Abstract
Peroxisomal membrane proteins (PMPs) traffic to peroxisomes by two mechanisms: direct insertion from the cytosol into the peroxisomal membrane and indirect trafficking to peroxisomes via the endoplasmic reticulum (ER). In mammals and yeast, several PMPs traffic via the ER in a Pex3- and Pex19-dependent manner. In Komagataella phaffii (formerly called Pichia pastoris) specifically, the indirect traffic of Pex2, but not of Pex11 or Pex17, depends on Pex3, but all PMPs tested for indirect trafficking require Pex19. In mammals, the indirect traffic of PMPs also requires PEX16, a protein that is absent in most yeast species. In this study, we isolated PEX36, a new gene in K. phaffii, which encodes a PMP. Pex36 is required for cell growth in conditions that require peroxisomes for the metabolism of certain carbon sources. This growth defect in cells lacking Pex36 can be rescued by the expression of human PEX16, Saccharomyces cerevisiae Pex34, or by overexpression of the endogenous K. phaffii Pex25. Pex36 is not an essential protein for peroxisome proliferation, but in the absence of the functionally redundant protein, Pex25, it becomes essential and less than 20% of these cells show import-incompetent, peroxisome-like structures (peroxisome remnants). In the absence of both proteins, peroxisome biogenesis and the intra-ER sorting of Pex2 and Pex11C are seriously impaired, likely by affecting Pex3 and Pex19 function.
Keywords: peroxin, PMP trafficking, pre-peroxisomal vesicle formation, peroxisome biogenesis, ER
Introduction
Peroxisomes are ubiquitous, single membrane-bound organelles present in almost all eukaryotic cells. They participate in a wide variety of metabolic processes, many of which are related to the metabolism of lipids and reactive oxygen species. Although great advances have been made in elucidating the mechanisms and players involved in the import of peroxisomal matrix proteins, the traffic of peroxisomal membrane proteins (PMPs) to peroxisomes has been the subject of much debate because evidence points to the direct, post-translational import of PMPs to the membrane of pre-existing peroxisomes, as well as to the trafficking of many PMPs to peroxisomes via the endoplasmic reticulum (ER) during de novo peroxisome biogenesis, especially when pre-existing peroxisomes are absent [1].
About 35 PEROXIN mutants are currently known across various species, and almost all of them contain peroxisome remnants, consisting of peroxisomal membranes (containing PMPs) but defective in the import of some or most matrix constituents. However, only mutations in genes encoding two peroxins in yeast (Pex3 and Pex19) and three in mammals (PEX3, PEX16, and PEX19) lack such peroxisomal membrane remnants [1, 2].
Strikingly, the re-expression of these missing peroxins in the appropriate mutant cells causes the reappearance of functional peroxisomes. These observations suggest that the cells can replenish pre-existing peroxisomes not only by growth and division but also via an alternative de novo peroxisome biogenesis pathway that does not require morphologically recognizable, pre-existing peroxisomal membranes. These findings establish PEX3, PEX16, and PEX19 as key factors in early peroxisome membrane synthesis.
In Saccharomyces cerevisiae, the size and number of peroxisomes depend on the Pex11 family proteins, which include Pex11, Pex25, Pex27, and Pex34 [3–5]. Several studies in yeast and mammals have clearly demonstrated the role of Pex11 in peroxisomal division, but the functions of Pex25, Pex27, and Pex34 remain more elusive.
Pex3 and Pex19, along with PEX16 in mammals, are key factors in peroxisome biogenesis and are implicated in both the growth and division and the de novo pathways of peroxisome biogenesis [6–8]. In the growth and division pathway, these proteins function in the direct peroxisomal traffic of PMPs from the cytosol. Pex19 is a cytosolic chaperone and an import receptor for PMPs, Pex3 is the peroxisome docking proteins for Pex19, and PEX16 is an integral membrane-bound receptor for PEX3. PEX16 is mostly present in higher eukaryotes, with the exception among yeasts being Yarrowia lipolytica. However, surprisingly, Y/Pex16 is not essential for peroxisome biogenesis; it functions in peroxisome division and not in PMP import [9]. During de novo peroxisome biogenesis, Pex3 and Pex19, plus PEX16 from mammals, function in the indirect traffic of PMPs to peroxisomes via the ER. Despite some controversy about the contribution of indirect PMP trafficking to the de novo peroxisome pathway (formation of new peroxisomes) or to the growth and division pathway (replenishing with PMPs and membrane for newly divided peroxisomes), convincing evidence exists that the ER contributes to the biogenesis of peroxisomes.
Recent findings suggest that at least a subpopulation of PMPs in yeast, plant, and vertebrate cells are targeted first to the ER, and sort from there to a punctate ER subdomain (pER), from which ppVs bud to form peroxisomes. In S. cerevisiae and humans, independent studies suggest that Pex3, and in yeast probably Pex13 and Pex14 as well, insert into the ER, post-translationally via the Sec61 translocon [10–12]. In the same yeast, two ER-resident peroxins, Pex30 and Pex31, contribute to the generation of the pER [13]. In Komagataella phaffii (formerly called Pichia pastoris), two intra-ER sorting routes to the pER are described [14]. In K. phaffii cells, the RING-domain proteins Pex2, Pex10, and Pex12 sort to the pER dependent on Pex3 and Pex19, although Pex2 is packaged in a different ppV than Pex10 and Pex12. The docking subcomplex protein, Pex17 (and probably its interacting partners, Pex13 and Pex14), sorts to the pER independent of Pex3 and Pex19, but it is co-packaged together with Pex10 and Pex12. Finally, both ppVs contain Pex3. All tested PMPs require Pex19 to bud from the pER as shown in K. phaffii and S. cerevisiae cells [14–16]. However, in K. phaffii cells, Pex3 is required for budding of Pex2, but is dispensable for the budding of Pex17- and Pex11-containing ppVs.
Furthermore, the ER-to-peroxisome trafficking of PMPs in mammals appears to be dependent on PEX16, whereby PEX16 itself targets initially to the ER and does so in a co-translational manner. Thereafter, at the ER, PEX16 appears to recruit other PMPs, and together, they traffic to peroxisomes in a yet-to-be identified manner.
However, the de novo model was challenged by a recent study that revealed the existence of pre-peroxisomal vesicles (ppVs) and reticular structures near the perinuclear ER (pn-ER) in Ogataea polymorpha (formerly called Hansenula polymorpha) cells lacking Pex3 or Pex19 [17]. These ppVs had eluded detection because they were degraded by autophagy [18–20]. Cells that lack Pex3 and Atg1, which is required for autophagy, contain ppVs that can mature into functional peroxisomes when Pex3 is re-expressed [17, 20]. This study also showed the presence of ppVs in cells lacking Pex19 and Atg1, but it does not explain how cells devoid of ppVs can form new peroxisomes [17, 21], or how mother cells, devoid of peroxisomes due to forced asymmetrical peroxisome segregation into daughter cells, can also form new peroxisomes [22]. In addition, this study did not eliminate the possibility that Pex3 itself could be trafficked via ER-derived ppVs and delivered to these pre-peroxisomal structures. The authors also speculated that these pre-peroxisomal structures could be derived either from pre-existing peroxisomes or from the ER. If the latter is true, it would be in agreement with their previous study in O. polymorpha demonstrating the de novo formation of peroxisomes from the pn-ER compartment [23].
One possible explanation for the disparate results seen with the PMP import to peroxisomes could be that an individual PMP may not be confined to a single pathway and might be sorted either directly to pre-existing peroxisomes or indirectly through the ER. However, the mechanism and factors that regulate and mediate when, where, and how a PMP will follow a particular route are unknown.
Our data describe a new PMP, Pex36, which shares some functional homology with PEX16 family proteins and ScPex34. We found that the pex36 mutant cells have a serious growth defect in peroxisome proliferation media, and when combined with the pex25 mutation, the phenotype of the double mutant becomes synthetic lethal. Pex25 and Pex36 play redundant roles in bridging the interaction between Pex3 and Pex19, and their absence mimics most phenotypes observed in pex3 mutant cells, which display an intra-ER sorting defect for Pex2 and Pex11C.
Results
Pex36 is a new PMP
An unknown ORF (PAS_chr1-1_0326, PEX36 hereafter) from K. phaffii was initially identified from a collection of peroxisome degradation-defective (pexophagy) mutants generated by UV mutagenesis [24]. The screened pexophagy phenotype of the UV-generated mutant PEX36 (pex36UV) was confirmed by the slow rate of alcohol oxidase 1 (AOX1) degradation in the mutant, in comparison to wild-type (WT) cells, after shifting cells from methanol to glucose medium without nitrogen (SD-N, pexophagy conditions) (Fig. S1 A). However, this defect in pex36UV cells was indirectly caused by a peroxisomal matrix protein import defect (described later) in which the AOX1 protein was mostly cytosolic and thereby escaped degradation by pexophagy. However, separate from AOX1, the peroxisome membrane protein Pex12 was normally degraded. Surprisingly, pex36Δ cells expressing Pex3 fused to the monomeric red fluorescent protein (Pex3–mRFP) were proficient for pexophagy, as judged by the red vacuolar staining (Fig. S1B) and by the appearance of free mRFP (Fig. S1C, due to vacuolar processing of Pex3-mRFP) during conditions suitable for peroxisome proliferation such as methanol medium (SM). In S. cerevisiae and mammals, such phenotypes have also been recently described in cells deleted for, or mutated in, the PEX1, PEX6, or PEX15/26 genes [25, 26]. Similarly, in K. phaffii, deletion of these genes also induced peroxisome degradation, and deleting the autophagy gene, ATG5, blocked peroxisome degradation caused by the deletion or mutation of the PEX1, PEX6, or PEX36 genes, confirming that the phenotype of the single, autophagy-proficient PEROXIN mutants tested was due to induced pexophagy (Fig. S1B, C).
Homologs of Pex36 protein are restricted to members of Saccharomycetaceae, particularly to the genus Komagataella, Ogataea, and Candida, but are absent in Saccharomyces. A low sequence similarity with Pex11 and Pex25 from yeasts and PEX11γ from higher eukaryotes was found after a second iteration of PSI-Blast (Fig. S2A), suggesting that Pex36 might be a member of the Pex11 family of proteins, comprising S. cerevisiae Pex11, Pex25, Pex27, and Pex34. K. phaffii contains Pex11, Pex11C, and Pex25, but lacks the Pex27 and Pex34 homologs.
