Abstract
TROSY-based triple resonance experiments are essential for protein backbone assignment of large biomolecular systems by solution NMR spectroscopy. In a survey of the current Bruker pulse sequence library for TROSY-based experiments we found that several sequences were plagued by artifacts that affect spectral quality hampering data analysis. Specifically, these experiments produce sidebands in the 13C(t1) dimension with inverted phase corresponding to 1HN resonance frequencies, with approximately 5% intensity of the parent 13C crosspeaks. These artifacts originate from the modulation of the 1HN frequency onto the resonance frequency of 13Cα and/or 13Cβ and are due to 180° pulses imperfections used for 1H decoupling during the 13C(t1) evolution period. These sidebands can become severe for CAi, CAi-1 and/or CBi, CBi-1 correlation experiments such as TROSY-HNCACB. Here, we implement three alternative decoupling strategies that make TROSY-based experiments artifact free and, depending on the scheme employed, boost the sensitivity up to 14% for Bruker pulse sequences. A class of comparable Agilent/Varian pulse sequences that use WALTZ16 1H decoupling can also be improved by this method resulting in up to 60–80% increase in sensitivity.
Keywords: NMR, TROSY-based triple resonance, Backbone Assignment, Pulse Imperfection, Sideband Artifacts
Introduction
Since its introduction in 1997,1 TROSY (Transverse Relaxation-Optimized Spectroscopy) based 3D experiments, such as TROSY-HNCA, TROSY-HN(CO)CA, TROSY-HNCACB, TROSY-HN(CO)CACB etc., have been widely used for backbone chemical shift assignment2–6, dynamic study7,8, and hydrogen bond measurements9 in medium-sized, uniformly labeled proteins (>20 kDa). Due to the increased sensitivity and resolution, TROSY-based experiments replaced HSQC-based pulse sequences.10 TROSY experiments can give improved data when using extensive protein deuteration.4 However, for technical reasons, high yield and complete deuteration for large proteins can be hard to achieve and spurious protonation as high as 10–15% is often present. In these cases, TROSY-based pulse sequences require decoupling of both 1H and/or 2H during the 13C(t1) evolution period to avoid signal losses (Fig. S1).4 Currently, the two most popular spectrometer brands (Bruker and Varian/Agilent) provide convenient libraries of triple resonance pulse sequences that are run as-is in a large number of protein NMR laboratories. In the library of Bruker Topspin up to version 4.0, 1H decoupling for 3D TROSY-based pulse sequences4 is routinely obtained using two 180° hard pulses during 13C(t1) evolution period. The corresponding Agilent/Varian ghn_*.c pulse sequences (VnmrJ Version 4.2 Revision A, issued on May 22, 2014) utilize a 180° hard pulse to refocus 1H antiphase coherence during the first 2T period (total four T delays, and T = 12.5 ms) and a 1H waltz16 decoupling throughout the 13C(t1) evolution (Fig. S2).11
In this study, we tested both Bruker and Varian/Agilent TROSY-based pulse sequences on a U-2H 15N,13C-labeled human heat shock protein 90 (Hsp90) sample (25 kDa), which is 85% deuterated as assessed by mass spectrometry. We found that Bruker experiments generate 3D data with intense side-bands artifacts in the 13C(t1) dimension. These artifacts are particularly problematic for heterogeneously broadened spectra where unstructured regions with very intense HN resonances are present, and are amplified at ultra-high fields spectrometers equipped with cryogenic probes, which are more prone to pulse imperfections. By propagating throughout the 3D planes, the side bands hamper backbone resonance assignments. In contrast, Varian/Agilent experiments do not give rise to sideband artifacts. However, we found that the 1H waltz16 decoupling sequence perturbs 1H spin state, dramatically reducing the sensitivity. In addition, 15N steady state magnetization (v), which is about 11% of 1H steady state magnetization (u), cannot be utilized in these experiments, leading to a further loss in sensitivity.2
Here, we propose three different strategies that improve 1H-decoupling during 13C(t1) evolution time and give rise to artifact-free 3D TROSY-based experiments. Artifacts are eliminated by either applying two-step phase cycling to the two 1H 180° pulses, by adjusting the positions of the two 1H 180° pulses in combination with the application of PFG (Pulsed Field Gradient), or by replacing the two 1H 180° pulses with a phase-modulated rectangular BIP pulses.12,13 These three strategies were successfully applied to 3D TROSY-HNCA and TROSY-HNCACB experiments and are applicable to all TROSY-based 3D experiments utilizing two 1H 180° pulses for 1H decoupling during 13C(t1) evolution period. These approaches enabled us to purge sideband artifacts from the spectra while maintaining or enhancing signal sensitivity up to ~14%. Furthermore, we implemented one of these approaches for the pulse sequences in the Agilent/Varian Biopack library, replacing the 1H waltz16 decoupling during the 13C(t1) period boosting the sensitivity up to 60–80%.
