SUMMARY
In mammals during the early postnatal period the environment plays a critical role in promoting the final steps in the neuronal development. While epigenetic factors are thought to contribute to this process, the underlying molecular mechanisms remain poorly understood. Here we show that in the brain during early life the DNA methyltransferase DNMT3A transiently binds across transcribed regions of lowly expressed genes, and its binding specifies the pattern of DNA methylation at CA sequences (mCA) within these genes. We find that DNMT3A occupancy and mCA deposition within the transcribed regions of genes is negatively regulated by gene transcription and may be modified by early-life experience. Once deposited, mCA is bound by the methyl-DNA-binding protein MECP2 and functions in a rheostat-like manner to fine-tune the cell type-specific transcription of genes that are critical for brain function.
INTRODUCTION
The methylation of cytosine is a classical epigenetic modification that plays critical roles in regulating gene expression and maintaining cellular identity (Bird, 2002; Jaenisch and Bird, 2003). The de novo methyltransferases DNMT3A and DNMT3B initiate methylation of cytosines in DNA at the 5’ position of the pyrimidine ring, and DNMT1 maintains methylation of these sites during DNA replication.
In most cell types, DNA methylation occurs at CG sequences (mCG). The mCG mark recruits the methyl-CpG-binding domain (MBD) proteins MBD1, MBD2, MBD4 and MECP2, together with co-repressors that mediate histone deacetylation and transcriptional repression, thus facilitating diverse biological processes such as genomic imprinting, X chromosome inactivation, and cellular differentiation (Bird, 2002). Recent studies have revealed the presence of methylation at non-CG sequences, primarily occurring in CA sequences (mCA), in many tissues and cell types, with neurons appearing to be unique in that they accumulate relatively high levels of mCA specifically during postnatal development (He and Ecker, 2015). While in a given population of neurons specific CA sites are methylated at a lower frequency than CG sequences, across the genome there are many more CA than CG sites so that ultimately CA and CG methylation reach similar levels in adult neurons. The timing of mCA accumulation coincides with an increase in DNMT3A expression in the developing postnatal brain (Lister et al., 2013), and deletion of DNMT3A in the brain results in reduction in mCA across the genome (Gabel et al., 2015; Guo et al., 2014), suggesting that DNMT3A is the primary enzyme that mediates mCA in neurons.
The functional significance of mCA in neurons is underscored by the fact that brain-specific deletions of DNMT3A, or the mCA-binding protein MECP2, give rise to significant neuronal and behavioral deficits (Chahrour and Zoghbi, 2007; Lister and Mukamel, 2015; Nguyen et al., 2007; Sztainberg and Zoghbi, 2016). Consistent with these findings, in humans, DNMT3A is mutated in some individuals with autism spectrum disorder (ASD) (Sanders et al., 2015) as well as those with intellectual disabilities (Tatton-Brown et al., 2014). In addition, loss-of-function mutations in MECP2 give rise to Rett Syndrome (RTT), an ASD that is one the most common causes of severe cognitive impairments in females (Chahrour and Zoghbi, 2007).
To better understand the specific functions of mCA during nervous system development and how its dysregulation contributes to neurological disorders such as RTT, we investigated the mechanisms regulating the deposition of the mCA mark, and how mCA contributes to the control of gene expression as the brain matures. Previous whole genome methylation analyses have shown that the presence of mCA is correlated with decreased gene expression in the brain and in neurons (He and Ecker, 2015), suggesting that mCA might recruit transcription factors to the DNA that function to negatively regulate gene expression. Consistent with this idea, in the cerebellum, the absence of Dnmt3a leads to the subtle up-regulation of many neuronal genes, with long genes that have a high density of mCA across their transcribed regions being affected the most (Gabel et al., 2015). These findings suggest that DNMT3A, by mediating the deposition of mCA within the transcribed regions of genes, functions as a repressor that dials down the level of gene expression.
Here we show that in the mouse brain during early life the DNA methyltransferase DNMT3A preferentially binds transiently to intergenic regions and across transcribed regions of lowly expressed genes, and that this binding primarily determines the pattern of mCA in the adult brain. We find that at sites where mCA is deposited in early life this methyl mark recruits MECP2, which then functions to restrain the transcription of genes in the maturing brain in a neuronal subtype-specific manner. These findings suggest a rheostat model of gene regulation in which DNMT3A, mCA, and MECP2 collaborate to fine control the genes whose precisely tuned expression maybe critical for normal brain development and function.
RESULTS
DNMT3A transiently interacts with the genome during early life
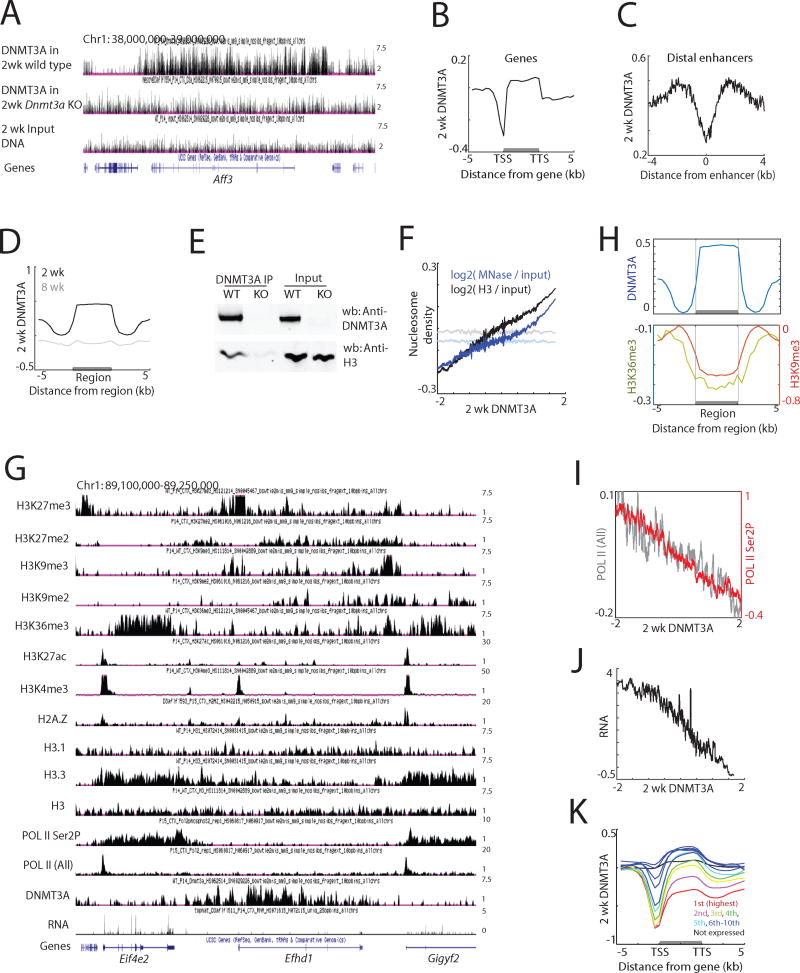
A previous study has shown that the patterns of mCA are at least partly conserved through evolution (Lister et al., 2013) and are neuronal subtype-specific (Mo et al., 2015), suggesting that there must be a conserved mechanism by which DNMT3A is recruited to particular sites in the genome. To identify this mechanism we investigated where across the genome DNMT3A binds, speculating that the sites of DNMT3a binding might provide insight into to the mechanism by which DNMT3A is recruited to DNA, and ultimately how DNMT3A regulates mCA to control gene expression. Given that in neurons the increased expression of the Dnmt3a mRNA and protein peaks two weeks after birth (Feng et al., 2005; Lister et al., 2013), we assessed DNMT3A binding to DNA in two-week-old mouse brain by chromatin immunoprecipitation followed by sequencing (ChIP-seq) using an anti-DNMT3A-specific antibody to enrich for sites of DNMT3A binding (Figures 1A and S1A, B). The specificity of the DNMT3A ChIP-seq analysis was validated using cortical and hippocampal extracts from two-week-old mice in which Dnmt3a expression was conditionally disrupted in the brain (Nestin-cre; Dnmt3afl/fl, hereafter designated Dnmt3a cKO) (Figures 1A and S1A–C). These DNMT3A ChIP-seq experiments revealed that in both brain regions DNMT3A binds broadly across the genome, and that DNMT3A binding is significantly decreased at gene promoters and enhancers (Figures 1B, C and S1D, E). Moreover, consistent with the reduction in DNMT3A expression following early postnatal development (Figure S1F), DNMT3A ChIP-seq using cortical extracts prepared from eight-week-old adult cortex showed that DNMT3A binding across the genome is significantly reduced relative to that in two-week-old mice (Figures 1D and S1G). Thus, we conclude that during the early postnatal period DNMT3A is transiently recruited to and broadly coats the neuronal genome.
Figure 1. Genomic binding of DNMT3A in the brain during early life.
(A) Genome browser views of DNMT3A ChIP-seq data in wild type and Dnmt3a cKO cortex.
(B) Average DNMT3A distribution across all genes. DNMT3A signal was normalized to input DNA. TSS, transcription start site. TTS, transcription termination site.
(C) Average DNMT3A distribution across putative distal enhancers. H3K27ac data from the forebrain over multiple developmental points was used (Nord et al., 2013), and H3K27ac peaks more than one kilobase apart from annotated TSS were analyzed (N=38,620).
(D) Average distribution of 2-week and 8-week DNMT3A across DNMT3A-enriched regions in 2-week cortex (N=22,223).
(E) Immunoprecipitation of DNMT3A under non-denaturing conditions from 2-week cortical extracts from wild type and Dnmt3a cKO (KO) mice.
(F) DNMT3A binding and nucleosome density in two-week cortex across the genome in 5 kilobase tiles. Both MNase-seq and H3 ChIP-seq reads were normalized to sonicated input DNA (log2 ratio). The average nucleosome density was binned according to DNMT3A enrichment relative to input DNA. Nucleosome densities after random grouping of genomic tiles are shown as controls in faded lines (P<0.001, permutation test).