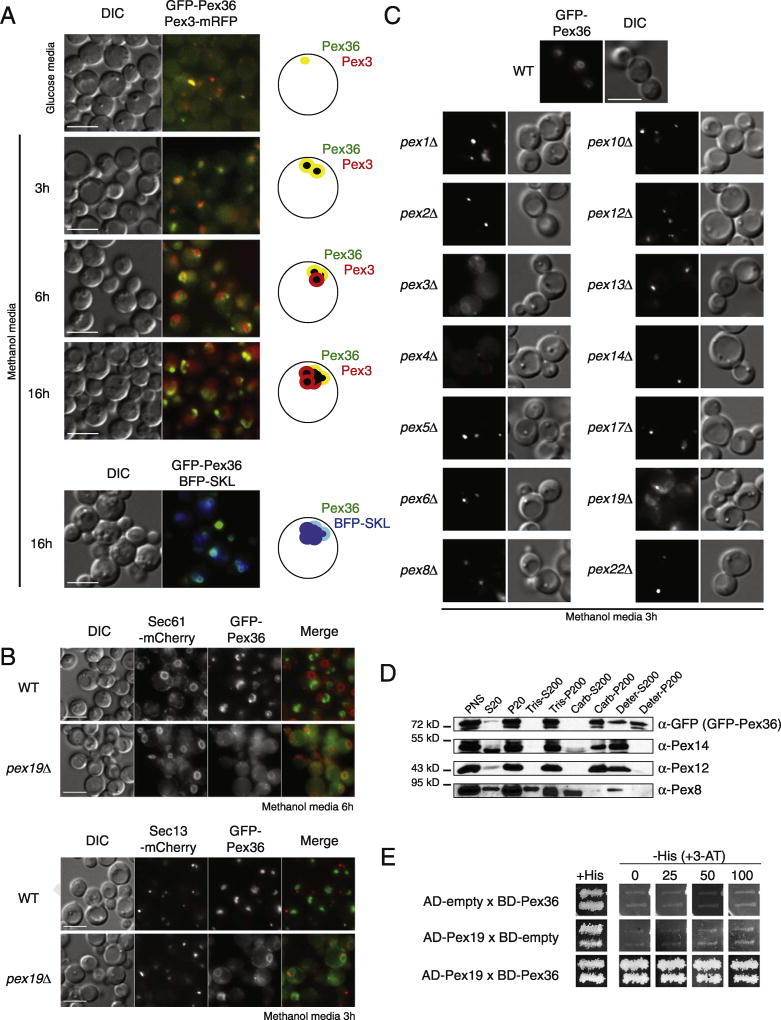
Pex36 tagged at its N-terminus with GFP (GFP-Pex36) was functional (see later), colocalizing with peroxisomes labeled with Pex3-mRFP and with the peroxisomal matrix protein marker, BFP tagged with a PTS1 peptide (BFP-SKL) (Fig. 1A, 16 h). However, unlike Pex3-mRFP, after 3 h in methanol medium, GFP-Pex36 was not evenly distributed around the peroxisome cluster; rather it accumulated where the peroxisomes face the cell periphery. In pex19Δ cells lacking peroxisomes, GFP-Pex36 localized mostly at the ER, as seen by the partial colocalization with the resident ER protein Sec61 (Sec61-mCherry) (Fig. 1B). Most of the GFP-Pex36 localized at the pn-ER, but in some cells, a single-dot structure was often observed near the pn-ER, which did not colocalize with the transitional ER protein, Sec13 (Sec13-mCherry) (Fig. 1B). These results suggest that Pex36 is a bona fide PMP that traffics via the ER to peroxisomes. Like most PMPs, GFP-Pex36 trafficked normally to peroxisome remnants, known to be present in mutants lacking key peroxins implicated directly in the import of peroxisomal matrix proteins, such as Pex1–2, 4–6, 8, 10, 12–14, 17, and 22 (Fig. 1C).
Fig. 1. KpPex36 is a PMP.
(A) GFP-Pex36 colocalizes with the chimeric PMP, Pex3–mRFP, and with the peroxisomal matrix protein, BFP-SKL, to punctate structures. The bar represents 5 µm. (B) Fluorescence microscopy images of WT and pex19Δ cells expressing GFP-Pex36, Sec61-mCherry (ER), and Sec13–mCherry (transitional ER). The bar represents 5 µm. (C) Fluorescence microscopy images of GFP-Pex36 in WT and pex mutant cells. The bar represents 5 µm. (D) GFP-Pex36 localizes to the P20 organelllar fraction enriched for peroxisomes. Immunoblot analysis of equivalent portions of the S20 and P20 fractions from cells expressing GFP-Pex36 was performed with antibodies to GFP-Pex36, the peroxisomal peripheral membrane protein, Pex14, and to the peroxisomal matrix protein, Pex8. Organelles in the P20 fraction from cells expressing GFP-Pex36 were treated with 10 mM Tris–HCI, pH 8.0, to lyse peroxisomes and were processed to yield the supernatant (Tris-S200) fraction enriched for matrix proteins and the pellet (Tris-P200) fraction enriched for membrane proteins. The P20 fraction was also treated with 0.1 M Na2CO3, pH 11.3, and separated into the supernatant (Carb-S200) fraction enriched for matrix and peripheral membrane proteins, and the pellet (Carb-P200) fraction enriched for integral membrane proteins. Finally, the P20 fraction was treated with detergents and separated into the supernatant (Deter-S200) fraction enriched with most of the peroxisomal proteins and the pellet (Deter-P200) fraction enriched with detergent-insoluble proteins. (E) Pex36 (BD-Pex36) interacts with Pex19 (AD-Pex19) in yeast-two-hybrid. 3-AT, 3-amino-1,2,4-triazole; AD, activation domain; BD, DNA binding domain.
Organelle extraction after 6 h of peroxisome induction in oleate medium was used to determine the sub-organellar location of Pex36. Organelles in the P20 (20,000g pellet) fraction were subjected to hypotonic lysis in Tris buffer, followed by ultracentrifugation at 200,000g to yield a supernatant (Tris-S200) fraction enriched for soluble proteins and a pellet (Tris-P200) fraction enriched for membrane proteins (Fig. 1D). GFP-Pex36 co-fractionated with the PMP Pex12, and the peroxisomal peripheral membrane protein Pex14, to the Tris-P200 fraction, whereas a considerable fraction of the soluble peroxisomal matrix protein, Pex8, was in the Tris-S200 fraction.
The P20 fraction was also extracted with alkaline Na2CO3 and subjected to ultracentrifugation. This treatment releases proteins in the matrix and those associated with, but not integral to, membranes [27]. GFP-Pex36 was detected exclusively in the pellet (Carb-P200) fraction enriched for integral membrane proteins, like Pex12, and no GFP-Pex36 was released into the supernatant (Carb-S200) fraction, like the peripheral membrane protein, Pex14. Finally, the integral membrane proteins, together with the rest of the peroxisomal proteins, were extracted from the P20 fraction with a detergent mix (TritonX-100/Chaps) and subjected to ultracentrifugation. As expected, GFP-Pex36 co-fractionated to the supernatant (Deter-S200) fraction, with all peroxisomal proteins tested. These data suggest that Pex36 is an integral PMP, consistent with the predictions of two topology prediction programs HMMTOP† and TMpred‡ that predict two trans-membrane spanning regions in Pex36, whose membrane topology is described later.
Since the Pex36 protein sequence does not provide any functional information regarding its role in peroxisome biogenesis, a GAL4-based yeast two-hybrid screen was performed to discover any putative interacting partners. To test whether our laboratory's collection of yeast two-hybrid (Y2H) functional peroxins interacted with Pex36, plasmids containing a binding domain (BD)-PEX36 and activation domain (AD)-PEX36 were transformed into the reporter strain AH109, in combination with a range of BD-peroxin and AD-peroxin constructs. Double transformants were analyzed for reporter gene expression by assaying the growth in -His + 3-amino-1,2,4-triazole (3-AT) plates (Figs. 1E and S3). Only a single double-transformant, BD-PEX36 and AD-PEX19, grew in the -His + 3-AT in considerable amounts. No obvious function could be elucidated from these data, as most of the PMPs interact with Pex19, a PMP receptor, as part of their own traffic to the peroxisomes. However, the interaction with Pex19 also confirmed Pex36 as a new K. phaffii PMP.
Phenotype of cells lacking the PEX36 gene
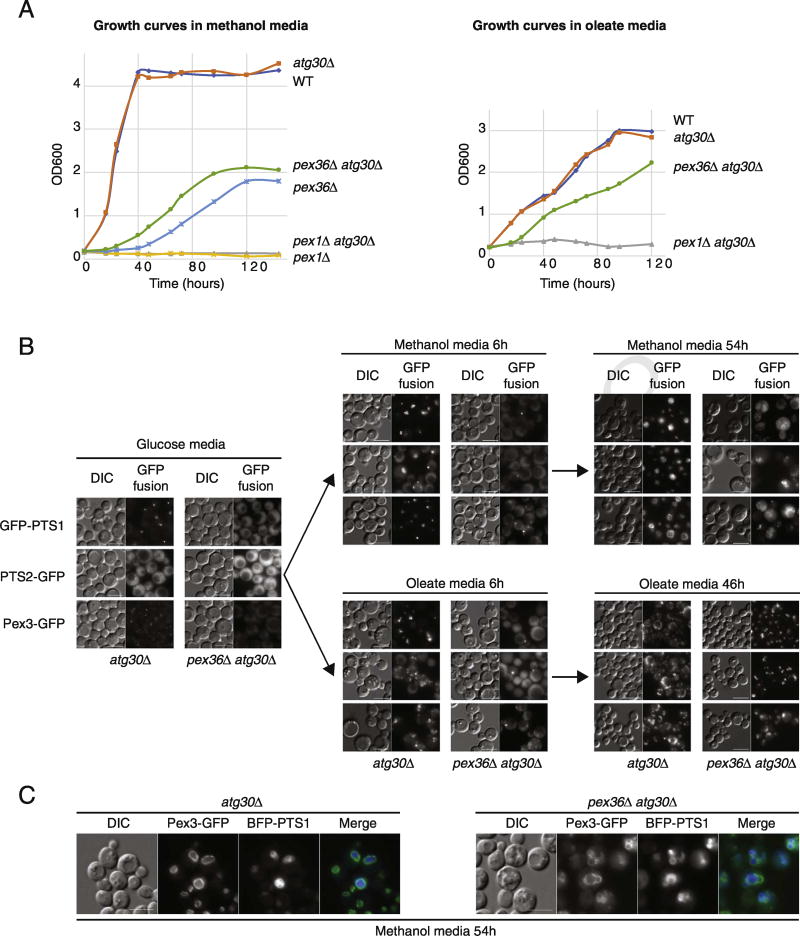
Peroxisome biogenesis was assessed by cell growth in media (methanol and oleate) requiring peroxisomal metabolic pathways in K. phaffii. Due to the induced pexophagy phenotype of K. phaffii pex36Δ cells (pex36Δ), we did a growth analysis in two different background strains, WT and atg30Δ (a specific pexophagy mutant). As expected, WT cells grew in both media and pex1Δ cells did not, and no difference was observed in both media when the ATG30 gene was deleted because this gene is not required for peroxisome biogenesis (Fig. 2A). Cells lacking PEX36 showed a serious growth defect in carbon sources requiring peroxisome biogenesis, with an extended lag phase and slow doubling time, at least in methanol medium. A slight growth improvement was observed for pex36Δ cells in methanol medium when pexophagy was blocked by ATG30 deletion. Nevertheless, the growth of the double mutant pex36Δ atg30Δ cells was still seriously compromised relative to that of WT cells, being delayed for more than 40 h, indicating that deficient peroxisome biogenesis, and not the slightly enhanced pexophagy, is the main cause of the growth defect.
Fig. 2. Phenotypes of cells lacking Pex36.