Experimental
NMR data were acquired at 298 K with Bruker 900 MHz spectrometers equipped with a TCI CryoProbe. A U-[85% 2H 15N,13C]-labeled human heat shock protein 90 (Hsp90) sample (25 kDa) in 20 mM phosphate buffer at pH 7.0, 100 mM KCl and 5mM BME was used for testing all of the pulse sequences. All comparisons were conducted using identical acquisition and processing parameters. The 3D TROSY-HNCA comparison was performed with the standard Bruker pulse sequence trhncagp2h3d24 and the updated one (described below). The 3D TROSY-HNCACB comparison was conducted between the reference Bruker pulse sequence (trhncacbgp2h3d2) and the updated one (described below).
The first 2D H-C planes of 3D ghn_ca, ghn_caB, and ghnca_trosy_3DA were acquired with an Agilent/Varian 800 MHz spectrometer equipped with an HCN coldprobe at NMRFAM. The ghn_ca and ghnca_trosy_3DA are pulse sequences included in the Biopack software, and the ghn_caB is a modified version of the ghn_ca sequence (Fig. S3). As test sample, we used a triply labeled (2H,13C,15N, >95% deuterated) protein of molecular weight 37 kDa at a concentration of 0.2 mM in 10 mM Tris buffer at pH 6.5, 50 mM NaCl and 10% D2O.
Results and Discussion
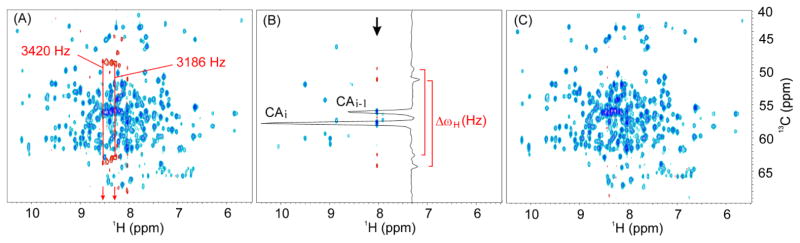
3D TROSY-HNCA spectra acquired using the Bruker standard and optimized trhncagp2h3d2 pulse sequence on a triply labeled protein of 25 kDa are shown in Fig. 1. When using the original Bruker pulse sequence, we found that the CAi or CAi-1 peak shows a pair of sidebands (Fig. 1A,B). The intensity of these sidebands is approximately 5% of the parent peak and are clearly visible above the noise level. Changing the phase cycling of 15N or 13C pulses and removing the steady state magnetization of 15N or 13C with 15N or 13C 90° pulses immediately followed by PFG at the end of d1 had no effect on the artifacts. Upon a close inspection, we noted that the difference in frequency of these satellite bands depends on 1HN chemical shifts (Fig. 1A). These artifacts are symmetric and appear as a cosine modulation of the chemical shift expressed as cos(Δωt) = 0.5(eiΔωt + e−iΔωt), with two peaks per parent peak separated by +/− Δω. In fact, imperfections of the two 1H 180° pulses cause the HzCx or HzCy antiphase coherences (where H and C represent spin operator of 1HN and 13Cα, respectively) evolving during the 13C(t1) time to be transformed into multiple quantum coherences, i.e., HyCx and HyCy, that generate symmetric spectral artifacts.
Fig. 1.
(A) 2D projection of a 3D TROSY-HNCA spectrum acquired with the conventional pulse sequence. Acquisition parameters: recycle delay d1 was 2 s, the number of scans was 8, the raw data size was 20480×64×80 points. The sideband artifacts display inverted phases. The difference in frequency between sideband artifacts decreases from left to right proportionally with the 1H chemical shift (e.g., 3,420 and 3,186 Hz). (B) 2D plane from the 3D TROSY-HNCA spectrum (A) showing a 1D trace featuring the artifacts with inverted phase. The differences in frequency (ΔωH) between two pairs of sideband artifacts arising from two peaks with the same 1HN chemical shift are identical. (C) 2D projection of a 3D TROSY-HNCA spectrum acquired with our modified pulse sequence in which the two 1H 180° hard pulses during 13C(t1) evolution time are phase-cycled with (x, -x) and (x, x, -x, -x), respectively. Blue and red colors denote peaks with positive and negative intensity, respectively. Negative peaks (red) in both (A) and (B) are the sideband artifacts.