(G) Genome browser view of ChIP-seq data in the two-week cortex.
(H) Average distribution of H3K9me3 and H3K36me3 across defined DNMT3A-enriched regions.
(I) Correlation between DNMT3A binding and RNA POL II occupancy across gene bodies in the two-week cortex. The average POL II (all) or POL II (Ser2P) across gene bodies were binned according to DNMT3A density. Promoter regions were excluded from the analyses.
(J) Correlation between DNMT3A binding and gene expression in the two-week cortex. The average gene expression level was binned according to DNMT3A density.
(K) Average DNMT3A distribution over genes of different expression levels in the wild type cortex.
See also Figure S1.
DNMT3A binds across the transcribed regions of lowly expressed genes
We next investigated what features of the neuronal genome determine where DNMT3A will bind. Previously, the mechanisms of DNMT protein targeting to the genome in vivo have been challenging to characterize, and most advances in our understanding of this process have been made using in vitro approaches. Prior work in other cell types has suggested that DNMT proteins are targeted to the genome through their interaction with specific accessory factors that associate with nucleosomes by recognizing specific histone modifications, or by binding specific genomic sequences (Rose and Klose, 2014). On the basis of these findings we hypothesized that in neurons DNMT3A recruitment might also be dictated by specific features of chromatin that are present during early postnatal development.
We asked whether DNMT3A binds to nucleosomes. Consistent with this possibility, we found that DNMT3A preferentially co-precipitates with histones (Figures 1E and S1H). In addition, micrococcal nuclease digestion of chromatin isolated from the cortex followed by sequencing (MNase-seq) as well as histone H3 ChIP-seq revealed that DNMT3A binding positively correlates with the density of nucleosomes across the genome (Figure 1F). These results suggest that in the brain during early postnatal life DNMT3A interacts with histones within nucleosomes.
To determine if DNMT3A recruitment is correlated with specific features of nucleosomes, we employed ChIP-seq analysis to generate genome-wide maps of a variety of chromatin modifications in two-week-old mouse cortex, a developmental time point during which the brain epigenome has not previously been well characterized. Our analysis included an assessment of seven histone marks associated with active transcription (H3K4me3, H3K36me3), active enhancers (H3K27ac), heterochromatin (H3K9me2, H3K9me3), and Polycomb targeting (H3K27me2, H3K27me3), as well as binding data for three well-characterized histone variants (H3.1, H3.3 and H2A.Z) (Figures 1G and S1I). We also carried out ChIP-seq on total RNA polymerase II (POL II) and elongating forms of POL II (phosphorylated in Ser2 of POL II), as well as RNA sequencing (RNA-seq) analysis to identify the regions of the neuronal genome that are transcribed two weeks postnatally and to assess the level of transcription of each gene.
While no single chromatin feature was sufficient to predict the sites of DNMT3A binding, several broad trends were readily apparent (Figure S1J). DNMT3A is for the most part excluded from genomic regions marked by chromatin features associated with active regulatory elements (e.g. active enhancers and promoters). The exclusion of DNMT3A from these sites is in part consistent with in vitro evidence suggesting that H3K4me3 blocks DNMT3A binding (Ooi et al., 2007). In addition, somewhat surprisingly given the finding that the homologous protein DNMT3B interacts with H3K36me3 in embryonic stem cells (Baubec et al., 2015), DNMT3A was found to be reduced in regions of chromatin that are associated with H3K36me3, a mark that is typically present within the gene bodies of actively transcribed genes (Figures 1H and S1K–M).
We found that DNMT3A is largely excluded from the bodies of genes that are transcribed at a high level, and also absent from genes that are silent due to their inclusion in H3K9 methylation-marked heterochromatin (Figures 1H–J and S1L, N). Instead, DNMT3A appears to bind to most other regions of the genome and perhaps most importantly within the transcribed regions of neuronal genes that are expressed at a relatively low level (Figures I–K), such that the more lowly expressed the gene, the higher the level of DNMT3A binding across the gene. Taken together these findings indicate that DNMT3A associates with nucleosomes and binds broadly across the genome, but is excluded from highly transcribed genes and silenced genes that reside in heterochromatin. Thus, DNMT3A displays preferential binding across genes that are transcribed at a low but detectable level.
Genomic DNMT3A targeting specifies mCA patterns
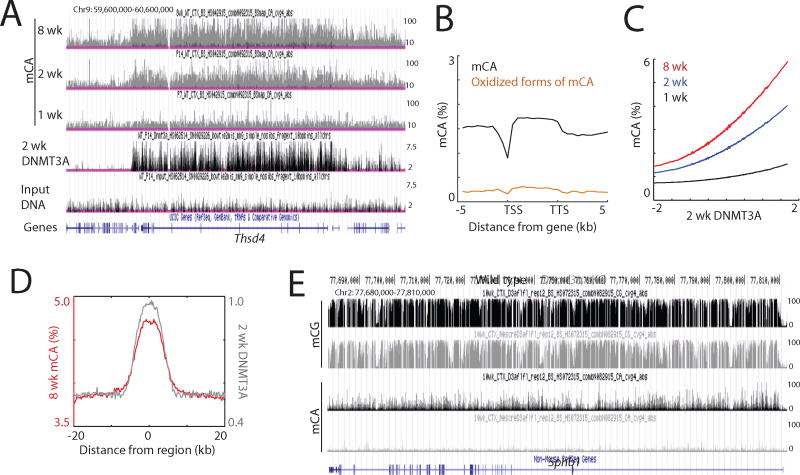
We next asked if DNMT3A binding across the genome determines the sites of mCA deposition. It is unclear, for example, if the sites of mCA deposition are determined primarily by where across the genome DNMT3A is bound, or if once bound to DNA DNMT3A’s enzymatic activity is regulated such that only a subset of DNMT3A-binding sites become methylated (Guo et al., 2015; Li et al., 2011). Alternatively, mCA may be catalyzed by DNMT3A at all sites to which DNMT3A binds, but the mCA mark might be actively removed at specific sites. Having first confirmed the ability of DNMT3A to directly catalyze mCA in vitro (Figure S2A), we performed whole genome bisulfite sequencing (WGBS) to generate base-resolution maps of DNA methylation in both the mouse cortex and hippocampus at one, two and eight weeks of age to determine if the genomic sites that DNMT3A preferentially binds become methylated. We observed a marked genome-wide increase in mCA in the cortex and hippocampus between one week and eight weeks after birth, consistent with a previous study in the frontal cortex (Lister et al., 2013) (Figures 2A and S2B–D).
Figure 2. DNMT3A binding specifies mCA patterns.
(A) Genome browser view of mCA and DNMT3A ChIP-seq data.
(B) Average distribution of mCA and oxidized forms of mCA over genes.
(C) Correlation between two-week DNMT3A binding and mCA in the cortex. mCA was binned according to DNMT3A densities within 5 kilobase tiles across the genome.
(D) Average distribution of 8-week mCA levels over 2-week DNMT3A-enriched regions. Distribution of DNMT3A is also shown.
(E) Genome browser view of DNA methylation in wild type and Dnmt3a cKO cortex.
See also Figure S2.
WGBS does not distinguish between cytosine methylation and oxidized forms of methylation at cytosine that are known to be present at high levels in the brain (Kriaucionis and Heintz, 2009). Because oxidized forms of methylation have been suggested to form as intermediates during the active removal of methyl groups in the CG context (Pastor et al., 2013), we employed oxidative bisulfite sequencing (oxBS-seq)(Booth et al., 2012), a method that enables direct measurement of DNA methylation, to accurately assess the level of mCA (Figure S2E). This analysis confirmed that in the 12-week cortex the percentage of mCA that is oxidized is relatively low, suggesting that most CA sites that become methylated on cortical DNA in early postnatal life are not oxidized later in life (Figures 2B and S2F). This result, together with the finding that DNMT3A primarily binds the genome during early development (Figure 1), and the fact that genome-wide mCA levels in the adolescent mouse and human brains appear to be similar to those in mature brains (Lister et al., 2013), suggest that once CA methylation has occurred in the brain a large proportion of the mCA mark is likely stable and persists throughout the lifetime of the animal.
Comparison of the DNMT3A genomic binding profile with the pattern of mCA revealed a strong correlation between DNMT3A binding to chromatin at two weeks after birth and the mCA pattern later in life (Figures 2A, C and S2C, G). Notably, DNMT3A-bound sites are highly enriched for mCA (Figures 2D and S2H). Moreover, genomic regions that differed in their level of DNMT3A binding between the cortex and hippocampus also display differential patterns of mCA such that regions that had a high density of DNMT3A become highly methylated and those that bind low amounts of DNMT3A are lowly methylated (Figure S2I). Thus, the loci and density of DNMT3A binding are highly correlated with the pattern of mCA with respect to position and density across the neuronal genome.
DNMT3A appears to be the sole enzyme responsible for postnatal CA methylation. By WGBS we found that mCA is essentially eliminated in the cortex of Dnmt3a cKO mice (Figures 2E and S2J, K). By contrast, mCG is partially reduced across the neuronal genome in Dnmt3a cKO mice compared to wild type mice specifically at sites that are bound by DNMT3A in the wild type cortex at two weeks after birth (Figure S2L). This suggests that DNMT3A controls the genome-wide postnatal increase in mCG observed between young and adult brains. However, we note that since the majority of mCG is deposited early in brain development prior to when DNMT3A expression was disrupted, our experiments do not identify which methyltransferases catalyze this early wave of mCG. Collectively, our results indicate that in the early postnatal period a transient physical interaction of DNMT3A with chromatin primarily determines the pattern of mCA, and in part the mCG pattern in the adult brain.
Experience-dependent transcriptional activation disrupts local binding of DNMT3A and mCA deposition
Given that DNMT3A binds across the neuronal genome during the time postnatally when sensory experience first affects neuronal gene expression, we asked if sensory experience/ synaptic activation has an effect on DNMT3A binding to DNA or on deposition of the mCA mark which if affected might ultimately lead to a change in neuronal gene expression.