(A) Growth curves of several strains pre-grown up to mid-log phase in YPD liquid medium, washed, and added to methanol or oleate medium in shaking cultures at 30 °C, using starting inocula at an optical density at 600 nm (OD600) of ~ 0.2. (B) Fluorescence microscopy of atg30Δ and pex36Δ atg30Δ cells expressing GFP-PTS1, PTS2-GFP, or Pex3-GFP in glucose, methanol, and oleate media. (C) Fluorescence microscopy of atg30Δ and pex36Δ atg30Δ cells co-expressing BFP-PTS1 and Pex3-GFP in methanol media. The bar represents 5 µm.
Next, we used fluorescence microscopy of single focal planes to analyze protein import via the main peroxisomal targeting signal pathways, which are PTS1, PTS2, and mPTS-containing proteins. K. phaffii WT cells (atg30Δ for this study) showed one small peroxisome per cell in glucose medium with the three different PTS-fusion proteins used—the PTS1-tagged GFP (GFP-PTS1) and the PTS2-tagged GFP (PTS2-GFP) peroxisomal matrix proteins expressed from the strong, constitutive promoter of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene and the mPTS containing protein, Pex3-GFP, expressed from the PEX3 gene promoter (Fig. 2B, C). However, in the same conditions and with the same PTS-fusion reporters, no peroxisome-like structures were observed in pex36Δ cells (pex36Δ atg30Δ). Instead, the PTS1 and PTS2 reporters were mislocalized to the cytosol and Pex3-GFP was not detected. After a short incubation (6 h) in methanol or oleate to induce peroxisome proliferation, most WT cells showed many large import-competent peroxisomes. However, in some of the pex36Δ cells, Pex3-GFP was seen localizing in one small, bright, dot-like structure and less than 4% of the pex36Δ cells contained an import-competent peroxisome when observed with PTS1 or PTS2 fusion proteins. After long periods of peroxisome induction in methanol or oleate media, most pex36Δ cells showed import-competent peroxisomes, although their morphology and numbers were compromised compared to WT cells (Fig. 2B, C). The morphology of peroxisomes in K. phaffii changes dramatically upon growth in different carbon sources. In oleate medium, peroxisomes are small and numerous and spread around the cell, whereas in methanol medium, peroxisomes are big, less numerous, and clustered. Interestingly, peroxisome morphology in pex36Δ cells was affected in opposite ways in both media, being less numerous, clustered, and bigger than oleate-induced peroxisomes relative to WT cells, and more numerous, clustered, and smaller than methanol-induced peroxisomes, in comparison with WT cells.
Peroxisome formation in pex36Δ cells was seriously delayed and only prolonged induction under peroxisome proliferation conditions could rescue this defect (Fig. 2A). The mislocalization of PTS1- and PTS2-containing proteins in this mutant was often observed with the absence of peroxisomes (Fig. 2B), suggesting that the peroxisomal matrix protein import defect was likely a consequence of insufficient peroxisomes. The phenotypic absence of peroxisomes in pex36Δ cells in glucose medium, and even after short induction under peroxisome proliferation conditions, resembles the phenotypes seen in pex3Δ and pex19Δ mutants in yeast [28] and pex3, pex16, and pex19 mutants in mammals [29–31].
Pex36 is the functional homolog of ScPex34 and Pex16 proteins
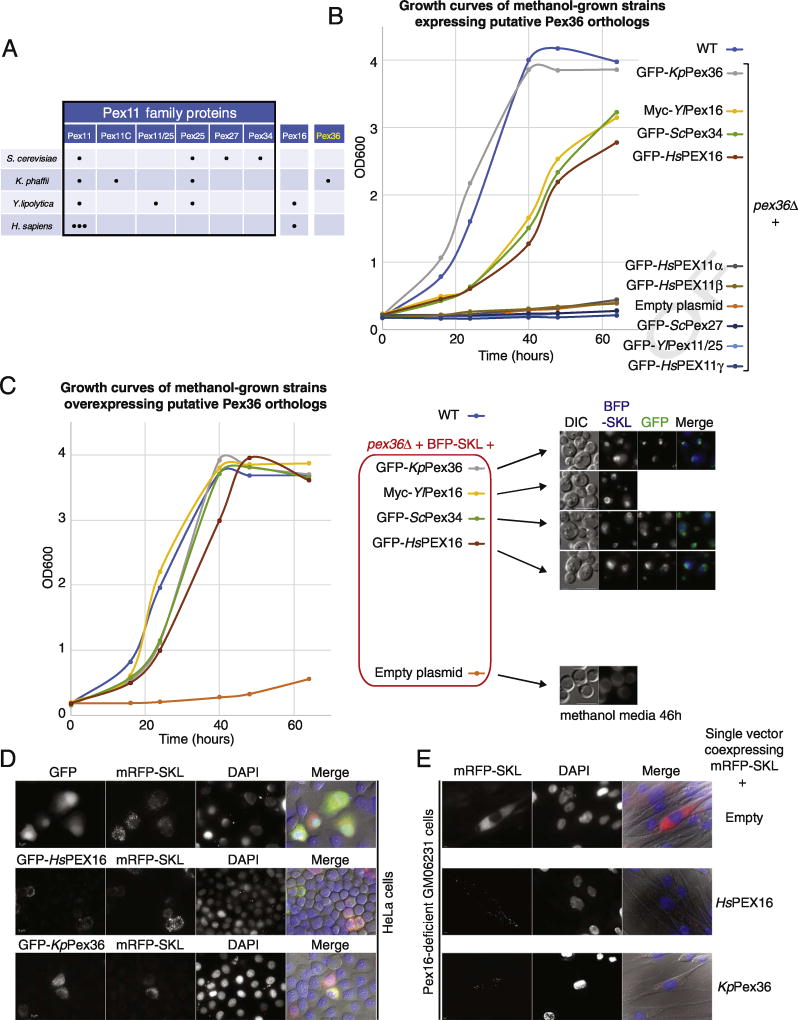
In order to gain information about the functions of K. phaffii Pex36 (KpPex36), we set up several complementation assays using pex36Δ cells and putative KpPex36 homologs. To assess the complementation, we focused on the simple, and probably the more reliable, assay for peroxisome function, which is cell growth in media requiring peroxisomal metabolism. For these assays, we used a few PEX11 family proteins, because Pex11 and Pex25 from yeast and PEX11γ from birds share a slight homology with KpPex36 (Fig. S2A). In addition, K. phaffii has Pex11, Pex11C and Pex25, but lacks proteins such as S. cerevisiae Pex27 (ScPex27), ScPex34 and Y. lipolytica Pex11/25 (Y/Pex11/25) (Fig. 3A). We also included Pex16 proteins because they are the only PMPs absent in most yeast species, except Y. lipolytica and filamentous fungi (Fig. 3A). Moreover, although we did not find protein sequence homology with Pex16 proteins using PSI-Blast (several iterations), this family was of interest because the human PEX16 mutants also lack peroxisomes, like cells lacking Pex3 and Pex19. Consistent with this interest, when we aligned sequences using the multiple-sequence alignment prediction (Clustal Omega), although we found only weak protein sequence homology between KpPex36, ScPex34, and PEX16 proteins (Homo sapiens PEX16 [HsPEX16] and Y/Pex16) (Fig. S2B, C), a strong similarity between the predicted protein structures was detected (Fig. S2D).
Fig. 3. Pex36 is the functional homolog of ScPex34 and Pex16 proteins.
(A) A survey of the Pex11 protein family, Pex16 and Pex36 (adapted from Ref. [32]). (B) Growth curves of WT and pex36Δ strains expressing putative Pex36 orthologs under the control of the PEX36 promoter in methanol medium. (C) Growth curves in same conditions as panel B of WT and pex36Δ strains expressing BFP-SKL and putative Pex36 orthologs from the strong GAPDH promoter. Fluorescence microscopy of some of the strains used for the growth curves after 46 h in methanol media. BFP-SKL and GFP fluorescence of the putative orthologs is shown when present. The bar represents 5 µm. (D) fluorescence microscopy of HeLa cells 48 h after transfection with plasmids expressing RFP-SKL and either GFP, GFP-HsPEX16, or GFP-Pex36, along with DAPI staining. The bar represents 5 µm. (E) Fluorescence microscopy of Pex16-deficient fibroblast cell line (GM06231) 48 h after transfection of a single plasmid co-expressing RFP-SKL, and either HsPEX16 or KpPex36. The bar represents 5 µm.
All these putative orthologs were cloned in a K. phaffii integration vector, expressed from the KpPEX36 promoter, and transformed into pex36Δ cells. As expected, the expression of GFP-KpPex36 complemented the methanol growth defect of pex36Δ cells and the growth was comparable to that of WT cells (Fig. 3B). Among the putative orthologs tested, GFP-ScPex34, Myc-Y/Pex16, and GFP-HsPEX16, but not GFP-ScPex27, GFP-Y/Pex11/25, and GFP-HsPEX11 proteins, clearly improved the growth of pex36Δ cells in methanol medium. We confirmed that all of the genes, encoding the GFP fusion proteins that did not complement pex36Δ cells (GFP-ScPex27, GFP-Y/Pex11/25, GFP-HsPEX11α, GFP-HsPEX11β, and GFP-HsPEX11γ), were properly integrated in the genome and expressed (Fig. S4A, and B). At least one of them, GFP-HsPEX11γ, complemented the peroxisome morphology defect of Kppex11Δ cells (Fig. S4), while some others (GFP-HsPEX11α, GFP-ScPex27, and GFP-Y/Pex11/25) showed some partial complementation.
The partial complementation of GFP-ScPex34, Myc-Y/Pex16, and GFP-HsPEX16 in pex36Δ cells might be due to the low expression of some of these heterologous genes. Because the tRNA pools vary between different organisms, the rates of transcription and translation of a particular coding sequence can be less efficient in a non-native context. To rule out the possibility that lower protein levels caused the partial complementation, we expressed the putative ortho-logs from the strong, constitutive GAPDH promoter, and performed growth curves and PTS1 import assays (Fig. 3C). Cells overexpressing the fusion proteins, GFP-ScPex34, Myc-Y/Pex16, and GFP-HsPEX16, but not those transfected with the empty plasmid control, behaved similarly to the overexpressed endogenous GFP-KpPex36 and fully complemented the growth defect of pex36Δ cells, as well as the PTS1 import defect, as seen by the appearance of import-competent peroxisomes labeled with BFP-PTS1, which colocalized with the GFP-tagged proteins. These results suggest that KpPex36 is the functional homolog of ScPex34, and more significantly, that these two proteins, ScPex34 and KpPex36, might be the yeast homologs of mammalian PEX16.
To independently confirm this conclusion that KpPex36 is the functional homolog of human PEX16, we expressed GFP-KpPex36 in HeLa cells and found that like GFP-HsPEX16, it was properly targeted to peroxisomes, as seen by colocalization with PTS1-tagged mRFP (mRFP-SKL) (Fig. 3D). Moreover, in cells from the human fibroblast cell line GM06231, lacking peroxisomes because of a mutated PEX16 gene, introduction of KpPEX36, co-expressing untagged KpPEX36 and mRFP-SKL from a single plasmid, led to the appearance of new import-competent peroxisomes labeled with mRFP-SKL, indicating that KpPex36 can complement at least some HsPEX16 functions, such as de novo peroxisome formation and PTS1 import (Fig. 3E).