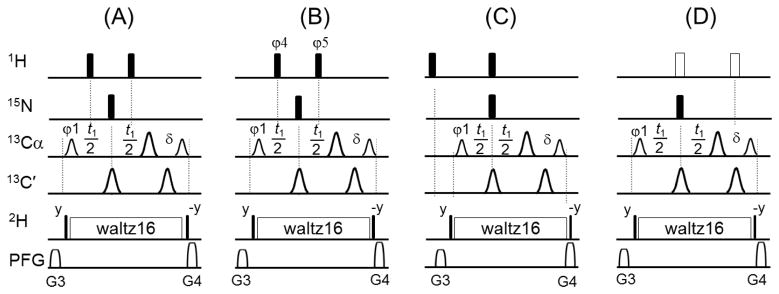
The 13C(t1) evolution period in the pulse sequence of a typical 3D TROSY-HNCA (Fig. S1) is given in Fig. 2A. It consists of two 180° 1H hard pulses given at the midpoint of each t1/2 period that are not phase-cycled in the reference Bruker sequence and result in the sideband artifacts. In the HSQC-based 3D experiments, only one 180° 1H hard pulse or a 1H waltz16 decoupling sequence is used to decouple 1H from 13C during the 13C(t1) evolution time and these elements do not produce artifacts. On the other hand, in the 3D TROSY-based experiments, two 180° 1H hard pulses are necessary for 1H decoupling from 13CA/13CB nuclei. Preventing the exchange between the Hα = 0.5 + Hz and Hβ = 0.5 − Hz spin states of 1HN such that only the slowly relaxing component (before and after the 13C(t1) evolution period) will contribute to the final 15N line shape. These two 180° 1H decoupling pulses are necessary even for deuterated proteins as spurious protonation can be present. However, experimental imperfections of the two 180° 1H hard pulses cause sideband artifacts. In fact, the magnetization at the beginning of Fig. 2A can be described by the HαNzCz and HβNzCz operators, in which H, N and C represent spin operators for 1HN, 15N and 13Cα, respectively, with Hα = 0.5 + Hz and Hβ = 0.5 − Hz. At the end of this pulse sequence, only the slow relaxing component, Hβ = 0.5 − Hz, will be observable. We reasoned that the side band artifacts originate from the fraction of the Hz component of Hβ that is converted to transverse magnetization (Hy) due to imperfections in the first 180° 1H hard pulse. During the 13C(t1) period, Hy evolves and is flipped back to Hz by the second, and also imperfect, 180° 1H hard pulse, resulting in the artifacts. The evolution of Hz magnetization in the pulse scheme of Fig. 2A is given by:
where ΔωH is the resonance frequency of 1HN, and α1 and α2 are the flip angles of the two 180° 1H hard pulses (full spin-evolution in Supplementary Materials). During the 13C(t1) evolution period, Hz is independent from N and C, thus these terms are not included in the expression. The first term, Hz cosα1 cosα2, will contribute to the central main or parent peaks, i.e. CAi and CAi-1 peaks in 3D TROSY-HNCA spectrum or CAi, CAi-1, CBi and CBi-1 peaks in 3D TROSY-HNCACB spectrum. Its intensity depends on cosα1cosα2, e.g., if α1 = 180° and α2 = 180°, the intensity is 1.0; if, for example, we assume a 20° degradation in pulse performance that results in α1 = 160° and α2 = 160°, the intensity is reduced to 0.88. The second term, , will contribute to the sideband artifacts. Note that the negative sign indicates the sideband artifacts has inverted phase with respect to the parent resonance (Fig. 1A) and intensity 0.5sinα1 sinα2. This analysis shows that the position of the two side-band artifacts should be symmetric with respect to the parent peak and the peak separation equal to ΔωH of 1HN. In fact, Fig. 1A shows two examples of peaks with pairs of sidebands with 3,420 and 3,186 Hz, respectively. The position of these sidebands perfectly matches ΔωH, i.e. the distance between the corresponding parent peaks and the center (4.77 ppm) of the 1H dimension. In fact, the parent peaks have 1HN chemical shifts of 8.57 and 8.31 ppm, which correspond to ΔωH values of 3,420 [ΔωH = (8.57–4.77)*900] and 3,186 Hz [ΔωH = (8.31–4.77)*900] respectively, as measured at 900 MHz. The product operator analysis describes artifacts originating from the imperfections of the two 180° 1H hard pulses consistent with the experiment. For example, if α1 = 160° and α2 = 160°, 0.5sinα1 sinα2 = 0.06 the intensity of the artifacts is approximately 7% (0.06/0.88) that of the central parent peaks. For the 1H 90° pulse width calibration we used Bruker standard ‘pulsecal’ routine14; however, accurate calibration of 1H 180° hard pulses is still problematic due to off-resonance effects.15 At higher field magnets (≥700 MHz) with salty biomolecular samples, off-resonance effects for 1H 180° pulse is further exacerbated.