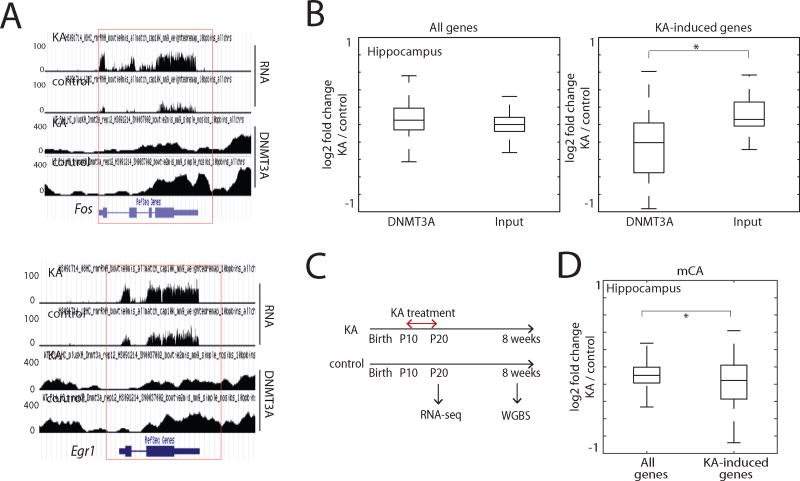
To investigate this possibility we administered two-week-old mice the glutamate receptor agonist kainic acid (KA), a treatment that triggers seizure activity, and is accompanied by a large increase in activity-dependent gene transcription throughout the hippocampus (Bloodgood et al., 2013). Hippocampi were then isolated for DNMT3A ChIP-seq. While compared to vehicle control KA treatment had no obvious effect on the average amount of DNMT3A across the genome, we observed a significant reduction in DNMT3A binding in KA-treated compared to control mice across the transcribed region of genes whose expression is induced by KA treatment, including key immediate-early genes such as Fos and Egr1 (Figures 3A, B and S3A, B). A more modest, but significant decrease in the binding of DNMT3A within the transcribed regions of activity-induced genes was also observed in the visual cortex in response to a more natural stimulation paradigm in which dark-reared mice were acutely exposed to light (Figures S3D–F). Importantly, deletion of DNMT3A had no effect on the level of expression of activity-regulated genes (Figure S3C), suggesting that rather than facilitating activity-dependent gene transcription the decrease in DNMT3A binding and mCA deposition in KA-treated relative to untreated neurons is a consequence of increased expression of the activity-regulated genes upon KA treatment (or light exposure).
Figure 3. Experience-driven gene transcriptional induction disrupts local DNMT3A binding.
(A) Genome browser views of DNMT3A binding and gene expression data in kainic acid-treated (KA) and PBS-treated (control) two-week hippocampi.
(B) Effect of KA treatment on DNMT3A binding over all genes and KA-induced genes (N=51). DNMT3A ChIP-seq and input DNA read densities of two-week hippocampi within gene bodies were calculated, and log2 ratios between KA and control samples are shown. *P<0.0001, Wilcoxon rank-sum test.
(C) Scheme illustrating the KA treatment from P10 to P20.
(D) Boxplots of the difference between 10-week mCA in KA treated and control hippocampi within all genes and KA-induced genes. *P<0.05, Wilcoxon rank-sum test.
See also Figure S3.
To determine if these changes in DNMT3A binding alter the adult mCA pattern at the relevant genes, we exposed mice to a daily injection of KA for ten days beginning on day ten after birth, and then allowed the mice to mature into adults (eight weeks)(Figure 3C). While this treatment does not cause a persistent induction of activity-dependent gene transcription over the ten-day period, we reasoned that the daily exposure would induce transient increases in activity-dependent gene expression over a period of several hours each day (Figure S3G), resembling the effects of persistent seizures on the developing postnatal brain observed in children with epilepsy. WGBS on DNA extracted from the adult hippocampi of KA-treated or untreated mice revealed a modest, but statistically significant decrease in mCA specifically across the bodies of activity-regulated genes in the KA-treated mice (Figure 3D), suggesting that perturbing gene expression during early postnatal brain development has the potential to alter the distribution of mCA across the transcribed regions of genes later in life and in this way may have a long lasting effect on gene expression in adult mice (Figure S3H).
Genetic mutation-driven transcriptional activation disrupts DNMT3A binding and mCA deposition
To assess further whether changes in the transcription of genes during early life can alter the neuronal methylome, we next sought to test whether genetic mutations that disrupt the function of a transcriptional repressor in early life and thus lead to aberrant upregulation of specific genes during early development might lead to changes in mCA levels across the mis-regulated genes. Such a mechanism of gene mis-regulation could in principle contribute to the etiology of neurological disorders that are known to be due to the mutation of a transcriptional repressor (Vissers et al., 2016). One such disorder is Weaver syndrome, a congenital overgrowth disorder associated with intellectual disabilities that is due to mutation of EZH2, one of the two catalytic subunits of the Polycomb repressive complex 2 (PRC2)(Gibson et al., 2012). EZH2 catalyzes H3K27 methylation and functions as a transcriptional repressor in many cell types (Margueron and Reinberg, 2011) including neurons (von Schimmelmann et al., 2016).
To investigate the possible effects of EZH2 loss on neuronal mCA patterning, we crossed Nestin-cre mice to Ezh2fl/fl mice (Shen et al., 2008) to delete Ezh2 in the brain (Ezh2 cKO). As might be predicted, the disruption of EZH2 function in the brain led to severe growth retardation (data not shown)(Zhang et al., 2014), and ChIP-seq analyses of brain extracts from these mice at two weeks of age indicated a genome-wide reduction in H3K27me3, a chromatin signature of gene repression (Figure S4A). By RNA-seq, we found that the genome-wide decreased presence of the H3K27me3 mark in Ezh2 cKO brains was accompanied by an up-regulation in expression of small subsets of both neuronal and non-neuronal genes.
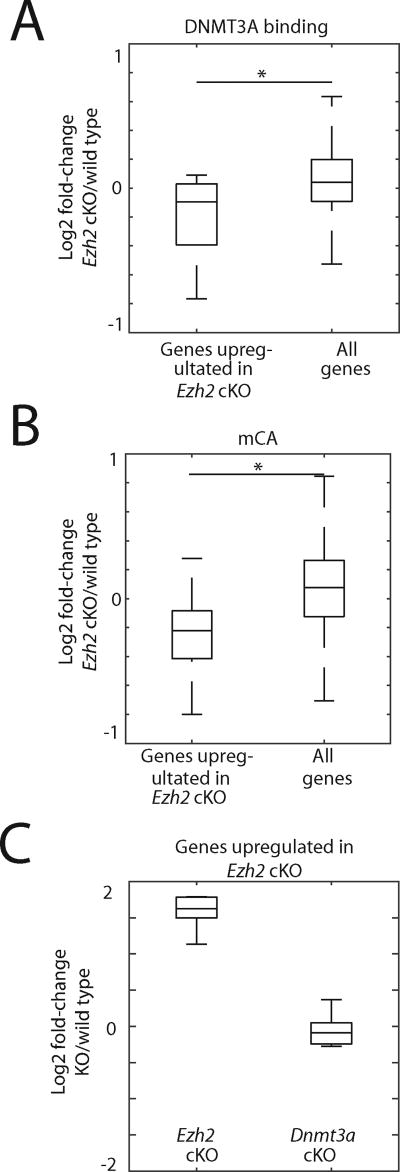
To determine whether DNMT3A binding and mCA patterns are altered across transcribed regions of the genes whose expression is up-regulated in the Ezh2 cKO cortex, we performed DNMT3A ChIP-seq and WGBS. When all genes were analyzed in aggregate, the disruption of EZH2 function was found to have no substantial effect on DNMT3A binding to DNA or mCA levels when Ezh2 cKO and wild type mice were compared. This suggests that EZH2 function is not required for DNMT3A binding to chromatin per se (Figures 4A, B and S4A–E). However, when we focused our analyses in the vicinity of genes whose expression is selectively up-regulated in Ezh2 cKO cortex, we found that DNMT3A binding and mCA levels were significantly reduced within the transcribed regions of these genes in the Ezh2 cKO compared to wild type mice (Figures 4A and S4B).
Figure 4. Genetic mutation-driven gene transcriptional induction disrupts local DNMT3A binding.
(A) Effect Ezh2 mutation on DNMT3A binding over all genes and genes up-regulated by at least 2-fold in Ezh2 cKO (N=11). DNMT3A and H3 ChIP-seq read densities of two-week corticies within gene bodies were calculated, and log2 ratios between Ezh2 cKO and wild type control samples are shown. *P<0.05, Wilcoxon rank-sum test.
(B) Boxplots of the difference between two-week mCA in Ezh2 cKO and wild type control corticies within all genes and genes up-regulated in Ezh2 cKO. *P<0.05, Wilcoxon rank-sum test.
(C) Gene expression levels of genes defined as genes up-regulated in Ezh2 cKO. RNA-seq data for two-week Dnmt3a cKO and respective littermate wild type controls are also shown.
See also Figure S4.
Importantly the disruption of DNMT3A function has no effect on the expression of the genes whose transcription is up-regulated in the Ezh2 cKO mice (Figure 4C). This suggests that the mis-regulation of gene expression in Ezh2 cKO mice is not due indirectly to a reduction in mCA across the transcribed region of these genes. We conclude that rather than facilitating the initial activation of gene transcription, the decrease in DNMT3A binding and mCA is likely a consequence of increased expression of these genes due to the Ezh2 mutation.