These complementation results in yeast and mammalian cells were rather surprising and unexpected mainly because Pex16 functions, topology, and localization seem to be substantially different between species. In Y. lipolytica, for instance, Pex16 is an intraperoxisomal peripheral membrane protein that participates in peroxisome division [9, 33]. In contrast, human PEX16 is an integral membrane protein with its N- and C-termini facing the cytosol [34]. S. cerevisiae Pex34 is a PMP with three potential transmembrane domains [3].
With the goal of confirming or revoking at least the previously described protein topology, we investigated the conformation assumed by these proteins in K. phaffii using protease protection assays with GFP-tagged, N-terminal fusion proteins. First, we confirmed that all of these fusion proteins localized with peroxisomal membrane fractions (P27) by cellular fractionation in WT cells (Fig. S5A). Then, using the peroxisomal membrane fractions, we checked the sensitivity of these proteins and a few other peroxisomal proteins to a proteinase K/trypsin cocktail in the absence and presence of detergent (Fig. S5B, C). In this system, peroxisomal matrix proteins (e.g., Pex8) should be protected and peripheral proteins (e.g., Pex17) are expected to be susceptible to protease in the absence of detergent. Our data indicate that for GFP-HsPEX16, GFP-Y/Pex16, GFP-ScPex34, and GFP-KpPex36, N-terminal epitope tags were sensitive to protease in the absence of detergent, like Pex17, whereas Pex8 was only sensitive to protease in the presence of detergent (Fig. S5B). These results indicate that in K. phaffii, all of the Pex16-like proteins tested showed a common topology with their N-termini facing the cytosol.
Finally, we further characterized KpPex36 topology by tagging both its N- and C-termini (GFP-Pex36-HA) and subjecting these proteins to the same protease protection assay. We found that KpPex36 has both its N- and C-termini facing the cytosol (Fig. S5C), in agreement with the two predicted transmembrane domains, and similar to the topology reported for human PEX16 [34].
The pex25Δ pex36Δ cells have numerous, small import-incompetent peroxisome remnants
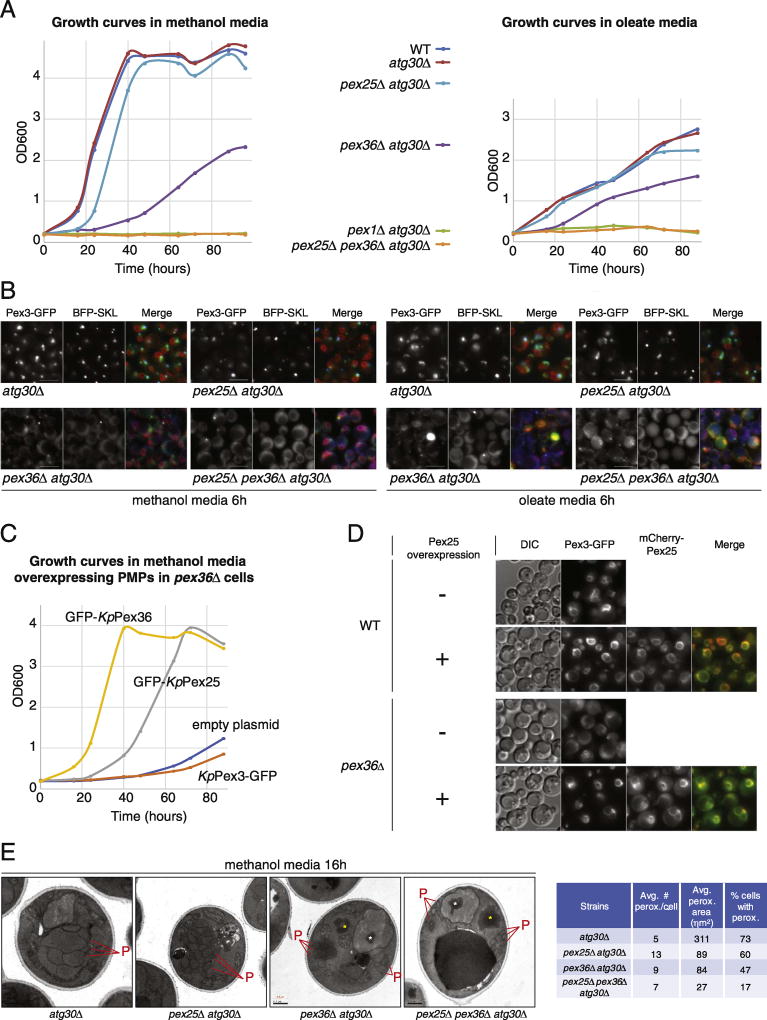
In S. cerevisiae, Pex34, the functional homolog of KpPex36, has genetic and physical interactions with several peroxisome division proteins [3]. ScPex34 interacts, in yeast two-hybrid, with ScPex11, ScPex25, ScPex27, and ScFis1. In addition, the absence of some of these proteins, when combined with cells unable to express ScPex34, enhanced the defect in peroxisome biogenesis when compared to cells lacking only one of these proteins. For example, Scpex25Δ and Scpex34Δ cells have import competent peroxisomes in glucose medium, but the deletion of both these genes results in a strain without observable functional peroxisomes in glucose medium, although peroxisome biogenesis is restored by inducing peroxisomes with oleate medium [3]. The Scpex34Δ cells share the same peroxisome morphology as Kppex36Δ cells grown in oleate media (fewer and larger peroxisomes), although Kppex36Δ cells had a lot more in common with the phenotype described for the S. cerevisiae double-deletion mutant pex25Δ pex34Δ (no peroxisomes in glucose medium, and fewer and larger peroxisomes in oleate medium). This prompted us to investigate the phenotype of pex25Δ pex36Δ cells in K. phaffii. We used atg30Δ as our background in order to avoid induced pexophagy in our mutants, and we followed the phenotype by monitoring PTS1 protein import (BFP-SKL), growth in peroxisome proliferation conditions, and peroxisome morphology (Fig. 4A, B). Remarkably, for pex25Δ pex36Δ atg30Δ cells, growth was completely impaired in any analyzed condition (Fig. 4A) and cells were devoid of peroxisomes that imported PTS1 proteins (Fig. 4B). In addition, similar to pex36Δ atg30Δ cells, Pex3-GFP was detected only in half of the pex25Δ pex36Δ atg30Δ cells, and when present, it was in a small dot-like structures often at, or adjacent to, the pn-ER, labeled with Sec61-mCherry (red fluorescence displayed only in the merge panel). Surprisingly, the pex25Δ atg30Δ mutant did not show any noticeable defect in Pex3-GFP targeting to the peroxisomes or for PTS1 import (BFP-SKL) in methanol medium, despite a partial growth defect (Fig. 4A).
Fig. 4. The pex25Δ pex36Δ cells have numerous, small import-incompetent peroxisomes.
(A) Growth curves of WT and atg30Δ strains in combination with various pex mutants in methanol or oleate media. Deletion of both Pex25 and Pex36 results in a synthetic lethality phenotype, whereas deletion of only Pex25 or Pex36 results in delayed growth. (B) Fluorescence microscopy of several strains expressing BFP-SKL, Pex3-GFP, and Sec61-mCherry after 6h in methanol and oleate media. Sec61-mCherry is displayed only in the merge panel (red fluorescence). The bar represents 5 µm. (C) Growth curves in methanol medium of pex36Δ strains overexpressing GFP-tagged PMPs. The overexpression of Pex25 rescues the growth phenotype of pex36Δ cells, albeit not as quickly as Pex36 overexpression. (D) Fluorescence microscopy of WT and pex36Δ strains expressing Pex3-GFP and mCherry-tagged, overexpressed Pex25 after 24 h in methanol medium. The bar represents 5 µm. (E) Transmission electron microscopy pictures of WT and pex mutant cells, showing labeled peroxisomes and peroxisomal remnants, after 16 h induction in methanol medium. The asterisks indicate protein aggregates that accumulate in the pex36Δ atg30Δ and pex25Δ pex36Δ atg30Δ strains. The inset table shows the numbers and average sizes of peroxisomes per cell, as well as the number of cells with peroxisomes, in each strain. Additional data and statistics are available in Table S3. The bar represents 5 µm. The pex25Δ pex36Δ cells have numerous, small import-incompetent peroxisomes. (A) Growth curves of WT and atg30Δ strains in combination with various pex mutants in methanol or oleate media. Deletion of both Pex25 and Pex36 results in a synthetic lethality phenotype, whereas deletion of only Pex25 or Pex36 results in delayed growth. (B) Fluorescence microscopy of several strains expressing BFP-SKL, Pex3-GFP, and Sec61-mCherry after 6 h in methanol and oleate media. Sec61-mCherry is displayed only in the merge panel (red fluorescence). The bar represents 5 µm. (C) Growth curves in methanol medium of pex36Δ strains overexpressing GFP-tagged PMPs. The overexpression of Pex25 rescues the growth phenotype of pex36Δ cells, albeit not as quickly as Pex36 overexpression. (D) Fluorescence microscopy of WT and pex36Δ strains expressing Pex3-GFP and mCherry-tagged, overexpressed Pex25 after 24 h in methanol medium. The bar represents 5 µm. (E) Transmission electron microscopy pictures of WT and pex mutant cells, showing labeled peroxisomes and peroxisomal remnants, after 16 h induction in methanol medium. The asterisks indicate protein aggregates that accumulate in the pex36Δ atg30Δ and pex25Δ pex36Δ atg30Δ strains. The inset table shows the numbers and average sizes of peroxisomes per cell, as well as the number of cells with peroxisomes, in each strain. Additional data and statistics are available in Table S3. The bar represents 5 µm.
The synthetic lethality of PEX25 and PEX36 genes in peroxisome proliferation conditions suggested that the products of these genes might play a redundant role. We addressed this possible redundancy through the overexpression of KpPex25 (GFP-KpPex25 or mCherry-KpPex25) in cells lacking KpPex36 (Fig. 4C, D). A partial complementation of the growth defect was obtained when we overexpressed GFP-KpPex25, but not KpPex3-GFP, or upon transfection with the empty plasmid (Fig. 4C). After growth in methanol medium for 24 h, only a few pex36Δ cells had recognizable peroxisomes labeled with Pex3-GFP, but almost every pex36Δ cell overexpressing mCherry-KpPex25 contained recognizable peroxisome-like structures in which there was colocalization of Pex3-GFP with mCherry-KpPex25 (Fig. 4D). These results are consistent with the hypothesized redundant role of these proteins.
