vs. those who do not. Developmental Neurorehabilitation. 2010;13(1):37–45. doi: 10.3109/17518420903107984. [DOI] [PubMed] [Google Scholar]
- Meyer D, Bonhoeffer T, Scheuss V. Balance and stability of synaptic structures during synaptic slasticity. Neuron. 2014;82(2):430–443. doi: 10.1016/j.neuron.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Molloy CA, Keddache M, Martin LJ. Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Molecular Psychiatry. 2005;10(8):741–746. doi: 10.1038/sj.mp.4001691. [DOI] [PubMed] [Google Scholar]
- Molloy CA, Morrow AL, Meinzen-Derr J, Dawson G, Bernier R, Dunn M, Hyman SL, McMahon WM, Goudie-Nice J, Hepburn S, Minshew N, Rogers S, Sigman M, Spence MA, Tager-Flusberg H, Volkmar FR, Lord C. Familial autoimmune thyroid disease as a risk factor for regression in children with autism spectrum disorder: A CPEA study. Journal of Autism and Developmental Disorders. 2006;36(3):317–324. doi: 10.1007/s10803-005-0071-0. [DOI] [PubMed] [Google Scholar]
- Momoi T, Fujita E, Senoo H, Momoi M. Genetic factors and epigenetic factors for autism: Endoplasmic reticulum stress and impaired synaptic function. Cell Biology International. 2010;34(1):13–19. doi: 10.1042/CBI20090250. [DOI] [PubMed] [Google Scholar]
- Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:472–486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- Murata Y, Constantine-Paton M. Postsynaptic density scaffold SAP102 regulates cortical synapse development through EphB and PAK signaling pathway. The Journal of Neuroscience. 2013;33(11):5040–5052. doi: 10.1523/JNEUROSCI.2896-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl CW, Lange N, Li DD, Barnett LA, Lee A, Buonocore MH, Simon TJ, Rogers S, Ozonoff S, Amaral DG. Brain enlargement is associated with regression in preschool-age boys with autism spectrum disorders. Proceedings of the National Academy of Sciences of the Unites States of America. 2011;108(50):20195–20200. doi: 10.1073/pnas.1107560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit PA. Autism's false prophets. New York, NY: Columbia University Press; 2008. [Google Scholar]
- O’Roak B, Stessman H, Boyle E, Witherspoon K, Martin B, Lee C, Vives L, Baker C, Hiatt J, Nickerson D, Bernier R, Shendure J, Eichler E. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nature Communication. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B, Vives L, Fu W, Egertson J, Stanaway I, Phelps I, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt J, Turner E, Levy R, O’Day D, Krumm N, Coe B, Martin B, Borenstein E, Nickerson D, Mefford H, Doherty D, Akey J, Bernier R, Eichler E, Shendure J. Massively multiplex targeted sequencing identifies genes recurrently disrupted in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak B, Vives L, Girirajan S, Karakoc E, Krumm N, Coe B, Levy R, Ko A, Lee C, Smith J, Turner E, Stanaway I, Vernot B, Malig M, Baker C, Reilly B, Akey J, Borenstein E, Rieder M, Nickerson D, Bernier R, Shendure J, Eichler E. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B, Derizioti P, Lee C, Vives L, Schwartz J, Girirajan S, Karakoc E, MacKenzie A, Ng S, Baker C, Rieder M, Nickerson D, Bernier R, Fisher S, Shendure J, Eichler E. Exome sequencing in sporadic autism reveals severe de novo mutations. Nature Genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Iosif AM, Baguio F, Cook IC, Hill MM, Hutman T, Rogers SJ, Rozga A, Sangha S, Sigman M, Steinfeld MB, Young GS. A prospective study of the emergence of early behavioral signs of autism. Journal of the American Academy of Child & Adolescent Psychiatry. 2010;49(3):256–266. [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, Bryson S, Carver LJ, Constantino JN, Dobkins K, Hutman T, Iverson JM, Landa R, Rogers SJ, Sigman M, Stone WL. Recurrence risk for autism spectrum disorders: A Baby Siblings Research Consortium study. Pediatrics. 2011;128(3):e488–495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SK, Schwartz B, Todd J, Pickering LK. Thimerosal-containing vaccines and autistic spectrum disorder: A critical review of published original data. Pediatrics. 2004;114(3):793–804. doi: 10.1542/peds.2004-0434. [DOI] [PubMed] [Google Scholar]
- Parr JR, Lamb JA, Bailey AJ, Monaco AP. Response to paper by Molloy et al.: Linkage on 21q and 7q in autism subset with regression. Molecular Psychiatry. 2006;11:617–619. doi: 10.1038/sj.mp.4001833. [DOI] [PubMed] [Google Scholar]