Cell type-specific mCA patterns are shaped by early-life gene expression
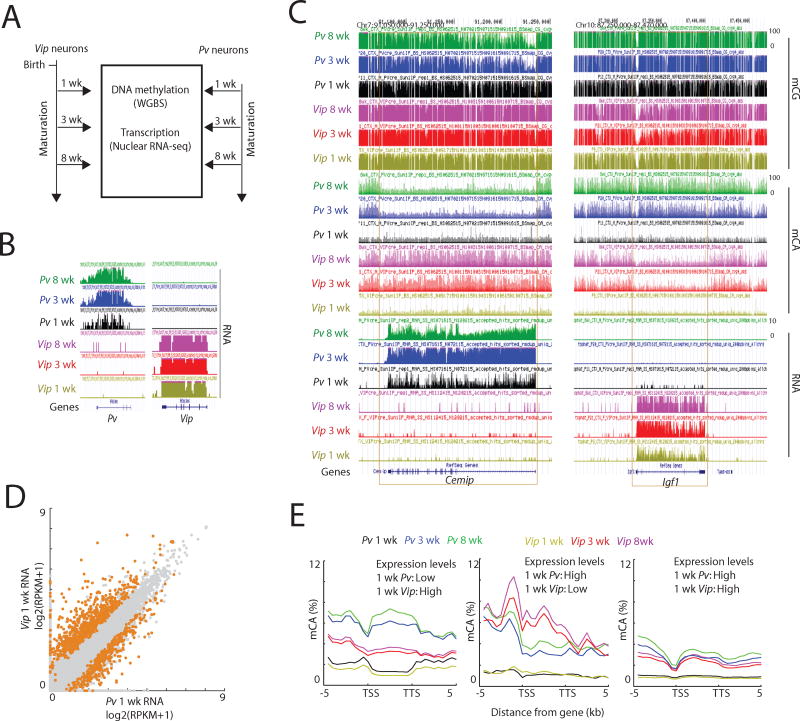
Several recent studies have suggested that the DNA methylation in the brain varies across different neuronal subtypes (Lister et al., 2013; Mo et al., 2015). However, it is not known how these different methylation patterns are established during postnatal brain development and whether the pattern of methylation determines the level of neuronal subtype-specific gene expression or visa versa. To address these issues we compared the developmental DNA methylome and transcriptome dynamics in two specific neuronal subtypes, Pv (parvalbumin)- and Vip (vasoactive intestinal peptide)-expressing interneurons.
Pv and Vip neurons are well-defined, relatively homogeneous, sparse cell types (<5% of total cells in the brain) that function in distinct ways to inhibit cortical circuit function (Markram et al., 2004). Pv neurons mediate somatic inhibition of excitatory pyramidal neurons, whereas Vip neurons have been shown to inhibit somatostatin interneurons which in turn mediate dendritic inhibition of pyramidal neurons. The distinct functions, Pv and Vip neurons may in part reflect the fact that these two neuronal subtypes display distinct patterns of mCA and mRNA expression (Mo et al., 2015). It is noteworthy that Pv neurons have substantially higher genome-wide levels of mCA compared to excitatory neurons (Mo et al., 2015), suggesting that mCA might play a particularly important role in this inhibitory neuronal subtype. By contrast, the level of mCA across the genome of Vip neurons is similar to that of excitatory neurons. The differences in the level of mCA across the genome between these two inhibitory neuron populations made them attractive choices for this analysis.
To determine if differential patterns of gene expression between Pv and Vip neurons in the early postnatal period lead to differential patterns of mCA in these neurons later in life we used the INTACT (isolation of nuclei tagged in specific cell types) method (see STAR Methods) to compare the mCA landscapes and gene expression profiles of Pv and Vip-expressing neurons in the mouse cortex at various stages of postnatal neuronal development. Pv or Vip nuclei were purified at postnatal weeks one, three, and eight (Figures 5A, B), and nuclear RNA-seq data obtained and compared to ribosome-bound mRNA-seq data for Pv- and Vip-expressing neurons to verify that the two approaches yielded similar results (Figure S5A). In addition, we performed WGBS on Pv and Vip purified nuclei to generate base-resolution maps of DNA methylation in the two cell types at each developmental time point.
Figure 5. Cell type-specific transcription during early life shapes mCA patterns across genes.
(A) The scheme of experimental design.
(B) Genome browser views confirming specific enrichment of Pv and Vip transcripts in INTACT-isolated Pv and Vip nuclei, respectively.
(C) Genome browser views of DNA methylation and transcription in cortical Pv and Vip neurons at the indicated developmental time points.
(D) Scatterplot comparing 1 wk Pv and Vip transcript levels across all genes. Genes with differential gene expression (FDR<0.05) are highlighted in orange. Pv > Vip genes: N=363; Vip > Pv genes: N=698.
(E) Average distribution of mCA over genes that were classified as lowly transcribed in one cell type and highly transcribed in the other, as well as genes classified as highly expressed in both cell types (see STAR Methods).
See also Figure S5.
In the one-week-old mouse cortex, Pv and Vip neurons exhibited relatively similar methylomes that lack detectable mCA and possess similar though not identical genomic patterns of mCG (Figures 5C and S5B, C). Importantly, we found that even before mCA accumulation, Pv and Vip neurons at one week of age exhibit pronounced differences in their gene expression profiles (Figures 5D and S5D). For example, young Pv neurons more highly express neuronal genes such as Cacna2d2 and Hapln4 compared to Vip neurons, and Vip neurons more highly expressed genes such as Kcnq4 and Accn4 compared to Pv neurons (Figure S5D).
We found that the density of mCA increases across the genome in both Pv and Vip neurons between postnatal week 1 and 3 (Figures 5C and S5B) and mCA is preferentially associated with lowly transcribed genes in both neuronal subtypes (Figure S5E). Specifically, by postnatal week three, genes that are transcribed at a high level in Pv neurons, but not in Vip neurons at week 1, have become highly methylated at CA sequences within their transcribed regions in Vip, but not Pv neurons (Figures 5C, E and S5F–I). A similar phenomenon was found to be the case for genes that are specifically transcribed at a high level in Vip neurons and a low level in Pv neurons; the lowly expressed genes in Pv neurons became highly methylated in Pv neurons but not Vip neurons. Thus, the decision to express a gene in a neuronal subtype-specific manner appears to be made prior to methylation at CA sequences. Once a gene has been specified to be lowly expressed the gene likely binds DNMT3A within its transcribed region and becomes methylated at CA sequences.
These findings suggest that neuronal subtype-specific patterns of gene expression are determined early during development and that the deposition of mCA occurs subsequently in a neuronal subtype-specific manner. In addition, these observations provide further support for the conclusion that highly expressed genes are refractory to DNMT3A binding and mCA deposition, as these genes only become methylated at CA sequences in neuronal subtypes where they are lowly expressed, not in subtypes where the genes are highly expressed.
mCA mediates cell type-specific fine-tuning of transcription
The function of intragenic mCA is not known and several possible functions can be envisioned. Methylation within the transcribed regions of lowly expressed genes could in principle modulate the expression of these genes either positively or negatively. Alternatively, mCA within gene bodies might preserve transcriptional fidelity by slowing transcriptional elongation, inhibiting spurious transcriptional initiation or preventing POL II from falling off the DNA. To begin to distinguish among these possibilities we blocked the deposition of mCA within the transcribed regions of neuronal genes by disrupting DNMT3A function as described above, and then assessed the effect on neuronal subtype-specific gene expression.
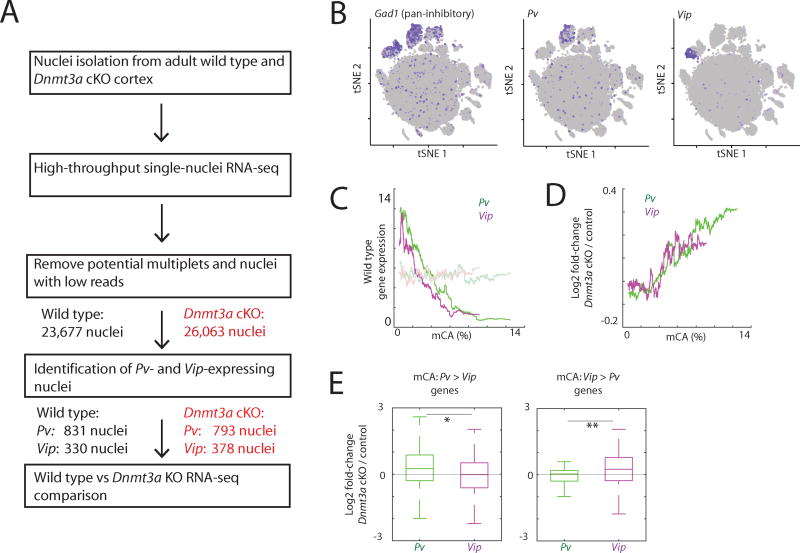
To assess the effect of DNMT3A disruption on gene expression in different neuronal subtypes, we adapted a protocol for high-throughput droplet-based microfluidic single-cell RNA-seq (Klein et al., 2015) so that the protocol could be applied to isolated nuclei. We reasoned that by measuring the level of nuclear RNA we would obtain a more accurate estimate of transcription than by measuring the level of cytoplasmic RNA. We performed single-nuclei RNA-seq using nuclei isolated from the cortices of Dnmt3a cKO or wild type mice, and analyzed the transcriptomes of a total of 26,063 nuclei from the Dnmt3a cKO and 23,677 nuclei from wild type controls (Figures 6A and S6A). Importantly, we found using this approach that we can efficiently distinguish nuclei of different inhibitory neuron subtypes (Figure 6B and S6B).
Figure 6. Single-nuclei sequencing reveals cell type-specific gene expression fine-tuning by mCA.
(A) The scheme of experimental and analysis design.
(B) Seurat t-SNE plots of all nuclei in samples. Nuclei expressing indicated marker genes are depicted in purple.
(C) Correlation between wild type mCA densities within the gene bodies and the expression levels determined by single-nuclei RNA-seq. Gene expression levels after random grouping of genes are shown as controls in faded lines.
(D) Correlation between wild type mCA densities within the gene bodies and transcriptional defects in Dnmt3a cKO cortex.
(E) Cell type-specific gene repression defects in Dnmt3a KO neurons. Genes were separated based on the mCA densities within the gene bodies in Pv and Vip neurons. *P=3.6e-4, **P=0.001, Kolmogorov-Smirnov test.