To confirm the fluorescence microscopy analysis, which suggested that K. phaffii pex25Δ pex36Δ cells were devoid of functional peroxisomes, electron microscopy of methanol-grown cells was used to confirm the status of peroxisomes (number, size, presence, or absence) (Fig. 4E and Table S3). At least 40 cells for each the different background were analyzed (Table S3). The atg30Δ, pex2Δ atg30Δ, and pex36Δ atg30Δ mutant cells contained numerous peroxisomes after 16 h in methanol medium. Peroxisomes in WT and pex25Δ atg30Δ cells were electron-dense, characteristic of import-competent peroxisomes, but only some peroxisomes in pex36Δ atg30Δ cells were electron-dense. Similar to our fluorescence microscopy observations, the peroxisomes were smaller and more numerous as compared to the WT cells. The WT cells had an average of 5 peroxisomes/cell with an average peroxisome area of 311 ηm2, pex25Δ atg30Δ cells contained an average of 13 peroxisomes/cell with an average peroxisome area of 89 ηm2, and pex36Δ atg30Δ cells contained an average of 9 peroxisomes/cell with an average peroxisome area of 84 ηm2. In addition, we observed that the peroxisomes of most pex36Δ atg30Δ cells were often grouped in two separate regions of the cell. Finally, most pex36Δ atg30Δ cells showed larger cytosolic and nuclear aggregates, most probably containing peroxisomal matrix proteins, such as AOX1, a very abundant protein during growth in methanol that is mislocalized for extended periods in this mutant. Unexpectedly, we also observed peroxisome-like structures in pex25Δ pex36Δ atg30Δ cells, which were on average 7/cell and 27 ηm2 in area. Similar to pex36Δ atg30Δ cells, the double mutant also contained the large aggregates in the cytosol and nucleus. However, the total number of cells containing peroxisomes or peroxisome-like structures was drastically different in pex36Δ atg30Δ and pex25Δ pex36Δ atg30Δ cells when compared to WT cells. Seventy-three percent of the WT cells, 60% of pex25Δ atg30Δ cells, 47% of pex36Δ atg30Δ cells, and only 17% for pex25Δ pex36Δ atg30Δ cells had peroxisome-like structures. The peroxisome-like structures in pex25Δ pex36Δ atg30Δ cells appeared as peroxisome clusters and also as dispersed vesicles in the cytosol often in proximity to the cell periphery. These peroxisome-like structures in the double mutant are reminiscent of import-incompetent peroxisome remnants because the mutant cells did not grow under peroxisome proliferation conditions and mislocalized BFP-SKL protein to the cytosol (Fig. 4A, B).
Role of Pex25 and Pex36 in peroxisome biogenesis
The presence of peroxisome remnants, and the defect in BFP-PTS1 import in pex25Δ pex36Δ cells prompted us to further investigate PMP import. In addition, the strong phenotype obtained by the double mutant and the functional redundancy of KpPex25 and KpPex36 motivated us to focus more on the phenotype of the double mutant (pex25Δ pex36Δ).
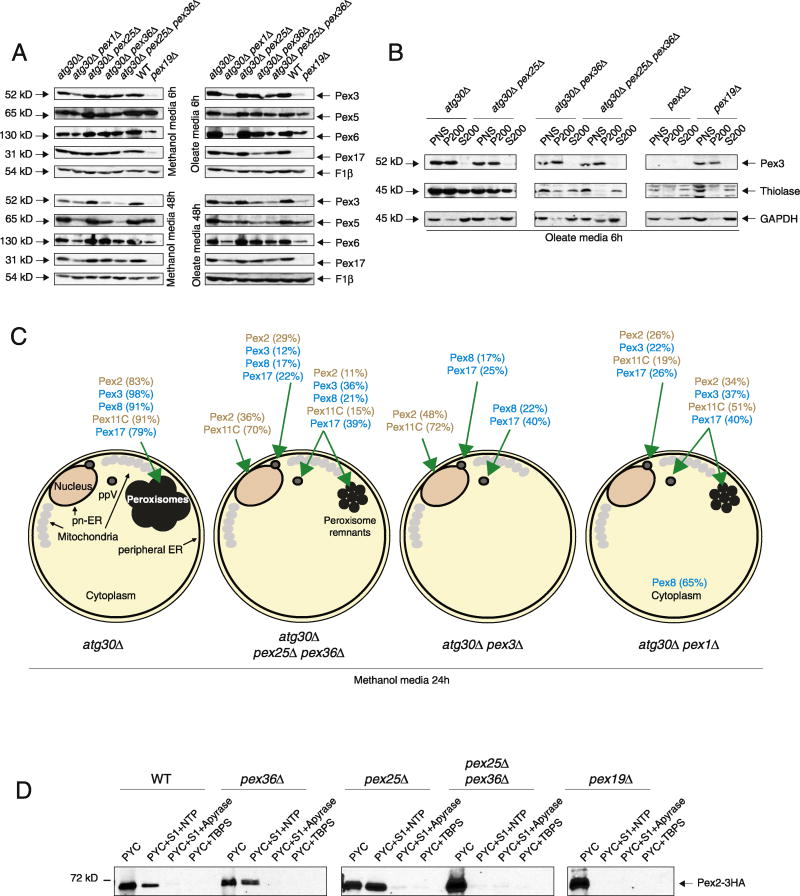
It is known that the absence of some peroxins destabilizes other PMPs and can provide some information about the function of the peroxin; for example, the absence of Pex3 and Pex19 destabilizes most PMPs [28] and mammalian PEX1 mutants destabilize the interacting proteins PEX5 and PEX6 [35]. We found that after 6 h of peroxisome proliferation, independent of the media, most of the PMPs we checked (Pex2, Pex3, Pex5, Pex6, and Pex17) were unaffected by the absence of Pex25, Pex36, or both in the atg30Δ background (Fig. 5A). When induction was prolonged to 48 h, some instability was observed for Pex3, Pex5, and Pex6 in pex25Δ pex36Δ atg30Δ cells, and to a lesser extent in pex36Δ atg30Δ cells. The low levels of Pex3 in pex25Δ pex36Δ atg30Δ and pex36Δ atg30Δ cells could be the reason for the peroxisome biogenesis defect, although the overexpression of Pex3-GFP in pex36Δ cells was unable to rescue the defect, ruling out this hypothesis (Fig. 4D). The low levels of Pex6, and probably Pex1, might be the reason for the induced-pexophagy observed in pex36Δ cells (Fig. S1). We also found that deletion of the ATG30 gene was unable to fully rescue PMP instability in the pex1Δ mutant. This is different from the result seen in S. cerevisiae [36], indicating that in K. phaffii pex1Δ cells, these PMPs are down-regulated by a mechanism other than just autophagy-related pathways.
Fig. 5. The pex25Δ pex36Δ atg30Δ cells affect peroxin stability, localization, and ppV budding from the ER.
(A) Western blot of various PMPs in a variety of strains after 6 and 48 h of methanol or oleate induction, respectively. F1β was used as a loading control. (B) Western blot of the membrane bound and cytosolic fractions of Pex3, thiolase, and GAPDH in various deletion strains after 6 h in oleate medium. (C) Schematic showing the localization of various GFP-tagged peroxins in various strains after 24 h of methanol induction. Percentages indicate the subset of the peroxin population that localizes to the indicated intracellular site. Percentages lower than 6% and cells without fluorescence were included in the analysis, but not presented. (D) Budding of Pex2-containing ppVs from the ER is inhibited in pex25Δ pex36Δ cells, but not in pex25Δ or pex36Δ cells. Western blot of Pex2-3HA visualized with anti-HA antibodies in WT and various mutants. PYC, permeabilized yeast cells; S1, cytosolic proteins; NTP, ATP-regenerating cocktail + GTP; TBPS, budding buffer.
In yeast and mammals, PMPs can be directly inserted into the peroxisome membrane from the cytosol or traffic through the ER to peroxisomes. A few steps of this traffic of PMPs through the ER have been elucidated, such as PMP insertion into the ER membrane, intra-ER sorting of PMPs to the pER, and ppV budding from the ER. Because mammalian PEX16 has been implicated in the direct insertion of PEX3 into the ER and peroxisomal membrane [1], and as pex36Δ cells can be complemented by human PEX16, we followed PEX3 localization by differential centrifugation assays. As shown in Fig. 5B, none of the mutants tested, including the pex25Δ pex36Δ atg30Δ cells, mislocalized Pex3 to the cytosolic fraction (S200), and all of the strains tested showed Pex3 associating with the membrane fraction. As expected, the peroxisomal matrix protein, thiolase, was partially mislocalized to the cytosolic fraction in atg30Δ pex36Δ cells and fully mislocalized in pex25Δ pex36Δ atg30Δ cells. These data confirm the fluorescence microscopy data for Pex3 and show that it still associates with the membrane fraction in pex36Δ afg30Δ and pex25Δ pex36Δ atg30Δ cells, ruling out a role for Pex36 in PMP insertion into membranes in K. phaffii.
After insertion at the ER, PMPs sort to the pER. Two routes have recently been described in K. phaffii, one dependent on Pex3 and Pex19, and another independent of these proteins [14]. To assess the role of Pex25 and Pex36 in intra-ER PMP sorting, the cellular localization of several peroxins (Pex2, Pex3, Pex8, Pex11C, and Pex17) was examined in detail (Figs. 5C and S6; Table S4). In addition to atg30Δ and pex25Δ pex36Δ atg30Δ cells, we included pex1Δ atg30Δ cells as a control because they contain peroxisome remnants and are not implicated in the import of PMPs [37]. We also included pex3Δ atg30Δ cells because they do not contain peroxisome remnants and are implicated in PMP import. All of the strains expressed the GFP fusions of the specific PMPs along with Sec61-mCherry to label the ER (cortical and perinuclear). BFP-PTS1 was not included because none of the mutants import peroxisome matrix proteins, and in the WT strain, methanol-induced peroxisomes are easily differentiated from other structures. As expected, in atg30Δ cells grown in methanol medium (6 and 24 h), all of the PMP-GFP fusions tested decorated the cluster of peroxisomes, which were well segregated from the cortical and pn-ER (Figs. 5C and S6; Table S4). In the same conditions, pex25Δ pex36Δ atg30Δ cells, similar to the pex3Δ atg30Δ cells, showed most of the Pex2-GFP and GFP-Pex11C in a ring-like structure, which fully colocalized with the pn-ER (Figs. 5C and S6; Table S4), in agreement with the role of Pex3 in the intra-ER sorting of Pex2 [14]. Such localization of Pex2-GFP or GFP-Pex11C was never found in the pex1Δ atg30Δ cells. However, distinct from pex3Δ atg30Δ cells, not all of the pex25Δ pex36Δ atg30Δ cells showed a Pex2-GFP and GFP-Pex11C intra-ER sorting defect and some cells displayed dot-like structures, some colocalized with the pn-ER, and some showed cytosolic labeling, but often near the pn-ER.
Next, we followed the localization of Pex17-GFP in the same mutant strains and found the same localization profile, dot-like structures mostly in the cytosol, irrespective of the mutant strains. These results indicate that none of these PMPs (Pex1, Pex3, Pex25, and Pex36) are implicated in the Pex17 ER-to-peroxisome remnant trafficking.