- Parr JR, Le Couteur A, Baird G, Rutter M, Pickles A, Fombonne E, Bailey AJ. Early developmental regression in autism spectrum disorder: Evidence from an international multiplex sample. Journal of Autism and Developmental Disorders. 2011;41(3):332–340. doi: 10.1007/s10803-010-1055-2. [DOI] [PubMed] [Google Scholar]
- Pickles A, Simonoff E, Conti-Ramsden G, Falcaro M, Simkin Z, Charman T, et al. Loss of language in early development of autism and specific language impairment. Journal of Child Psychology and Psychiatry. 2009;50:843–852. doi: 10.1111/j.1469-7610.2008.02032.x. [DOI] [PubMed] [Google Scholar]
- Pierce K, Eyler LT. Structural and functional brain development in ASD: The impact of early brain overgrowth and considerations for treatment. In: Fein DH, editor. The neuropsychology of autism. New York, NY: Oxford University Press, Inc.; 2011. pp. 407–450. [Google Scholar]
- Richler J, Luyster R, Risi S, Hsu W-L, Dawson G, Bernier R, et al. Is there a regressive phenotype of autism spectrum disorder associated with the Measles-Mumps-Rubella vaccine? A CPEA study. Journal of Autism and Developmental Disorders. 2006;36(3):299–316. doi: 10.1007/s10803-005-0070-1. [DOI] [PubMed] [Google Scholar]
- Rutter M, LeCouteur A, Lord C. Autism Diagnostic Interview-Revised. Los Angeles, CA: Western Psychological Services; 2003. [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, State MW. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway S, Thurm A, Swedo SE, Deprey L, Barnett LA, Amaral DG, Rogers SJ, Ozonoff S. Brief report: Symptom onset patterns and functional outcomes in young children with autism spectrum disorders. Journal of Autism and Developmental Disorders. 2011;41(12):1727–1732. doi: 10.1007/s10803-011-1203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro J, Beaudet AL, Goin-Kochel RP, Bernier R, Faucett WA, Sherr E, Hanson E, Chung WK. Simons Variation in Individuals Project (Simons VIP): A genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. doi: 10.1016/j.neuron.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren I, et al. A twin study of autism in Denmark, Finland, Iceland, Norway, and Sweden. Journal of Child Psychology and Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Steinberg J, Webber C. The roles of FMRP-regulated genes in autism spectrum disorder: single-and multiple-hit genetic etiologies. The American Journal of Human Genetics. 2013;93(5):825–839. doi: 10.1016/j.ajhg.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman H, Bernier R, Eichler E. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156(5):872–877. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Kubota Y, Forest A, Waxham MN. Structure and composition of the postsynaptic density during development. Journal of Comparative Neurology. 2010;518(20):4243–4260. doi: 10.1002/cne.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Zwaigenbaum I, MacLean JE. The genetics of autism: An overview and new directions. Journal of Autism and Developmental Disorders. 1998;28:351–368. doi: 10.1023/a:1026096203946. [DOI] [PubMed] [Google Scholar]
- Taylor LE, Swerdfeger AL, Eslick GD. Vaccines are not associated with autism: An evidence-based meta-analysis of case-control and cohort studies. Vaccine. 2014;32(29):3623–3629. doi: 10.1016/j.vaccine.2014.04.085. [DOI] [PubMed] [Google Scholar]
- Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Current Opinion in Neurology. 2013;26(2):146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, Dawson G, Munson J, Osterling J. Variation in early developmental course in autism and its relation with behavioral outcome at 3—4 years of age. Journal of Autism and Developmental Disorders. 2005;35(3):337–350. doi: 10.1007/s10803-005-3301-6. [DOI] [PubMed] [Google Scholar]
- Zhang C, Milunsky JM, Newton S, Ko J, Zhao G, Maher TA, Tager-Flusberg H, Bolliger MF, Carter AS, Boucard A, Powell CM, Sudhof TC. A Neuroligin-4 missense mutation associated with autism impairs Neuroligin-4 folding and ER export. Journal of Neuroscience. 2009;29(35):10843–10854. doi: 10.1523/JNEUROSCI.1248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu Q, Liu J, Li SC, Xu X. Risk factors for autistic regression: Results of an ambispective cohort study. Journal of Child Neurology. 2012;27(8):975–981. doi: 10.1177/0883073811430163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.