See also Figure S6.
We found by single-nuclei RNA-seq that wild type Pv and Vip neurons exhibit gene profiles that are similar to those observed in the INTACT experiments, indicating that single-nuclei RNA-seq enables accurate cell type-specific analyses of nuclear RNA (Figure S6C). In wild type Pv and Vip neurons, genes with a high density of mCA within their transcribed regions were lowly expressed, thus confirming our findings using the INTACT approach (see above) (Figure 6C). When we compared Dnmt3a cKO to wild type mice, we found that genes that are normally lowly expressed and have a high density of mCA across their gene bodies are expressed on average at a higher level in Dnmt3a KO neurons compared to wild type neurons (Figures 6D and S6D). Importantly, while the magnitude of the increases in transcription observed in Dnmt3a KO neurons are relatively modest, a large number of neuronal genes are affected, suggesting that mCA likely functions at a genome-wide level to restrain the transcription of lowly expressed neuronal genes. Notably, the more lowly expressed a gene is, the higher the density of mCA, and the greater the degree of mis-regulation when DNMT3A function is disrupted.
We next asked if the disruption of DNMT3A function differentially affects gene expression in Pv and Vip neurons. Remarkably, we observed substantial differences in gene mis-regulation in the Dnmt3a cKO cortex between neuronal subtypes, such that genes with a high density of mCA in Pv neurons, but not in Vip neurons, were preferentially up-regulated in Pv neurons when DNMT3A function is disrupted (Figure 6E). Likewise, when DNMT3A function is disrupted, genes with a relatively high density of mCA selectively in Vip neurons are up-regulated in Vip neurons but not in Pv neurons. These findings collectively suggest that DNMT3A, by catalyzing the methylation of CA sequences within the transcribed regions of lowly expressed genes, dampens the transcription of these genes in a neuronal subtype-specific manner. Given that the pattern of neuronal subtype-specific gene expression is determined prior to DNMT3A binding and the deposition of mCA, these findings are most consistent with a model in mCA functions to restrain the expression of lowly expressed genes in the same manner across various neuronal subtypes, with the differences in gene expression between neuronal subtypes being determined prior to DNMT3A binding and mCA deposition.
mCA controls cell type-specific gene expression by recruiting MECP2 to gene bodies
To begin to investigate how DNMT3A catalyzed mCA functions to restrain the expression of lowly expressed genes we next investigated the neuronal subtype-specific binding patterns of MECP2 and the effect of disrupting MECP2 function on neuronal subtype-specific gene transcription. Mutations in MECP2 cause RTT, and this disorder has been suggested to be due to a loss of binding of MECP2 to mCG and mCA sequences across the neuronal genome (Chen et al., 2015; Gabel et al., 2015; Guo et al., 2014; Lagger et al., 2017). However, whether the selective binding of MECP2 to mCA sequences in different neuronal subtypes accounts for neuronal subtype-specific gene mis-regulation in RTT is not known. In mixed neuronal populations MECP2 has been shown to bind ubiquitously across the genome accumulating at near histone-octamer levels (Skene et al., 2010), and studies have suggested that MECP2 might bind to DNA to some extent regardless of the DNA methylation status (Baubec et al., 2013; Rube et al., 2016). Because accumulating evidence indicates that MECP2 binds avidly to mCA sequences in neurons to recruit the transcriptional co-repressor NCOR and thereby temper gene transcription, we asked whether MECP2 might function to enforce the neuronal subtype-specific dampening of the transcription of mCA-marked genes.
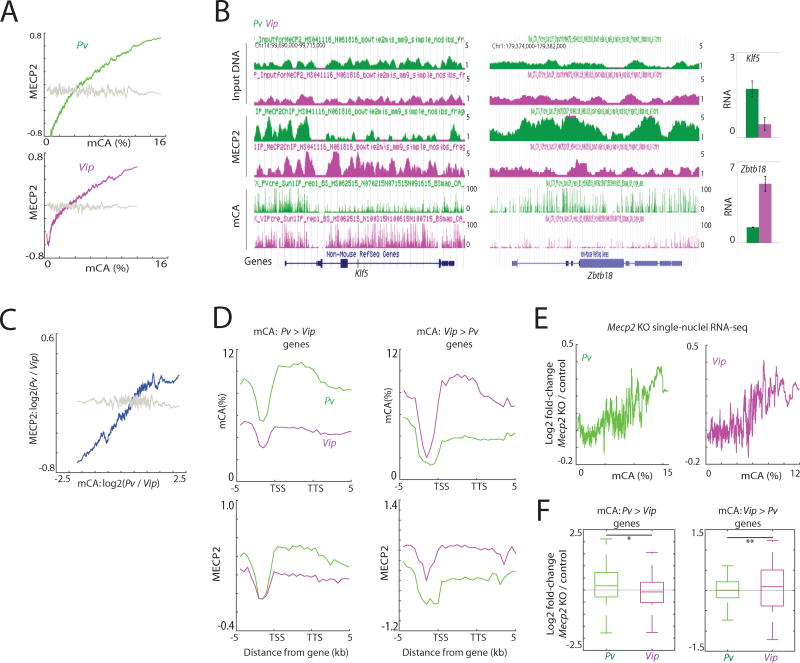
To investigate this possibility, we performed MECP2 ChIP-seq using anti-MECP2 specific antibodies to map MECP2 binding across the genomes of Pv and Vip neurons that we isolated from the adult cortex. By aligning the Pv and Vip MECP2 ChIP-seq data with the Pv and Vip mCA data, respectively (Figure 7A), we found that MECP2 binding within the transcribed regions of genes is significantly correlated with the mCA distribution in both Pv and Vip neurons, providing further evidence that MECP2 binds to mCA. Moreover, we found that genes enriched for mCA specifically in Pv versus Vip neurons are likewise enriched for MECP2 binding and visa versa (Figures 7B–D and S7A, B). We conclude that the binding pattern of MECP2 is at least in part neuronal subtype-specific, and that the subtype-specific pattern of MECP2 binding is associated with cell type-specific differences in intragenic mCA patterns.
Figure 7. Cell type-specific mCA directs MECP2-mediated gene repression.
(A) Correlation between mCA and MECP2 binding in Pv and Vip neurons in the 8-week cortex within genes. The average MECP2 density relative to input DNA was binned according to mCA density within genes. MECP2 densities after random grouping of genes are shown as controls in grey (P<0.001, permutation test).
(B) Genome browser views of mCA and MECP2 profiles in Pv and Vip neurons. Gene expression level determined by INTACT-purified RNA-seq is shown on right. Error bars represent s.d. between two biological replicates.
(C) Correlation between the difference in mCA between Pv and Vip neurons, and MECP2 binding. MECP2 densities after random grouping of genes are shown as controls in grey (P<0.001, permutation test).
(D) Average distribution of MECP2 over genes differentially CA-methylated. (mCA Pv > Vip genes: N=872; Vip > Pv genes: N=84). Distributions of mCA over the genes are also shown.
(E) Correlation between mCA densities within the gene bodies and transcriptional defects in Mecp2 KO cortex (P<0.001 for Pv and P=0.001 for Vip, permutation test). (F) Cell type-specific gene repression defects in Mecp2 KO neurons. Genes were separated based on the mCA densities within the gene bodies in Pv and Vip neurons. *P=3.6e-5, **P=0.04, Kolmogorov-Smirnov test.
See also Figure S7.
To test whether the differences in MECP2 binding between neuronal subtypes might explain the distinct patterns of gene mis-regulation observed in Pv and Vip neurons from the Dnmt3a cKO cortex, we performed high-throughput single-nuclei sequencing of adult Mecp2 KO and wild type control cortices (Figure S7C). By comparing the patterns of gene mis-regulation in Pv and Vip-expressing nuclei from wild type and Mecp2 KO mice, we found that Mecp2 KO neurons exhibit defects in gene expression such that genes with a high density of mCA within their transcribed region (and thus a high density of bound MECP2 in the same region) were preferentially up-regulated (Figures 7E, F and S7D), with the pattern of gene mis-regulation being quite similar to that detected in the same subtype of neurons obtained from Dnmt3a cKO mice (Figure 6D). These findings collectively suggest that mCA contributes to the fine-tuning of genes, including those with critical neuronal functions, in a neuronal subtype-specific manner at least in part by differentially recruiting MECP2 to neuronal gene bodies. Once bound to mCA, MECP2 appears to restrain gene transcription to a level of expression that is directly correlated with the number of mCA marks/MECP2 binding sites per gene thus preferentially regulating some of the longest genes in the genome (Kinde et al., 2016; Lagger et al., 2017; Sugino et al., 2014). We note that the long genes that are up-regulated when MeCP2 function is disrupted are typically lowly expressed in wild type neurons (Figure S7E). These results suggest that transcriptional dysregulation of lowly expressed genes might contribute to RTT and possibly other neurodevelopmental disorders.
DISCUSSION
Our findings suggest that in the mammalian nervous system DNMT3A binds across the genome transiently during early life, in a neuronal subtype-specific manner thereby catalyzing the methylation of nearby CA sequences thus laying down a pattern of neuronal mCA that potentially lasts the life-time of the animal. The probability that a given cytosine within a gene is methylated by DNMT3A is in part dependent on the level of expression of the gene, such that the less the gene is expressed, the more likely cytosines within its transcribed region are to become methylated. While this appears to be a general trend, we note that a small number of high-mCA genes are highly expressed (Figure S7F).
mCA within a gene recruits MECP2 to fine-tune of the expression of the gene, such that the more mCA and MECP2 recruited to the gene body, the more restrained expression of the gene becomes (Figure S7G). In contrast to enhancer-based mechanisms that provide coarse control of gene expression thus augmenting the expression of actively transcribed genes over a wide range of expression levels anywhere between 2 and 1000- fold, the fine control of gene expression by DNA methylation within gene bodies appears to provide precise regulation of lowly expressed genes with a range of 1–2 fold at most.