Remarkably, GFP-Pex8, a peroxin whose import depends on PTS1 and/or PTS2 signals, showed the same distribution as Pex17-GFP in pex3Δ atg30Δ and pex25Δ pex36Δ atg30Δ cells, suggesting that the punctate structures might also contain Pex14, as well as the PTS1 and/or PTS2 receptors. Lastly, and in agreement with the role of Pex1 in import of peroxisomal matrix proteins, no punctate structures were observed for GFP-Pex8 in pex1Δ atg30Δ cells and the entire fluorescent signal was cytosolic. Finally, we found that ER-to-peroxisome traffic of Pex3-GFP was not affected in pex25Δ pex36Δ atg30Δ, when compared to pex1Δ atg30Δ cells, and it localized and distributed like Pex17.
To confirm the defect in ER-to-peroxisome remnant traffic of Pex2 and Pex11C, we performed the in vitro budding assay described previously [15] (Fig. 5D). To force ppV budding in yeast, we used strains devoid of peroxisomes in glucose media by replacing the PEX19 promoter with the tightly regulated, methanol-inducible AOX1 promoter, which makes cells dependent on methanol to generate new, import-competent peroxisomes. Cells expressing Pex2-3HA were induced for peroxisome biogenesis by growing them in methanol for 3 h. The assay consists of permeabilized yeast cells (PYCs) expressing Pex2-3HA combined with an S1 (cytosolic) fraction lacking the HA-tagged PMP (crude cytosol), with concomitant addition of an ATP-regenerating system and incubation at 20 °C for 90 min. In the controls, PYCs were pretreated with apyrase to deplete ATP before the addition of the S1 fraction. PYCs were separated from the released vesicular fraction via a brief centrifugation step; the supernatant was then analyzed by Western blot with an antibody against the HA tag. As expected from our previous results [14], we detected Pex2-3HA in the budded fraction of WT cells. However, in control reactions with apyrase, or when TBPS buffer was substituted for cytosol, the budding of Pex2-3HA was dramatically decreased or absent. In PYCs prepared from pex19Δ cells, no budding of Pex2 was observed, confirming its essential role in the trafficking of RING-domain PMPs to ppVs. As in WT cells, pex36Δ and pex25Δ single mutants showed normal budding of Pex2-3HA. However, in pex25Δ pex36Δ cells, Pex2-3HA was not detected in the budded fraction. These results confirm the defect in ER-to-peroxisome remnant trafficking observed by fluorescence microscopy in pex25Δ pex36Δ cells, showing a role for these proteins during the budding of ppVs. The total absence of ppVs containing Pex2 in the double mutants, but the presence of some Pex2-GFP in dot-like structures in the cytosol in pex25Δ pex36Δ atg30Δ cells, suggests either that some Pex2 might bud at an efficiency below levels of detection of this assay, or that some Pex2 may be imported to the peroxisome remnants through a different pathway.
The observed defect in intra-ER sorting and ppV budding in cells lacking Pex25 and Pex36 suggests that these proteins are directly implicated in this process. Very little is known about these mechanisms in K. phaffii, other than that Pex19 is an essential component for this process and Pex3 is indirectly required through its role in intra-ER sorting of Pex2. It is well known that Pex19 directly interacts with most of the PMPs, and it is possible that Pex25 and Pex36 assist in the interactions between PMPs and Pex19 at the ER.
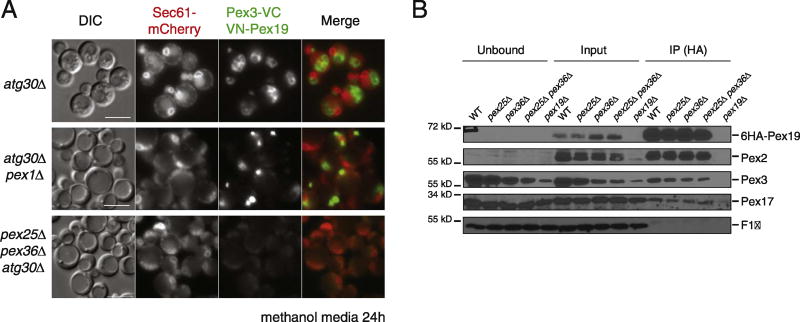
We studied Pex3–Pex19 interaction in vivo by using the bimolecular fluorescence complementation (BiFC) assay [38], in which the Venus yellow fluorescent protein (vYFP) is split into two fragments, VN (N terminus of vYFP) and VC (C terminus of vYFP), and the appearance of vYFP fluorescence is an indication of close proximity between the fusion proteins, which enables the formation of the full fluorescent protein. We fused VN to Pex19 and VC to Pex3 and tested for proximity and colocalization in several genetic backgrounds (Fig. 6 and Table S3). We tested the proximity of VN-Pex19 and Pex3-VC during peroxisome proliferation conditions. These fusion proteins were expressed from the inducible AOX1 promoter, and similar protein levels were observed in all analyzed strains (Fig. S6). When expressing both VN-Pex19 and Pex3-VC, 80% of atg30Δ cells showed vYFP decorating a characteristic peroxisome cluster distinct from Sec61-mCherry, which indicates that these proteins are close to each other at the peroxisome surface (Fig. 6 and Table S3). We studied their proximity in pex1Δ atg30Δ cells and found that at least 50% of cells showed vYFP fluorescence in a single dot-like structure which was distributed in a similar pattern as Pex3-GFP in pex1Δ atg30Δ cells (Figs. 5C, S6, and 6A; Table S4). However, in pex25Δ pex36Δ atg30Δ cells, vYFP fluorescence was not detected. This result suggested that Pex25 and Pex36 might facilitate the direct interaction of Pex3 and Pex19 or might affect the proper conformation of at least one of the proteins, which will affect the proximity of the two vYFP moieties. Independent of which of the two models (interaction or conformation) is the reason for the lack of vYFP fluorescence, this is probably how Pex25 and Pex36 affect the proper trafficking of Pex2 and Pex11C.
Fig. 6. Pex25 and Pex36 are required for the direct interaction of Pex3 and Pex19.
(A) fluorescence microscopy of atg30Δ and mutant cells expressing Sec61-mCherry, and the split Venus-tagged Pex3 (Pex3-VC) and Pex19 (VN-Pex19) after 6 h of methanol induction. The bar represents 5 µm. (B) Western blot of the immunoprecipitation assay in which 6xHA-tagged Pex19 was pulled down to determine its interaction with other PMPs proteins (Pex2, Pex3, and Pex17) in PMP mutant strains. F1β was used as a negative control.
Finally, we studied the interaction of Pex19 and Pex3 in pex25Δ pex36Δ atg30Δ cells by immunoprecipitation of Pex19-6HA after 6 h in methanol media (Fig. 6B). Surprisingly, the interaction between Pex19 and Pex3 was not affected by the absence of Pex25 and/or Pex36, nor were the interactions between Pex19 and either Pex2 or Pex17. The discrepancy between the BiFC and co-immunoprecipitation results might be due to direct versus indirect interactions, or a change in conformation without the loss of interaction.
Pex36 has non-overlapping functions to Pex16 and Pex34
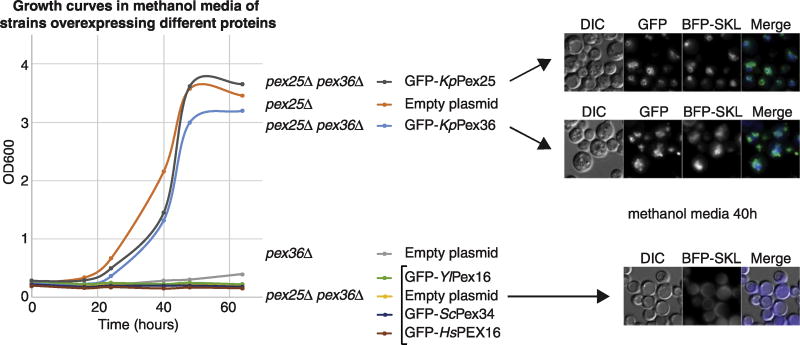
Because Pex25 and Pex36 play redundant roles during peroxisome biogenesis, we hypothesized that human PEX16 might be the functional ortholog of both PMPs. Surprisingly, when we overexpressed Y/Pex16, HsPEX16, Y/Pex34, and KpPex36 in pex25Δ pex36Δ cells, only KpPex36 partially complemented the growth defect in methanol medium, as well as the PTS1 import (Fig. 7). These results indicate that Pex36 has additional functions, absent in Y/Pex16, HsPEX16, and Y/Pex34 proteins. Hence, we named Pex36 as a new peroxin, rather than as a Pex16- or Pex34-like protein.
Fig. 7. Overexpression of Pex36 functional orthologs cannot rescue growth or peroxisome formation in cells lacking Pex25 and Pex36.
Growth curve of WT and mutant strains overexpressing KpPex25 and Pex36 functional orthologs in methanol medium, fluorescence microscopy of the GFP-tagged PMPs and BFP-SKL in some of the strains shown in the growth curves after 40 h in methanol medium. The bar represents 5 µm.
Discussion
We have identified a new peroxin in K. phaffii, Pex36, involved in peroxisome biogenesis. This protein is conserved among several yeasts, but is absent in S. cerevisiae and higher eukaryotes. Pex36 has a weak protein sequence homology with Pex11 family proteins, but it has strong, predicted structural homology with PEX16 proteins, despite the absence of primary sequence homology. We found that endogenous expression of Pex34 from S. cerevisiae and PEX16 from H. sapiens and Y. lipolytica partially complemented the peroxisome biogenesis defect of K. phaffii pex36Δ cells. Furthermore, in cells from the human fibroblast cell line GM06231, which lacks peroxisomes due to a mutated PEX16 gene, introduction of KpPex36 protein led to the appearance of new import-competent peroxisomes, indicating that KpPex36 is a functional homolog of PEX16. In addition, we found that Pex25 and Pex36 have redundant functions, overexpression of K. phaffii Pex25 partially rescues the lack of Pex36, and double deletion of PEX25 and PEX36 genes is synthetic lethal in peroxisome proliferation conditions. Surprisingly, none of the functional homologs of K. phaffii Pex36 (ScPex34, Y/Pex16, and HsPEX16) could complement the absence of both Pex25 and Pex36 upon overexpression, suggesting that Pex36 has additional function(s), which is not present in its functional homologs. These differences, in addition to the absence of an adequate sequence homology, persuaded us to name it as a new peroxin.
The pex36Δ cells have a growth defect in oleate and methanol media and cell growth is delayed more than 20 h when compared to WT cells, and peroxisome size and number are compromised. The pex36Δ cells grown for a short period in peroxisome proliferation conditions barely contain any import-competent peroxisomes (less than ~4%) but longer induction partially rescues this phenotype. Deletion of the PEX36 gene, like deletion of other peroxin genes such as PEX1, PEX6, and PEX15/26, induced pexophagy during peroxisome proliferation conditions and deletion of autophagy-related genes prevented this phenotype.