Fig. 2.
(A) Blocks of the 3D TROSY-HNCA pulse sequence including the 13C(t1) evolution period (details in Fig. S1). The filled narrow and wide bars represent 90° and 180° hard pulses, respectively. The smaller and larger shaped pulses on 13C channel represent 213 μs 90° Q5 and 170 μs 180° Q3 pulses, respectively.17 is a short delay to achieve zero evolution time with the first FID. The durations and strengths of the gradients are G3 = (1 ms, 13 G/cm) and G4 = (1 ms, 15 G/cm). By replacing block (A) with either (B), (C) or (D) in the pulse sequence, the sideband artifacts are completely removed. Note that in (B) φ4= (x, -x) and φ5= (x, x, -x, -x). In (C), the locations of the two 1H 180° hard pulses are changed compared to the block (A). In (D), the two open pulses represent phase-modulated rectangular BIP720,50,20.1 pulses with pulse width of 8*p1 (where p1 is the pulse width of 1H 90° hard pulse) and power level set to the corresponding power of 1H hard pulses.12 To further improve their performance, the two BIP720,50,20.1 pulses can also be phase-cycled as shown for the 1H hard pulses in (B).
Three strategies for artifact removal were implemented. First, sideband artifacts can be removed by phase cycling the two 1H 180° hard pulses as shown in Fig. 2B. The intensity of the artifacts is proportional to 0.5sinα1 sinα2, therefore the sign of the intensity is inverted by changing of the sign of α1 or α2. Reversing the phase of a pulse corresponds to changing the sign of the flip angle α. Therefore, setting the φ4 = (x, -x) and φ5 = (x) or φ4 = (x) and φ5 = (x, -x) results in the cancellation of the artifacts over the course of two scans. Increasing the phase cycling to four steps as φ4= (x, -x) and φ5= (x, x, -x, -x) results in almost a complete cancellation over four scans. The phase cycling does not affect the parent peak because its intensity is proportional to cosα1 cosα2. Note that the two or four steps phase cycling for the 1H decoupling pulses is performed simultaneously to the phase cycling of the other 13C and 15N pulses and thus will not increase the minimum number of phase cycling steps in the original pulse sequence. Fig. 1C shows the artifact-free 3D TROSY-HNCA spectrum acquired with the modified pulse sequence (Fig. 2B).
A second strategy uses pulse field gradients (PFG) and repositioning of the two 1H hard 180° re-focusing pulses as shown in Fig. 2C. Based on the evolution of the spin operator (Supplementary Materials), the first imperfect 1H 180° pulse produces a fraction of transverse magnetization Hy that evolves during t1/2 with frequency ΔωH, which will results in the observed artifacts after the second imperfect 1H 180° hard pulse flips a fraction of this transverse magnetization back to the z direction. If we remove the spurious transverse magnetization produced by the first 1H 180° pulse, the sideband artifacts are suppressed (see pulse scheme in Fig. 2C). According to this scheme, the PFG G3 dephases the spurious transverse magnetization produced by the first 1H 180° and PFG G4 will purge the residual magnetization caused by the second 1H 180° pulse. Thus, changing the positions of the two 1H 180° hard pulses removes the artifacts completely. Please note that if the first 1H 180° pulse is moved to any point after PFG G3, the sideband artifacts will still be present in agreement with the spin operator analysis.