The precise control of transcription is critical for proper neuronal function inasmuch as aberrant transcription of lowly expressed genes when either DNMT3A or MECP2 are mutated leads to significant defects in brain development and function. One possibility is that mCA/MECP2-dependent fine control of gene expression has evolved to control the expression of genes whose transcription is sufficiently low that typical transcription factor bound regulatory elements, which are generally capable of course control of transcription, would not be able to achieve the precise level of gene expression required for normal neuronal function.
Previous studies have provided evidence that dynamic changes in mCG also occur in response to neuronal activity at specific loci in the genome, although the mechanism and functional significance of the changes remain elusive (Lister and Mukamel, 2015). These findings appear to be consistent with the observation that mCG is prone to oxidation by TET proteins and thus active removal (Pastor et al., 2013). Here, we suggest a mechanism by which the deposition of mCA can potentially be affected by neuronal activity at a time during development when sensory input plays a key role in neuronal maturation. Given that once the mCA mark is deposited during this critical period in brain development it appears to largely remain stable, our findings suggest that patterns of activity in the brain, specifically early in life, may trigger lasting changes in the epigenetic states of genes. This then may have a long-term effect on gene expression and neuronal function much later in life. We note that KA treatment during early life results in subtle changes in mCA, and by single-nuclei sequencing we observe cell type-specific changes in expression of a few thousand genes in the adult hippocampus (not shown). While at this stage we cannot directly assess the causality between the changes in mCA and gene expression, future studies should elucidate the overall role of experience-dependent changes in mCA on gene expression and neuronal function.
Consistent with the findings that DNMT3A catalyzes the deposition of mCA, and MECP2 binds mCA and serves as a reader of mCA, humans and mice that have mutations in either DNMT3A or MECP2 appear to exhibit somewhat similar neurological deficits including intellectual disability in humans, and learning and memory deficits in mice (Chahrour and Zoghbi, 2007; Lister and Mukamel, 2015; Tatton-Brown et al., 2014). In addition, both adult Mecp2 KO and Dnmt3a cKO mice are less active, have weaker grip strength and reduced motor coordination (Figure S7H)(Chahrour and Zoghbi, 2007; Nguyen et al., 2007). In both Dnmt3a cKO and Mecp2 KO mice aberrant phenotypes are less apparent at birth and become more obvious as the mice mature (Figure S7I)(Chahrour and Zoghbi, 2007). Notably, to date Mecp2 KO mice have been more thoroughly characterized than Dnmt3a cKO mice. In the Mecp2 KO mice neurological symptoms appear around the time during the postnatal period when DNMT3A begins to catalyze mCA across the transcribed regions of lowly expressed genes suggesting that both RTT and the DNMT3A deficiency syndrome may be a consequence of the disruption of mCA-dependent gene regulation in neurons.
STAR METHODS
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for reagents should be directed to the Lead Contact Michael E Greenberg (michael_greenberg@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All wild type mice used in this study were in the C57BL/6J background. For conditional Dnmt3a knockout experiments, Nestin-cre mice (Tronche et al., 1999) were crossed to Dnmt3afl/fl mice (Kaneda et al., 2004), where Nestin-cre was always introduced from the male mouse. Littermate controls were used for all comparisons between wild type (Dnmt3afl/fl) and Dnmt3a cKO (Nestin-cre; Dnmt3afl/fl) mice. For conditional Ezh2 knockout experiments, similarly Nestin-cre mice were crossed to Ezh2fl/fl mice (Shen et al., 2008), and littermate controls were used for all comparisons between wild type (Ezh2fl/fl) and Ezh2 cKO (Nestin-cre; Ezh2fl/fl) mice. Homozygous SUN1-2xsfGFP-6xMYC mice were generated in a previous study (Mo et al., 2015) and were crossed with homozygous Pv-cre (Hippenmeyer et al., 2005) and Vip-cre mice (Taniguchi et al., 2011). Mecp2 KO mice have been previously described (Guy et al., 2001).
METHOD DETAILS
Kainic acid (KA) treatment
Postnatal day 14 (P14) mice were injected with either kanic acid (Sigma Aldrich K0250) (5mg/kg) or 1X PBS for controls. KA-injected animals began to seize ~10 minutes after injection, and hippocampi were dissected an hour post-seizure. Hippocampi of treated and untreated mice from the same litter were pooled for ChIP and RNA-seq experiments (two to three mice each). For daily KA administration, we exposed mice to daily injections of KA for ten days beginning day ten postnatally, and then allowed the mice to mature into adults without any injections (eight weeks). Littermate mice were used for all comparisons between KA-treated and control mice.
Dark-rearing/ light exposure experiments
Mice were housed in a standard light cycle until P10 (before eye opening). The mice were then dark-housed until P17. For light stimulation, animals were transferred to the light chamber for one hour prior to dissection of visual cortices. The remaining mice were sacrificed in the dark and not exposed to light (dark-housed controls). The eyes of all animals were enucleated before dissecting the visual cortices. Littermate mice were used for all comparisons between light-exposed and dark-reared mice.
DNMT3A immunoprecipitation
Cortices from two-week-old wild type mice or Dnmt3a cKO mice were homogenized in 8 ml buffer A (10 mM HEPES pH 8.0, 25 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 2M sucrose, 10% glycerol, 1mM DTT, 0.15 mM spermine, 0.5 mM spermidine, Roche protease inhibitor tablets, phosphatase inhibitors 2/3) using a Dounce homogenizer with the “Tight” pestle. Homogenized cortices were rotated at 4°C for 10 min then layered on top of a 2 ml cushion of buffer A and spun at 24,000 rpm for 40 min at 4°C. The nuclei pellet was resuspended in 1 ml buffer NE1 (20 mM HEPES pH 8.0, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, protease inhibitors, phosphatase inhibitors), and was spun at 800 g for 5 min at 4°C. The pellet was resuspended in an equal volume of NE1 (20–30 uL). Three uL of benzonase (Sigma) were added, and the sample incubated for one hour at 4°C. The volume of NE1 was raised t o 500 uL, and the concentration of Triton X-100 raised to 0.5%. NaCl was added to a final concentration of 150 mM. The sample was rotated at 4°C for 20 min, then spun at 16,000 g for 20 min. The supernatant was pre-cleared by incubating with 40 uL Dynabeads protein G (Thermo) for 1 hr, then transferred to a tube containing 40 ul Dynabeads protein G pre-bound with anti-DNMT3A antibodies (Abcam ab13888), and rotated for one hour at 4°C. After incubation, the beads were washed four times with NE1+150 mM NaCl+0.5% Triton X-100, and finally eluted off the beads by adding 50 ul 2X LDS buffer (Thermo) and incubating at 95°C for 10 min.
Chromatin immunoprecipitation sequencing (ChIP-seq)
Tissue was crosslinked 1% formaldehyde. The reaction was stopped with 0.125M glycine. After one wash with PBS, samples were lysed twice with L1 buffer (50mM Hepes pH7.5, 140mM NaCl, 1mM EDTA, 1mM EGTA, 0.25% Triton X-100, 0.5% NP40, 10% Glycerol, protease inhibitors). The samples were incubated in L2 buffer (10mM Tris-HCl pH8.0, 200mM NaCl), then lysed in L3 buffer (1M Tris-HCl pH8.0, 5M NaCl, 0.5 M EDTA, 0.5 M EGTA, 10% Na-Deoxycholate, 20% N-lauroylsarcosine), and sonicated with a Bioruptor (Diagenode). Sonicated chromatin was pre-cleared by incubating with Protein A Dynabeads for 2 hours. Pre-cleared chromatin was added to Dynabeads conjugated to specific antibodies and incubating overnight at 4°C, and 1 to 2.5% of chromatin were kept as input material. Antibodies used: DNMT3A (Santa Cruz 20703), H3.1/H3.2 (EMD Millipore ABE154), H3.3 (EMD Millipore 09-838), H2A.Z (EMD Millipore 07-594), H3K4me3 (EMD Millipore 07-473), H3K9me2 (Abcam ab1220), H3K9me3 (Abcam ab8898), H3K27me2 (Cell Signaling 9728), H3K27me3 (EMD Millipore 07-449), H3K36me3 (Abcam ab9050), H3 (Abcam ab1791), MECP2 (Chen et al., 2003), H3K27ac (Abcam ab4729), RNA Pol II (Abcam 817) and RNA Pol II Ser2P (Abcam 5095). Beads were washed twice with Low Salt buffer (0.1% SDS, 1% Triton X-100, 20mM Tris HCl pH8.0, 150mM NaCl, 2mM EDTA), High Salt buffer (0.1% SDS, 1% Triton X-100, 20mM Tris HCl pH8.0, 500mM NaCl, 2mM EDTA) and LiCl buffer (250mM LiCl, 1% NP40, 1mM EDTA, 10mM TrisHCl pH8.0, 1% Sodium Deoxycholate) at 4°C, and washed once with TE buffer at room temperature. Chromatin was eluted off the beads with TE+1%SDS at 65°C, and de-crosslinked by incubating overnight at 65°C. Samples were treated with RNaseA, and then with Proteinase K for 2 hours at 55°C. DNA was extracted by phenol-chloroform extraction and purified with a Minelute PCR cleanup column (Qiagen). Libraries were constructed by using Ovation Ultralow Library Systems (Nugen) following manufacturer instructions. Libraries were sequenced on an Illumina Hiseq 2500 or Nextseq 500. Reads were mapped to the mm9 genome by allowing up to two mismatches using Bowtie (Langmead et al., 2009). Reads mapping to identical locations were collapsed into one read. For the analysis, reads were normalized to the total number of uniquely mapping reads. To define DNMT3A-enriched regions in the genome, we used SICER (Zang et al., 2009) and compared two-week old DNMT3A-immunoprecipitated samples to input DNA (parameters: W = 1,000; G=2,000; FDR<0.05). For building linear models for DNMT3A binding, ChIP-seq densities were calculated within TSS+/−1kb for promoters, 5kb tiles within gene bodies (excluding 3 kilobases downstream of promoters) and 5kb tiles across intergenic regions (at least 10 kb away from annotated genes). To normalize for nucleosome occupancy, histone ChIP-seq data were normalized to histone H3, whereas DNMT3A and H3 were normalized to input DNA. The model was built using the linear regression function lm() in R. To define regions of the genome enriched in histone marks, we used SICER. For broadly enriched histone marks (i.e. H3.1, H3.3, H3K9me2, H3K9me3, H3K27me2, H3K27me3, H3K36me3) or Pol II Ser2P, parameters of W = 500; G=1,500; FDR<0.05) was used, and for histone marks present as peaks (i.e. H3K4me3, H3K27ac, H2A.Z) or Pol II, parameters of W = 500; G=500; FDR<0.001 was used. Input DNA was used as controls for H3.1, H3.3, Pol II and Pol II Ser2P, whereas H3 ChIP was used as controls of the other histone marks. For plotting average distributions of ChIP-seq reads across specific genomic elements, DNMT3A, POLII and POLII Ser2P were normalized to input DNA, and histone modifications were normalized to histone H3 (log2 ratio).