Pex36 is a PMP that traffics to peroxisomes via the ER, where it accumulates in the absence of Pex19, like some yeast PMPs and PEX16 in higher eukaryotes [16, 39, 40]. In addition, the lack of Pex36, together with its redundant protein Pex25, affects ER-to-peroxisome traffic of some PMPs, independent of the length of the peroxisome induction. The pex25Δ pex36Δ atg30Δ cells accumulated most of their Pex2 and Pex11C at the ER, mostly around the pn-ER and, in some cases, in a dot-like structure overlapping with the pn-ER. However, a small fraction of Pex2 and Pex11C could leave the ER and were localized in a dot-like structure in the cytosol adjacent to the pn-ER, likely representing peroxisome remnants which were observed by electron microscopy in a similar abundance. It is important to note that the expression levels of Pex2, Pex3, and Pex11C were seriously affected in pex25Δ pex36Δ atg30Δ cells, similar to pex19Δ cells, but this was not a consequence of induced pexophagy. We confirmed that the Pex2 intra ER-sorting defect resulted in a ppV budding defect in pex25Δ pex36Δ atg30Δ cells. In contrast, Pex3 and Pex17 trafficking, and presumably budding, was unaffected by the lack of Pex25 and Pex36.
Remarkably, most of the defects observed in pex25Δ pex36Δ atg30Δ cells resembled those previously reported for cells lacking Pex3. As mentioned before, the lack of Pex25 and Pex36 also affects the stability of several PMPs, including Pex3, which could explain the trafficking deficiency phenotypes. However, the overexpression of Pex3 was not able to rescue the defect of the triple mutant. In trying to understand the mechanistic basis of the impaired intra-ER sorting of Pex2 in the absence of Pex25 and Pex36, we found using BiFC that the proximity between the C-terminal of Pex3 and the N-terminal of Pex19 was altered, suggesting that Pex25 and Pex36 might bridge/facilitate the direct interaction between Pex3 and Pex19 or affect the conformation of at least one of the proteins during peroxisome biogenesis to preclude their interaction. These results are in line with our recent report about the role of Pex25 bridging the Pex3 and Pex19 interaction and implicate an additional factor to support their direct interaction [41].
In conclusion, we characterized a new PMP in K. phaffii named Pex36. Cells lacking Pex36 display a major growth defect in methanol medium and a slight defect in oleate medium. Methanol induced-peroxisomes in the pex36 mutant cells are twice as abundant, and one-fourth the size, when compared to those in WT cells. Overexpression of Pex25, a non-essential PMP by itself, partially restores the growth defect of the pex36 mutant cells, suggesting redundant roles for Pex25 and Pex36. Double deletion of PEX25 and PEX36 is synthetic lethal in all tested media requiring peroxisomal metabolic pathways. In mutant cells lacking both proteins, peroxisome biogenesis and the intra-ER sorting of Pex2 and Pex11C are seriously impaired, likely due to the loss of a direct interaction between Pex3 and Pex19 at the ER membrane. Despite the strong defect in peroxisome biogenesis in the pex25Δ pex36Δ double mutant, 20% of these cells contain peroxisome remnants, which are one-tenth of the area of peroxisomes in WT cells. Finally, Pex36 shares some functional and structural homology with human PEX16 and ScPex34 proteins.
Materials and Methods
Strains and plasmids are described in Supplemental Tables S1 and S2.
Media used to grow strains
YPD (2% glucose, 2% bacto-peptone, 1% yeast extract), YNB (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate), nitrogen starvation medium or SD-N (0.17% yeast nitrogen base without amino acids and ammonium sulfate; 2% glucose), oleate medium (2xYNB, 0.02 g histidine/L, 0.02 g arginine/L, 0.05 mM biotin, 0.02% Tween-40, 0.2% oleate), and methanol medium (2xYNB; 0.02 g histidine/L, 0.02 g arginine/L, 0.05 mM biotin, 1% methanol). All cultures were grown at 30 °C.
Isolation of K. phaffii pex36 UV-induced and construction of pex36 deletion mutant
A collection of UV-induced K. phaffii mutants defective in pexophagy was isolated using a plate-screening assay for elevated residual peroxisomal alcohol oxidase activity after shift of mutagenized colonies from methanol to glucose or ethanol-containing media [24].
The K. phaffii PEX36 gene was isolated from a genomic library by functional complementation of a UV-induced pdg1 (peroxisome degradation) mutant (named pex36 herein). To select for complementation, we made use of a partial growth defect of the initial pex36 mutant on methanol plates. From the pex36 mutant cells transformed with a genomic library, multiple transformants were obtained that restored WT phenotype with respect of methylotrophic growth and pexophagy. Several complementing plasmids were recovered after retransformation into Escherichia coli, and all were overlapping in the genomic DNA fragment carrying the unknown ORF encoding a protein of 363 amino acid residues, later designated as K. phaffii PEX36.
The shortest plasmid carrying the PEX36 gene, named pOS1, when retransformed into the original pex36 mutant, fully complemented the mutant phenotype as judged by growth and alcohol oxidase colony assay [42]. Next, the integrative plasmid pOS6 containing smaller subfragment of the genomic DNA insert from pOS1 was constructed by subcloning into the plasmid pBL-HIS [43], a 1.36-kb-long EcoRV fragment comprising entire PEX36 ORF with 105 nucleotides of the upstream promoter sequence and 161 nucleotides downstream of its terminator. pOS6 was shown to fully complement phenotype of the UV-induced pex36 mutant. Sequencing of the PEX36 genomic DNA isolated from this UV-induced mutant revealed that the PEX36 harbored a single-point mutation causing in-frame stop codon and producing a truncated Pex36 protein of only 71 N-terminal amino acids.
Biochemical studies of pexophagy
In K. phaffii, peroxisomes were induced by incubation of cells in methanol medium (starting OD600 of 0.2) for 15–16 h and transferred to SD-N medium at an OD600 of 2 to induce pexophagy. One milliliter of cells was collected at different times as described in the figures; trichloroacetic acid precipitated and analyzed by Western blot.
Electron microscopy
Cells were grown in YPD overnight and then switched to methanol medium. After 40 h of induction, 10 OD600 of cells were placed on a 0.45-µm filter (Millipore), washed with 10 mL 0.1 M cacodylate (pH 6.8), and fixed overnight at 4 °C. Then, they were washed with 50 mM KPi (pH 7.5) and spheroplasted with 0.5 mg Zymolyase-100 T for 40 min at 37 °C. After this, the cells were washed with 0.1 M cacodylate buffer and incubated in 1.5 mL 2% cacodylate-buffered OsO4 for 1 h on ice. Subsequently, they were incubated in 1.5 mL 2% UrAc for 1 h at room temperature. The cells were dehydrated with ethanol washes. Later, they were incubated in 50% acetone/50% SPURR. Finally, the cells were resuspended in 100% SPURR, baked at 80 °C overnight, and stained with lead citrate and UrAc. Images were captured on a transmission electron microscope (1200 EX II; JEOL, Peabody, MA) coupled to a digital camera (Orius 600; Gatan, Pleasanton, CA) and processed using the Gatan Digital Micrograph and Adobe Photoshop software (San Jose, CA). Areas of individual peroxisomes were measured using AxioVision software. Details of the EM analysis can be found in Supplemental Excel Table S3.
Fluorescence microscopy
Yeast cells were grown in YPD media (1% yeast extract, 2% bacto peptone, 2% dextrose) overnight at 30 °C to approximately 1 OD600/mL. The cells were then washed twice to remove dextrose and resuspended in methanol media [0.67% yeast nitrogen base without amino acids, 0.02 g histidine/L, 0.02 g arginine/L, 1% (vol/vol) methanol, 0.05 mM biotin]. Peroxisome biogenesis was induced via shaking at 250 rpm at 30 °C. At desired time points, cells were imaged using 100× magnification on an Axioskop fluorescence microscope. Images were taken on an AxioCam digital camera and processed on AxioVision software. Methodology to count the peroxins in different background strains is as follows: cells showing distinct ER labeling with Sec61-mCherry were first marked and counted. Then, these cells were analyzed for localization of GFP-tagged peroxins. Details of the fluorescence microscopy analysis for Fig. 5C can be found in Supplemental Excel Table S4.
BiFC
Plasmids containing both halves of the split Venus GFP were inserted into a plasmid containing two A0X1 promoters. The various peroxin genes were amplified from K. phaffii genomic DNA and inserted into these plasmids. The plasmids were linearized and used to transform WT and mutant strains already containing Sec61-mCherry (marker for ER). Pictures, acquisition, and analysis were performed as described in the fluorescence microscopy section.
Carbonate extraction
Cells were grown in YPD medium and then switched to oleate medium [0.67% yeast nitrogen base without amino acids, 0.02 g histidine/L, 0.02 g arginine/L, 0.2% (vol/vol) oleic acid, 0.02% (vol/vol) Tween-40]. After 6 h of induction, 250 OD600 of cells was spheroplasted in 3 mL Zymolyase solution [0.5 M KCI, 5 mM Mops/KOH (pH 7.2), 10 mM Na2SO3, 12.5 mg Zymolyase-100 T/mL] at 30 °C for 30 min. The cells were then spun down at 2200g for 8 min at 4 °C and resuspended in 1.5 mL homogenization buffer (5 mM Mes, 1 M sorbitol, 5 mM NaF, 20 mM EDTA). After this, they were lysed using a Dounce homogenizer (15 strokes). The PNS was generated by collecting the supernatant after the second centrifugation at 1000g for 10 min. 20kgS and 20kgP samples were formed from the further centrifugation of the PNS at 20,0000g for 30 min; the 20kgP sample was resuspended in the same amount of homogenization buffer. The 20kgP was then aliquoted into four tubes. In one tube, the pellet was switched to Tris buffer [10 mM Tris/HCl (pH 8)]. In another, the pellet was placed in carbonate solution [100 mM sodium carbonate, 10 mM Tris/HCl, (pH 11.5)]. The pellet in the third tube was dissolved in detergent solution [10 mM Tris/HCl (pH 8), 2% Chaps, 2% Triton-X100]. The last tube remained the 20kgP sample in homogenization buffer. After 30 min of incubation, the buffer, carbonate and detergent samples were centrifuged at 200,000g for 30 min, generating S200 and P200 samples for each condition. The samples were analyzed via SDS-PAGE and Western blot using anti-GFP antibody (Roche).
Growth curves
Cells were grown in YPD medium overnight to around 1–2 OD600/mL. Then, they were diluted to 0.2 OD600/mL with fresh YPD medium and left to grow to approximately 1 OD600/mL. They were then washed twice and resuspended in either methanol or oleate medium to a concentration of 0.2 OD600/mL. The growth of the cells was measured twice daily using a Beckman DU730 spectrophotometer, until a plateau in growth was reached.
Yeast two-hybrid assay
The GAL4-based matchmaker (Clontech) was used for yeast two-hybrid studies. K. phaffii PEX36 and PEX3 genes were inserted into the BD plasmid, pGBT9; PEX19 was inserted into the AD plasmid, pGAD-GH. S. cerevisiae strain AH109 was used in the transformation. Strains were selected on SD medium (Leu−, Trp−), then streaked on SD medium (His−, Leu−, Trp−) with various concentrations of 3-AT.
Subcellular fractionation
Cells were grown in YPD and then switched to oleate medium for 6 h. One hundred OD600 of cells was spheroplasted in 1.5 mL Zymolyase solution for 30 min at 30 °C. Cells were then resuspended in 1 mL homogenization buffer and lysed using a Dounce homogenizer (15 strokes). Intact cells and cellular debris were removed via centrifugation at 1000g for 10 min at 4 °C. The resulting PNS sample was then spun at 200,000g for 30 min to separate the : membrane (P200) from the cytosol (S200) fractions. The samples were TCA-precipitated using 12.5% TCA to concentrate the proteins. They were analyzed by SDS-PAGE and Western blot analysis with anti-Pex3 antibody.