Improving the excitation profile and avoiding off-resonance effects should in principle prevent the creation of artifacts. Indeed, replacing the two 1H 180° hard pulses with two phase-modulated rectangular BIP720,50,20.1 pulses12 (Fig. 2D) completely removes the sideband artifacts and, in addition, increases signal intensity by up to 14% compared to spectra obtained with the reference sequence and the other modified ones (Fig. 2A–C). This is easily explained by considering that the signal intensity of the central peak is proportional to cosα1 cosα2, and cosα1 cosα2 = 0.88 if α1 = 160° and α2 = 160°. By replacing the two 1H 180° hard pulses with two BIP720,50,20.1 pulses with a uniform inversion profile and a flip angle α close to 180°, we obtain a signal enhancement up to 14% (=1.00/0.88=1.14). Since the pulse width of 1H 90° hard pulse is 14.4 μs for our sample, BIP pulse length is 115.2 μs (=8×14.4), resulting in a uniform inversion bandwidth of approximately 20,000 Hz that is enough to cover the entire 1H spectrum. Other adiabatic shaped pulses may require longer pulse lengths (≥ 0.5 ms) with possible loss of signal, thereby we chose BIP pulse here. Of the three strategies depicted in Fig. 2, the implementation with BIP pulses gives the best result (Fig. 2D). Note that a similar scheme is used in the BEST-TROSY sequence, where two 180° BIP pulses are used to decouple 1H from 13C during 13C(t1) period.13 In addition, the BIP pulses can also be phase-cycled as that in Fig. 2B to further improve the performance of these experiments.
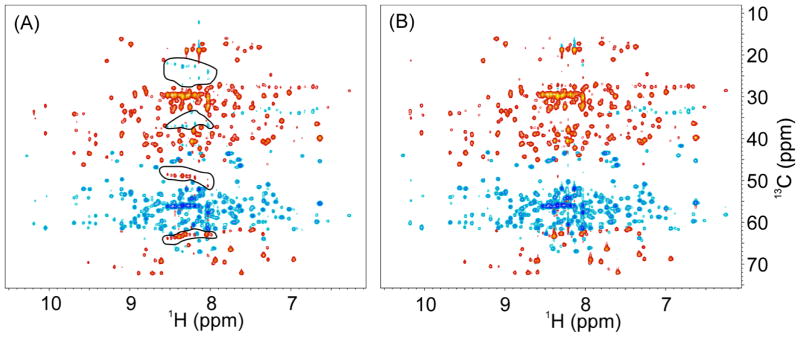
For the 3D TROSY-HNCACB experiment, the sideband problem is more severe since Cα and Cβ resonances have opposite phases and these artifacts can be easily mistaken for real correlation peaks. In fact, the sidebands of the Cα resonances have the same sign as Cβ correlations and may overlap with Cβ resonances of Ser and Thr residues, further complicating the spectra analysis. This problem is clearly illustrated in Fig. 3A, where the red peaks highlighted in two lower circles are sidebands of the intense Cα resonances and may be confused with some of red peaks also present in the lowest circle that are real peaks arising from the Cβ resonances of Ser and Thr residues. On the other hand, the blue peaks in two upper circles are sideband artifacts from the Cβ resonances. In agreement with our observations, the sideband peaks in the HNCACB experiment appear at ΔωH as they are modulated by 1HN frequency. By applying our decoupling strategies, these sideband artifacts were completely removed as shown in Fig. 3B.
Fig. 3.
2D projections of the 3D TROSY-HNCACB experiment. Acquisition parameters: recycle delay d1 was 2 s, the number of scans was 8, and the raw data size was 2048×64×128 points. (A) Bruker pulse sequence trhncacbgp2h3d; (B) our modified version in which the two 1H 180° hard pulses during 13C(t1) evolution time are phase-cycled with φ4= (x, -x) and φ5= (x, x, -x, -x) (see Fig. 2B). Artifactual peaks circled in (A) arise from intense signals from Cα and Cβ resonances. Some of the sideband artifacts overlap with Cβ peaks of Ser and Thr residues.
The standard TROSY pulse programs in the Bruker spectrometer library use two 1H 180° hard pulses in the 13C(t1) dimension for 1H-decoupling, whereas the TROSY versions of the ghn_*.c sequences (where * represents ca, cacb, coca, cocacb, caA, cacbA, cocaA, cocacbA, etc) from the Agilent/Varian spectrometer BioPack’s library use a 1H waltz16 pulse train to remove the scalar J-coupling between 1H and 13C. Although the 1H waltz16 decoupling approach does not generate sideband artifacts, it causes a significant loss in sensitivity. In principle, 1H spin state before and after 13C(t1) evolution time should be preserved so that a particular component of the TROSY multiplet, i.e., the slowly relaxing component, can be selected throughout all four T delays (Fig. S2). The TROSY version implemented in the ghn_*.c pulse sequences in the Agilent/Varian Biopack’s library is based on the work of Weigelt11 in which 1H waltz16 decoupling of 12.5 kHz RF amplitude on a 600 MHz spectrometer is used during 13C(t1) evolution time. The 1H waltz16 decouples 1HCA or 1HCA/1HCB from 13CA or 13CA/13CB, while acting as a low-power spin-lock to preserve 1H spin state. Theoretically, on a 900 MHz spectrometer, the 1H RF amplitude should be 12.5×900/600 kHz, i.e., 18.8 kHz, to have the same RF bandwidth as on a 600 MHz spectrometer. However, the 18.8 kHz RF amplitude exceeds the maximum allowable RF power on the 1H channel. We acquired a 2D H-C plane of the TROSY-HNCA with a pulse sequence described by Weigelt11 and we observed strong artifacts (Fig. S4). Note that regular RF amplitude for 1H waltz16 decoupling on a Bruker 900 MHz spectrometer is only 4.5 kHz that is much lower than the required 18.8 kHz or used 12.5 kHz. Although increasing the RF amplitude of the waltz16 may improve the quality of the spectrum, higher RF amplitude for the waltz16 decoupling is currently not technically feasible on either a Bruker or Varian/Agilent cryogenic probe.