Immunoblotting
Cortices were lysed in RIPA buffer, ran on a NuPAGE Novex 4–12% Bis-Tris gel (ThermoFisher), and transferred to a nitrocellulose membrane, and anti-DNMT3A (1:200, Santa Cruz 20703) and anti-H3 (1:1000, Abcam 1791) antibodies were used for detecting the proteins.
Micrococcal nuclease sequencing (MNase-seq)
Tissue were homogenized in L1 buffer and spun down. Pellets were resuspended in L2 buffer and rotated at room temperature for 10 min. Tubes were spun down and the pellet was resuspended in 100 µl MNase digestion buffer (20mM Tris-Cl, pH 7.5, 10mM NaCl, 2.5mM CaCl2, 0.01 mM PMSF, 1× Roche protease inhibitors) and incubated with 40µg of RNase A (Qiagen), and incubated at room temperature for 10 min. Treated samples with 1 µl MNase (NEB) for 4min at 37C. The reaction was stopped with addition of 2 µl 0.5M EDTA. Triton X-100 was added to a final concentration of 1%, and samples were rotated at 4°C for 30 min. DNA was subsequently pur ified using Qiagen PCR purification kit. Libraries were constructed by using Ovation Ultralow Library Systems (Nugen) following manufacturer instructions. Libraries were sequenced on a Hiseq 2500 or Nextseq 500. Reads were mapped to the mm9 genome by allowing up to two mismatches using Bowtie (Langmead et al., 2009). For the analysis reads were normalized to the total number of uniquely mapping reads. For analyses presented in Figure 1F, significance was assessed by repeating the random grouping and comparing the Spearman’s correlation coefficients between the actual data and randomized data.
Whole genome bisulfite-sequencing (WGBS)
WGBS libraries were constructed by using a similar method as previously described (Mo et al., 2015) except that NEXTflex methylated adapters (Bioo Scientific) were used. Briefly, DNA was sonicated to 200 bp using a Covaris S2. Bisulfite treatment was performed using EZ DNA Methylation-Gold kit (Zymo). Unmethylated lambda DNA (0.1–0.5%; Promega D1521) was spiked in to assess conversion rates. All datasets generated in this study were confirmed to have conversion error rates of less than 1%. Identical reads were collapsed, and reads were mapped to the mm9 genome using BSmap (Xi and Li, 2009) retaining uniquely mapping reads. Methylation levels were determined by calculating #C/(#C+#T). To calculate methylation within gene bodies, promoter regions, defined as transcription start site (TSS) to TSS+2 kb, were excluded from the analyses. Differentially methylated regions between the cortex and hippocampus were determined by comparing CA methylation levels in 5 kb tiles. To remove tiles with low sequencing coverage and/or with low frequency of CA sequences, tiles that had 100 or greater CA sites that are covered by four reads in both samples were kept. Tiles with absolute mCA difference of 0.02 and Benjamini-Hochberg corrected FDR<0.01 (Fisher’s exact test) were selected. Differentially methylated genes between the adult Pv and Vip neurons were determined by comparing CA methylation levels in within genes (excluding promoter regions), and genes that had 100 or greater CA sites that are covered by four reads in all samples were kept. Genes with absolute mCA difference of 4% and Benjamini-Hochberg corrected FDR<0.01 (Fisher’s exact test) in all biological replicate comparisons were selected. To identify 100 bp genomic tiles enriched in mCG or mCA, data from biological replicates were combined, and 100 bp tiles with >20 #C+#T calls for CG (4,466,873) or >150 #C+#T calls for CA (3,962,712) in all samples (Pv 1 wk, 8 wk, and Vip 1 wk, 8 wk) were selected. Tiles with greater than 70% mCG, or greater than 10% mCA were compared between the neuronal subtypes. For gene ontology analyses, genes within the top 5 percent in mCA densities within the gene bodies were analyzed using DAVID 6.8 (https://david.ncifcrf.gov/)(Huang da et al., 2009). For the analyses presented in Figures 6E and 7F the subtype defined with lower mCA densities, genes with mCA densities above 5% were excluded from the analyses. (mCA Pv > Vip genes: N=345; Vip > Pv genes: N=64).
Oxidative bisulfite sequencing (oxBS-seq)
OxBS-seq was performed on DNA extracted from 12-week-old cortex. DNA was sonicated with a Covaris S2 to 200 bp and ligated to NEXTflex methylated adapters (Bioo Scientific), then bisulfite conversions and oxidation reactions were performed using the TrueMethyl oxBS-seq kit following the manufacturer instructions (Cambridge Epigenetix). Half the sample was used for BS-seq without oxidation treatment and half the sample was used for oxBS-seq as recommended by the manufacturer.
In vitro DNA methyltransferase activity assay
100 ng of unmethylated lambda DNA (Promega D1521) was sonicated to 200 bp using a Covaris S2 and was incubated with 1 µg of full-length human DNMT3A (Abcam 170408) along with 5µM biotinylated histone H3 (amino acids 1–20; Epicypher 12-0001) and S-Adenosyl methionine (SAM) (NEB B9003S) in 0.5mg/mL BSA, 25mM Tris-Cl (pH 8). As a control, the same reaction except without the DNMT3A protein was performed in parallel. The reactions were incubated at 37°C for 1 hour. After the incubation, DNA was isolated by phenol-chloroform extraction followed by ethanol precipitation, and bisulfite sequencing libraries were prepared as described above.
RNA sequencing (RNA-seq)
Total RNA was extracted from cortices and hippocampi with TRIzol (Invitrogen) and purified with an RNeasy kit (with on-column DNase treatment) (Qiagen) following manufacturer instructions. Libraries were generated with either the Tru-Seq Strand Specific RNA-Sequencing kit (Illumina) or NEBNext Ultra Directional RNA Library Prep Kits (NEB). Reads were mapped to annotated genes and the genome using Tophat2 (Kim et al., 2013) and quantified with HTseq (Anders et al., 2015). Gene expression levels were quantified by calculating reads per kilobase of transcript per million mapped reads (RPKM). To define KA-induced genes and light-induced genes, gene expression in KA-treated or light-exposed mice were compared to PBS-injected or dark-reared littermate controls, respectively. Because DNMT3A binding levels primarily varied within genes of wild type expression levels between 1st and 4th deciles, we considered genes with gene expression levels greater than the 5th decile in the treated samples and gene expression levels less than the 1st decile in the control samples. We selected genes that were greater than two-fold in the KA-treated samples compared to control. To define lighted-induced genes, we selected genes that were greater than 1.5-fold in the lighted-exposed samples compared to dark-reared control and a FDR<0.1 cutoff using DEseq (Anders et al., 2015). To define genes upregulated in Ezh2 cKO compared to wild type, we used a two-fold cutoff and a FDR<0.1 cutoff using DEseq. For generating tracks for the genome browser, we tiled the genome into 20 bp bins, and computed number of reads mapping within the bins, divided this value by total mapping million reads to normalize for sequencing depth, and multiplied by an arbitrary constant.
INTACT nuclei isolation
The INTACT method employs a transgenic mouse that expresses in a CRE-dependent manner a SUN1-2xsfGFP-6xMYC fusion protein at the inner nuclear membrane (SUN1 mice). By crossing SUN1 mice with Cre driver lines that promote expression of the SUN1 fusion protein in Pv or Vip neurons we obtained progeny that express the SUN1 fusion protein on the nuclear membrane of Pv or Vip neurons, respectively. SUN1-GFP expressing nuclei were isolated as previously described (Mo et al., 2015) using GFP antibodies (Fisher G10362) coupled with Protein G Dynabeads (Invitrogen 10003D). RNasein Plus RNase Inhibitors (Promega N2611) were added to the buffers to prevent degradation of RNA. One to two mice were pooled for each experiment. Typically ~10 million nuclei were isolated from a cortex from one animal. After the anti-GFP immunoprecipitation, essentially all (~100%) nuclei were GFP positive.
Nuclear RNA-seq
RNA from SUN1-purified nuclei was extracted using Trizol and prepared as described for the standard RNA-seq (using random primers). For data analysis, duplicated reads were removed, and only uniquely mapping reads were retained. Differential gene expression was determined by comparing one week Pv and Vip neuron data by DEseq using a FDR cutoff of 0.05 (Anders et al., 2015). Genes transcribed at a high level in Pv neurons but not Vip neurons were defined as genes with expression levels in Pv neurons to be in the 1st quintile of expression among expressed genes (i.e. RPKM>0) and expression levels in Vip neurons to be in the bottom quintile among expressed genes (N=9). Genes transcribed at a high level in Vip neurons but not Pv neurons were defined using the same criteria (N=46). Genes highly expressed in both Pv and Vip neurons were defined as genes that are within the 1st quintile of expressed genes in both neuron subtypes, and differed in gene expression by less than 10% (FDR>0.1) (N=224).