To reduce the higher power requirement of the 1H waltz16 decoupling of the TROSY version of ghn_*.c Agilent/Varian pulse sequences (Fig. S2), it is necessary to modify Weigelt’s experiment.11 Specifically, (1) one 1H 180° hard pulse must be applied during the first T delay, (2) the phase of the second 15N 90° needs to be changed from y to x, and (3) the phase of the first 1H 90° shaped pulse needs to be changed from x to –x for water flipback.16 These modifications transfer 1H,15N antiphase coherence to in-phase 15N magnetization during the first two T delays. At that point, the waltz16 is used only for decoupling and not for spin-locking the magnetization, and a lower RF amplitude (e.g., 4.5 kHz at 900 MHz) can then be used for the waltz16-decoupling. Unfortunately, these modifications also disrupt the TROSY effect. In fact, the waltz16 decoupling disrupts the 1H spin state, and the 15N in-phase coherence (Nx and Ny) of average relaxation rate will be detected during the first two T delays, while the slow relaxing 15N component (HβNx or HβNy) will be detected only during the last two T delays. This indicates that the potential TROSY effect during the first two T delays is not utilized causing a significant loss in sensitivity. Furthermore, since the pulse sequence is HSQC-based before the 13C(t1) evolution, it does not take advantage of the 15N steady state magnetization, which is about 11% of 1H steady state magnetization.2
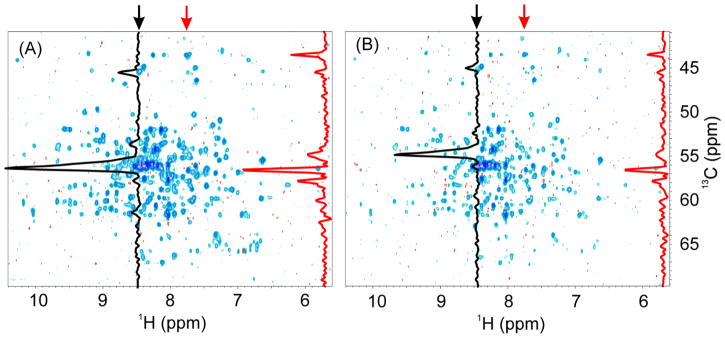
To corroborate our analysis, the Bruker pulse sequence was modified as described in Fig. 4 in a manner similar to the Agilent/Varian Biopack pulse sequence ghn_ca.c, and two experiments were performed back-to-back on Bruker 900 MHz spectrometer. In the experiment with properly implemented TROSY coherence transfers, the strong peaks indicated in Fig. 4A,B, which belong to the Hsp90 flexible termini, show about 30% sensitivity enhancement, while other peaks from the core of the protein show an average 2-fold and up to 2.9-fold higher sensitivity. This difference in signal enhancement is due to the different relaxation properties of the N-H groups that in the core region of the protein show stronger TROSY effect than those in flexible regions. The ratio of the average signal intensity of Fig. 4A,B over 200 well-resolved peaks in the 2D H-C plane is 1.8 ± 0.4 in favor of the sequence using two 1H 180° BIP pulses during 13C(t1) evolution period compared with the modified one that uses 1H waltz16 decoupling.
Fig. 4.
Comparison of the first 2D H-C planes of the 3D TROSY-HNCA spectra acquired with the original Bruker and our modified pulse sequences. (A) spectrum acquired with the pulse sequence shown in Fig. S1 with inset D, (B) corresponding spectrum acquired with a modified pulse sequence to mimic Agilent/Varian pulse sequence in Fig. S2. The modified pulse sequence was based on Fig. S1 in which a 1H 180° pulse was added during the first T delay as in Fig. S2 to refocus 1H antiphase coherence; the phase of the second 1H 90° shaped pulse was changed from -y to +y to achieve water flipback; the phase of the second 15N 90° pulse was changed from y to x; and the two 1H 180° pulses were replaced by 1H waltz16-decoupling as in Fig. S2. Acquisition and processing parameters for the two experiments were identical. On each 2D spectrum, two 1D traces taken at the two frequencies indicated by the arrows are shown. Acquisition parameters: recycle delay d1 was 2 s, the number of scans was 16, and the number of FIDs in 13C(t1) dimension was 128. The average ratio of signal intensities between the A and B panel over 200 peaks is 1.8 ± 0.4.
To further address our concerns with the 1H waltz16 decoupling used in the TROSY version of the 3D ghn_*.c experiments in the Biopack library, the ghn_ca sequence (Fig. S2) was modified to the ghn_caB version (Fig. S3). The first 2D H-C plane of a 3D TROSY-HNCA spectrum was then acquired on a Varian 800 MHz spectrometer equipped with a coldprobe using the ghn_ca and ghn_caB pulse sequences. 1D traces from the 2D planes show that the signal strength of ghn_caB is about 60% higher than that of ghn_ca (Fig. 5). The data acquired on a Varian 600 MHz spectrometer with a room temperature probe demonstrate that the signal strength of ghn_caB is about 30% higher than that of ghn_ca (data not shown). All these clearly show again that the implementation of the 1H waltz16 decoupling in the 3D TROSY ghn_*.c experiments present in the Biopack library of Agilent/Varian spectrometers reduces signal strength significantly. Conversely, modifying these experiments and employing one of the three decoupling strategies described in Fig. 2B, 2C or 2D in place of the 1H waltz16-decoupling will considerably increase their sensitivity. Indeed, the TROSY version of all Biopack pulse sequences with name ‘ghn_*.c’ in the Agilent/Varian library use 1H waltz16 decoupling during 13C(t1) evolution period and will benefit from the modifications outlined here. The signal enhancement resulting from these modifications is derived, in part, from the TROSY effect during the first two T delays, and in part from utilizing the 15N steady state magnetization.2
Fig. 5.
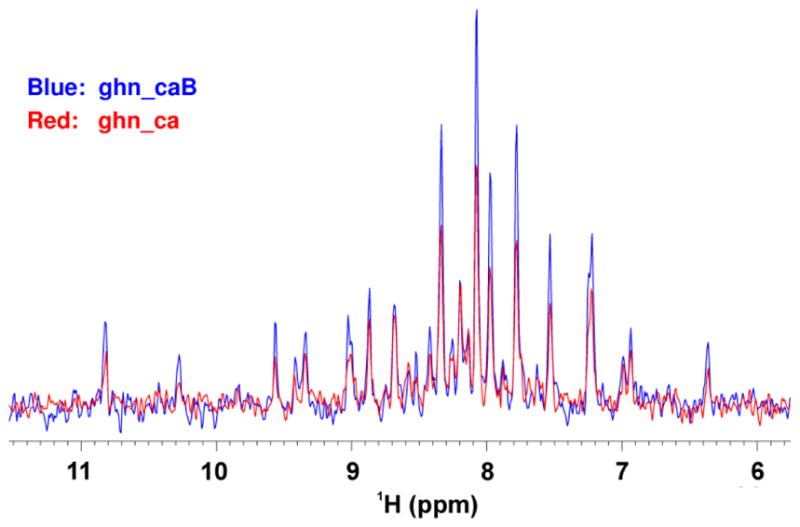
Comparison of the 1D traces extracted from the first 2D H-C planes of the 3D TROSY-HNCA spectrum acquired on a Varian 800 MHz spectrometer with two pulse sequences: ghn_ca and ghn_caB, reported in Fig. S2 and Fig. S3, respectively. Acquisition parameters: recycle delay d1 was 2 s, the number of scans was 128, and 64 complex points in the 13C dimension were used.