MECP2 ChIP-seq on SUN1 purified nuclei
ChIP-seq on purified nuclei was performed and analyzed as described above using endogenous antibodies against MECP2 (Chen et al., 2003). For gene ontology analyses, genes within the top 10 percent in MECP2 ChIP read densities relative to input DNA within the gene bodies were analyzed using DAVID 6.8 (https://david.ncifcrf.gov/)(Huang da et al., 2009). For the analyses presented in Figures 7A, C and E, significance was assessed by repeating the random grouping of genes and comparing the Spearman’s correlation coefficients between the actual data and randomized data.
Single-nuclei RNA sequencing (inDrops)
Adult mice were dark-reared for one week prior to dissecting the visual cortices to reduce variation in transcription caused by light exposure (Mardinly et al., 2016). Nuclei from Dnmt3a cKO and Mecp2 KO visual cortices along with those from the respective littermate wild type controls were isolated as previously described (Mo et al., 2015). 0.1% BSA was added to the buffer after nuclei were isolated. Co-encapsulation into microfluidic droplets of nuclei, polyacrylamide gels containing barcoded reverse transcription primers, reverse-transcriptase enzyme, and lysis buffer were performed as previously described to generate barcoded cDNA from single nuclei (Klein et al., 2015; Zilionis et al., 2017). We added 57µL of FD pre-mix (5µL 10X FD buffer; 45µL ddH20; 3µL ExoI (NEB M0293S, 20U/µL); 4µL HinFI (Thermo FD0804) to every 40 µl of sample. Samples were filtered through a 0.45 µm cellulose acetate filter, and incubated at 30 min at 37 °C. Samples were purified with 1.2X volume Agencourt AMPure XP beads (Beckman Coulter A63881). Second strand complementary DNA synthesis was performed by adding 10X NEB Second Strand Synthesis Buffer and NEB Second Strand Synthesis Enzyme Mix (New England Biolabs E6111S) and incubated at 16°C for 2.5 hours, then heat inactivated at 65 °C for 20min. In vitro transcription was performed with NEB T7 High Yield 10X Buffer, dNTPs, NEB T7 High Yield Enzyme Mix (NEB E2040S) and incubating at 37 °C for 13 hours. Samples were purified with 1.3X volume AMPure beads. RNA was fragmented with Fragmentation reagent (Ambion AM8740) by incubating at at 70°C for 3 min, stopped with the S TOP mix, and purified with the 1.2X volume AMPure beads. Reverse transcription was performed using random hexamers: Fragmented RNA were incubated with 10 µM primers (TCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNN) and 10mM dNTPs for 3 min at 70°C. Then added RNase Inhibitor, 5X PrimeSc ript Buffer, 0.5 µL PrimeScript Reverse Transcriptase (Takara Clontech 2680A), and incubated at 30 °C for 10 min, 42 °C for 1 hour, then 70 °C for 15 min. The product w as purified with 1.2X volume AMPure beads. The product was amplified using 2X KAPA HiFi HotStart ReadyMix (KAPA Biosystems KK2601), 5 µM primer mix (PE1: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA; PE2: CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAAC) using the following conditions 98°C 2 min, 2 cycles of (98°C 20sec, 55°C 30sec, 72°C 40sec), 9 to 14 cycles of (98°C 20sec, 65°C 30sec, 72°C 40sec), 72°C 5 min, 4°C hold. The final libraries were purified with 0.7X volume AMPure beads. The libraries were pair-end sequenced for 54 cycles and 21 cycles on a Nextseq 500. We used the published inDrops sequencing read mapping Python pipeline from Klein et al. (Klein et al., 2015)(https://github.com/indrops/indrops) with default parameters (umi quantification - m:10, u:1, d:600, split-ambigs:False, min_non_polyA:15, bowtie [v 1.1.1] - m:200, n:1, l:15, e:80, trimmomatic [v 0.33] - LEADING: "28", SLIDINGWINDOW: "4:20", MINLEN: "16", max_low_complexity_fraction : 0.50) on a custom built transcriptome (made with default parameters of same Python pipeline using the GRCm38.p5 mouse genome assembly with the Ensembl transcript annotation build 85) The data were clustered using Seurat (Satija et al., 2015). Nuclei with at least 500 unique molecular identifiers (UMIs) with at least 200 genes covered were retained for analyses and genes expressed in at least 3 nuclei were kept for analyses. Furthermore, nuclei with greater than 15,000 UMIs or nuclei in which >10% of UMI came from mitochrondial genes were excluded from the analyses. T-distributed stochastic neighbor embedding (t-SNE) plots were generated using the “TSNEplot” function. Gene expression overlays were visualized using the “FeaturePlot” function. We used known neuronal and non-neuronal markers “Slc17a7”, “Gad1”, “Aldoc”, “Olig1”, ”Cx3cr1”, “Cldn5”, “Mrc1” and “Vtn” genes, to identify potential doublets. All nuclei expressing more than one of the above markers were excluded from the analyses. Furthermore, nuclei that expressed more that one of either “Sst”, ”Pvalb” and “Vip” genes were marked as potential doublets of inhibitory neurons and excluded from the analyses. Average normalized gene expression in a given cell type was quantified using Monocle (Trapnell et al., 2014), and these values were used for all downstream analyses. For the analyses presented in Figure S7D, a p<0.05 cutoff was used to define up-regulated genes in either Dnmt3a KO and Mecp2 KO neurons.
QUANTIFICATION AND STATISTICAL AVAILABILITY
Statistical analyses were performed using software noted above, R or in MATLAB.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE103214.
Supplementary Material
Research highlights.
-
*
In the brain DNMT3A binds the genome during early life to specify CA methylation
-
*
DNMT3A preferentially binds across transcribed regions of lowly expressed genes
-
*
DNMT3A binding across genes is modulated by the transcription states of genes
-
*
mCA recruits MECP2 and fine-tunes gene expression in the adult brain
Acknowledgments
We thank J. Nathans and A. Mo for SUN1 mice and E. Griffith, E. Pollina and T. Cherry for editorial assistance, P. Zhang, J. Wang and K. Fazioli for assistance with the mouse colony, and G. Mandel, A. Bird and M. Morselli for helpful discussions. This work was supported by grants from the Rett Syndrome Research Trust and the NIH (RO1NS048276) to MEG, the NARSAD and Hearst Funds to HS. SS was supported by the NRSA training grant and fellowship (T32NS007484 and F32NS089186). HS is an HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG-2194-14).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes seven figures.
AUTHOR CONTRIBUTIONS
HS designed the research and performed most of the experiments. HS and SS performed nuclei isolations. SH, MAN and DH established the computational pipeline for inDrops RNA-seq analyses. AG performed DNMT3A IP experiments. WR and LB generated the Mecp2 KO inDrops data. BK and HWG provided Dnmt3afl/fl mice. MEG advised on all aspects of the study. HS and MEG wrote the manuscript.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015 doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, Zoghbi HY. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci U S A. 2015;112:5509–5514. doi: 10.1073/pnas.1505909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90:110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnersen JM, Kim MH, Fuller SJ, De Silva M, Britto JM, Hammond VE, Davies PJ, Petrou S, Faber ES, Sah P, et al. Sez-6 proteins affect dendritic arborization patterns and excitability of cortical pyramidal neurons. Neuron. 2007;56:621–639. doi: 10.1016/j.neuron.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nature neuroscience. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- He Y, Ecker JR. Non-CG Methylation in the Human Genome. Annu Rev Genomics Hum Genet. 2015;16:55–77. doi: 10.1146/annurev-genom-090413-025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde B, Wu DY, Greenberg ME, Gabel HW. DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc Natl Acad Sci U S A. 2016;113:15114–15119. doi: 10.1073/pnas.1618737114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger S, Connelly JC, Schweikert G, Webb S, Selfridge J, Ramsahoye BH, Yu M, He C, Sanguinetti G, Sowers LC, et al. MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 2017;13:e1006793. doi: 10.1371/journal.pgen.1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BZ, Huang Z, Cui QY, Song XH, Du L, Jeltsch A, Chen P, Li G, Li E, Xu GL. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA. Turning over DNA methylation in the mind. Front Neurosci. 2015;9:252. doi: 10.3389/fnins.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya PC, Hennesy Z, Zhou R, Wagner GC. Changes in attack behavior and activity in EphA5 knockout mice. Brain Res. 2008;1205:91–99. doi: 10.1016/j.brainres.2008.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, Tzeng CP, Mandel-Brehm C, Harmin DA, Adesnik H, Fagiolini M, et al. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature. 2016;531:371–375. doi: 10.1038/nature17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochimica et biophysica acta. 2014;1839:1362–1372. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube HT, Lee W, Hejna M, Chen H, Yasui DH, Hess JF, LaSalle JM, Song JS, Gong Q. Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nature communications. 2016;7:11025. doi: 10.1038/ncomms11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Okaty BW, Arnson HA, Kato S, Dani VS, Nelson SB. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J Neurosci. 2014;34:12877–12883. doi: 10.1523/JNEUROSCI.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, Zoghbi HY. Lessons learned from studying syndromic autism spectrum disorders. Nature neuroscience. 2016;19:1408–1417. doi: 10.1038/nn.4420. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, Zachariou A, Hanks S, O'Brien E, Aksglaede L, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- von Schimmelmann M, Feinberg PA, Sullivan JM, Ku SM, Badimon A, Duff MK, Wang Z, Lachmann A, Dewell S, Ma'ayan A, et al. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nature neuroscience. 2016;19:1321–1330. doi: 10.1038/nn.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. Multiple roles of EPH receptors and ephrins in neural development. Nat Rev Neurosci. 2001;2:155–164. doi: 10.1038/35058515. [DOI] [PubMed] [Google Scholar]
- Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZL, Jiang JM, Wu DH, Xie HJ, Jiang JJ, Zhou L, Peng L, Bao GS. Febrile seizures are associated with mutation of seizure-related (SEZ) 6, a brain-specific gene. J Neurosci Res. 2007;85:166–172. doi: 10.1002/jnr.21103. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji F, Liu Y, Lei X, Li H, Ji G, Yuan Z, Jiao J. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014;34:5184–5199. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.