Another implementation of the TROSY pulse sequence5,6 available in the Biopack library is named ghn*_trosy_3DA.c, such as ghnca_trosy_3DA.c and ghncacb_trosy_3DA.c. These pulse sequences do not use either 1H waltz16 decoupling or a 1H 180° hard pulses during 13C(t1) evolution time and thus do not have the shortcomings of the ghn_*.c experiments, but take full advantage of the TROSY effect and the 15N steady state magnetization. When tested on a Varian 800 MHz spectrometer equipped with a coldprobe against the ghn_ca and ghn_caB experiments described earlier, the ghnca_trosy_3DA experiment yielded a signal strength comparable to that of the ghn_caB experiment (data not shown). However, since no 1H decoupling of any kind is used during 13C(t1) evolution, proteins with incomplete deuteration will display the 13C-1H moieties doublets with 1JCH splitting, while the 13C-2H will appear as singlets, resulting in signal loss and poor spectral quality. Thus, if 1H decoupling was to be implemented for these experiments, the three strategies described in Fig. 2B–D should be applied to yield clean spectra with improved signal sensitivity for partially deuterated samples.
Finally, we tested the effects of the water signal for our new pulse schemes at high fields (see section titled ‘Effects of the new 1H decoupling schemes on water magnetization’ in Supplementary Information and Fig S5–S10). We found that for conventional evolution times used for large proteins in the indirect dimensions, the water signal does not interfere with the signal acquisition. Only at very long and impractical evolution times in the 13C dimension, the water signal relaxes and affects the FID.
Conclusions
We outlined simple modifications to the TROSY triple resonance experiments that eliminate sideband artifacts along the key 13C(t1) dimension and enhance signal strengths by up to 14%. The artifacts result from imperfections in the 1H decoupling during 13C(t1) evolution time as demonstrated by product operator formalism. Significant improvements are demonstrated for the 3D TROSY-HNCA and TROSY-HNCACB experiments in which artifacts are completely removed by applying one of the three different strategies with no impact on experimental time. Introducing the modification into the Agilent/Varian Biopack-based TROSY triple resonance pulse sequences (ghn_*.c) enhances signal strength by 60 to 80% on 800 MHz and 900 MHz spectrometers, respectively.
Supplementary Material
Acknowledgments
This work is financially supported by the NIH grants GM 100310 to G. V. and AI094623 to C.G.K.. The experiments were carried out at the Minnesota NMR Center (MNMR) and at the National Magnetic Resonance Facility at Madison (NMRFAM) [NIH support: P41GM103399 (formerly P41RR002301); P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220. NSF support: DMB-8415048, OIA-9977486, BIR-9214394]. Many thanks to Prof. E. Komives and Dr. T. Kromann-Tofting at UCSD for providing testing sample.
References
- 1.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–90. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc. 1999;121:844–848. [Google Scholar]
- 4.Eletsky A, Kienhofer A, Pervushin K. TROSY NMR with partially deuterated proteins. J Biomol NMR. 2001;20:177–180. doi: 10.1023/a:1011265430149. [DOI] [PubMed] [Google Scholar]
- 5.Yang DW, Kay LE. Improved (HN)-H-1-detected triple resonance TROSY-based experiments. J Biomol NMR. 1999;13:3–10. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- 6.Yang DW, Kay LE. TROSY triple-resonance four-dimensional NMR spectroscopy of a 46 ns tumbling protein. J Am Chem Soc. 1999;121:2571–2575. [Google Scholar]
- 7.Loria JP, Rance M, Palmer AG. A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J Biomol NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 8.Zhu G, Xia YL, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 9.Wang YX, et al. Measurement of (3h)J(NC ′) connectivities across hydrogen bonds in a 30 kDa protein. J Biomol NMR. 1999;14:181–184. doi: 10.1023/a:1008346517302. [DOI] [PubMed] [Google Scholar]
- 10.Bodenhausen G, Ruben DJ. Natural Abundance N-15 Nmr by Enhanced Heteronuclear Spectroscopy. Chemical Physics Letters. 1980;69:185–189. [Google Scholar]
- 11.Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments (vol 120, pg 10778, 1998) J Am Chem Soc. 1998;120:12706–12706. [Google Scholar]
- 12.Smith MA, Hu H, Shaka AJ. Improved broadband inversion performance for NMR in liquids. J Magn Reson. 2001;151:269–283. [Google Scholar]
- 13.Solyom Z, et al. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 14.Wu PSC, Otting G. Rapid pulse length determination in high-resolution NMR. J Magn Reson. 2005;176:115–119. doi: 10.1016/j.jmr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Levitt MH. eMagRes. John Wiley & Sons, Ltd; 2007. Composite Pulses. [Google Scholar]
- 16.Grzesiek S, Bax A. The Importance of Not Saturating H2o in Protein Nmr - Application to Sensitivity Enhancement and Noe Measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 17.Emsley L, Bodenhausen G. Optimization of Shaped Selective Pulses for Nmr Using a Quaternion Description of Their Overall Propagators. J Magn Reson. 1992;97:135–148. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.