Budding assay
Cytosol fraction preparation (S1)
K. phaffii strains were grown in YPD medium overnight at 30 °C to approximately 2.0 OD600/mL. The cells were switched to YYHR medium (1.7 g/L yeast nitrogen base, 1 g/L yeast extract, 5 g/L ammonium sulfate, 0.02 g/L l-His, 0.02 g/L l-Arg, 0.5% methanol) medium for 12 h. Two thousand OD600 of cells was spheroplasted in 650 mL spheroplasting medium [1% yeast extract, 1 M sorbitol, 0.05 M Kpi (pH 7.5), 0.05 M ß-mercaptoethanol, 20 mg Zymolyase-100 T] at 37 °C for 30 min. After this, the cells were pelleted at 3000 rpm for 5 min at 4 °C and resuspended in 1.3 L recovery medium (1% yeast extract 1 M sorbitol). The cells were then incubated 37 °C for 90 min. The cells are pelleted and resuspended in 3.5 mL 20 mM Hepes/KOH (pH 7.6) to release the contents from the membrane fraction. The solution was centrifuged three times at 3000 rpm for 10 min, and the supernatant was collected.
Membrane Fraction Preparation (PYC)
K. phaffii strains expressing Pex2-3HA and Pex17-3HA proteins were grown in YPD medium overnight at 30 °C to approximately 1 OD600/mL. The cells were induced in YYHR medium for 3 h. Seventy-five OD600 of cells was washed and resuspended in 25 mL spheroplasting solution. After incubation at 37 °C for 30 min, the cells were transferred to 50 mL recovery medium and left at 37 °C for 30 min. Then, they were pelleted and resuspended in permeabilization buffer [100 mM KOAc, 200 mM sorbitol, 20 mM Hepes/KOH (pH 7.2), 2 mM MgCl2]. After this, the cells were pelleted and resuspended in 50 µL column buffer [20 mM Hepes/KOH (pH 7.4), 250 mM sucrose, 250 mM DTT, 1 mM EGTA]. Finally, the cells were washed and resuspended in TBPS buffer [250 mM Hepes/KOH (pH 7.2), 1 M KOAc, 25 mM Mg(OAc)2, 250 mM sorbitol, PIC yeast] to a concentration of 4.5 OD600/25 µL.
The ppV budding reaction
Each 80 µL reaction comprised 1–1.5 mg S1 and 4.5 OD600 PYC. In the NTP condition, an ATP-regenerating cocktail was added. In the apyrase condition, apyrase was added to deplete the source of energy. As a control for the PYC, 4.5 OD600 of PYCs was mixed with apyrase and brought up to a total volume of 80 µL with TBPS buffer. The samples were incubated at 20 °C for 90 min. After this, the samples were centrifuged at 13,000 rpm for 1 min, and the supernatant was saved. The samples were analyzed via SDS-PAGE and Western blot with HA-tag antibodies.
Co-immunoprecipitation assay
Cells were grown in YPD medium overnight to approximately 2 OD600/mL and then switched to methanol medium. After 6 h of induction, 250 OD600 of cells was harvested and resuspended in 4 mL Zymolyase solution. The cells were spheroplasted at 30 °C for 30 min. Next, they were spun down and resuspended in 1.5 mL lysis buffer [20 mM Hepes/KOH (pH 7.4), 0.15 M NaCl, 5 mM NaF, 20 mM EDTA (pH 8.0), 50 µg leupeptin/mL, 50 µg aprotinin/mL, 1 mM PMSF, PIC yeast]. The cells were lysed by vortexing in the presence of acid-washed glass beads. The lysate was centrifuged at 20,000g for 20 min at 4 °C to remove the cytosolic fraction. The pellet was resuspended in 1.5 mL lysis buffer (with 1% Chaps) and placed on a rotator at 4 °C for solubilization. The samples were then spun at 20,000g for 20 min. The supernatant was collected and mixed with specified antibody affinity matrix (Sigma-Aldrich EZview Red Anti-HA) to pull down HA-Pex19. They were left to incubate overnight before washing the beads four times with lysis buffer. One hundred twenty microliters of sample buffer was added to the beads and placed in boiling water for 5 min. The eluate was analyzed via SDS-PAGE and Western blotting for anti-HA, anti-Pex3, anti-Pex2, and anti-Pex17 antibodies.
Protease protection assay
Cells were grown in YPD and then switched to oleate medium for 6 h. Three hundred fifty OD600 of cells was washed and spheroplasted in 5.4 mL Zymolyase buffer for 30 min at 30 °C. They were then transferred to 2 mL homogenization buffer and lysed using a Dounce homogenizer (15 strokes). Intact cells and cellular debris were removed via centrifugation at 1000g × 10 min at 4 °C. The resulting PNS was then spun at 20,000g × 15 min to isolate the membrane fraction. The pellet was resuspended in 2 mL homogenization buffer and aliquoted in three tubes. For the protease condition, 80 µg proteinase K and 40 µg trypsin were added. For the detergent condition, 0.5% Triton X-100 was added in addition to the proteinase K and trypsin. For the buffer condition, additional homogenization buffer was added. Samples were taken at various time points and preserved using liquid nitrogen. The samples were TCA-precipitated using 12.5% TCA to concentrate the proteins. They were analyzed via SDS-PAGE and Western blotting for anti-GFP and anti-HA antibodies.
Mammalian cell culture and transfection
HeLa (human cervical carcinoma) cells were grown in Dulbecco's modified Eagle's medium high-glucose medium supplemented with 10% fetal calf serum and antibiotics in a humidified atmosphere containing 5% CO2 at 37 °C. HeLa cells were transfected with mammalian expression plasmids using Fugene6 (Roche Diagnostics Corp., Indianapolis, IN) as follows: Cells were seeded onto glass coverslips in one well of a 6-well plate until they reached 60% confluency, followed by lipofection. Per well, the lipofection mix contained 1 µg total DNA and 3 µL Fugene6 in 100 µL serum-free medium. After 15-min incubation time, the lipofection mix was added dropwise onto the cells. Cells were analyzed by fluorescence microscopy 24–48 h after transfection. Primary fibroblasts of patients lacking peroxisomes because of a mutated PEX16 gene (cell line GM06231 obtained from Coriell Institute for Medical Research) were grown in Dulbecco's modified Eagle's high-glucose medium supplemented with 2 mM glutamine, 10% fetal calf serum, and antibiotics in 5% CO2 at 37 °C. Transfections of primary human fibroblasts were performed using the Basic Nucleofector Kit for Primary Mammalian Fibroblasts (Cat No. VPI-1002, Lonza Group Ltd., Basel, Schweiz). In short, 1 × 106 cells were electroporated with 1 µg total plasmid DNA in 100 µL electroporation buffer using the Nucleofector (Lonza Group Ltd.) with program A-024 and seeded onto cover slips in pre-warmed medium directly after electroporation. Cells were analyzed by microscopy not earlier than 48 h after lipofection.
Supplementary Material
Acknowledgments
We thank members of our team for helpful discussions. We wish to thank Drs. Einat Zalckvar and Maya Schuldiner (Weizmann Institute, Rehovot, Israel) for providing the BiFC plasmids. Funded by grants from National Institutes of Health (5 RO1DK41737 to S.S.), a seed grant by the San Diego Center for Systems Biology/SDCSB (to A.T. and S.S.). The funders had no input in the study design or in the collection, analysis or interpretation of data.
Abbreviations used
- BiFC
bimolecular fluorescence complementation
- ER
endoplasmic reticulum
- pER
punctate endoplasmic reticulum subdomain/pre-peroxisomal ER
- PMP
peroxisomal membrane protein
- pn-ER
perinuclear endoplasmic reticulum
- ppV
pre-peroxisomal vesicle
- PTS
peroxisome targeting signal
- PYC
permeabilized yeast cells
Footnotes
Accession numbers
GenBank accession number: HsPex11α, EAX02062; HsPex11β, EAW71423; HsPex11γ, EAW69038; HsPex16, EAW68023; KpPex2, CAY69969; KpPex3, CAY71136; KpPex11C, CAY68384; KpPex17, CAY71882; KpPex19, CAY69364; KpPex25, CAY70171; KpPex36, CAY67701; Y/Pex11/25, CAG81480; Y/Pex16, CAG79622; ScPex27, DAA10968; ScPex34, DAA07430.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2O17.10.009.
Authors' contributions: K.C. and J.C.F. performed most the growth curves, fluorescence microscopy, and proteins assays. A.T. did the mammalian complementation study. G.A. and K.C. performed the ER budding experiments. Z.H. performed growth curves assays and fluorescence microscopy experiments. O.V.S., O.S., J.C., and A.A.S. isolated the PEX36 genes and participated in the discovery of the functional orthologs. M.P. performed the peroxin stability assays and EM. J.C.F. designed the studies, analyzed the data, and wrote the paper. S.S. supervised the study and participated in data interpretation and writing the paper. All authors read, proofread, and approved the final manuscript.
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Kim PK, Hettema EH. Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 2015;427:1176–1190. doi: 10.1016/j.jmb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schliebs W, Kunau WH. Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 2004;14:R397–9. doi: 10.1016/j.cub.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Tower RJ, Fagarasanu A, Aitchison JD, Rachubinski RA. The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol. Biol. Cell. 2011;22:1727–1738. doi: 10.1091/mbc.E11-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan M, Rayapuram N, Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, et al. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 2010;123:3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- 6.Hua R, Kim PK. Multiple paths to peroxisomes: mechanism of peroxisome maintenance in mammals. Biochim. Biophys. Acta. 2016;1863:881–891. doi: 10.1016/j.bbamcr.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal G, Subramani S. De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta. 2016;1863:892–901. doi: 10.1016/j.bbamcr.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theodoulou FL, Bernhardt K, Linka N, Baker A. Peroxisome membrane proteins: multiple trafficking routes and multiple functions? Biochem. J. 2013;451:345–352. doi: 10.1042/BJ20130078. [DOI] [PubMed] [Google Scholar]
- 9.Guo T, Gregg C, Boukh-Viner T, Kyryakov P, Goldberg A, Bourque S, et al. A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J. Cell Biol. 2007;177:289–303. doi: 10.1083/jcb.200609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayerhofer PU, Bano-Polo M, Mingarro I, Johnson AE. Human peroxin PEX3 is co-translationally integrated into the ER and exits the ER in budding vesicles. Traffic. 2016;17:117–130. doi: 10.1111/tra.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoms S, Harms I, Kalies KU, Gartner J. Peroxisome formation requires the endoplasmic reticulum channel protein Sec61. Traffic. 2012;13:599–609. doi: 10.1111/j.1600-0854.2011.01324.x. [DOI] [PubMed] [Google Scholar]
- 12.van der Zand A, Braakman I, Tabak HF. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 2010;21:2057–2065. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi AS, Huang X, Choudhary V, Levine TP, Hu J, Prinz WAA. Family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 2016;215:515–529. doi: 10.1083/jcb.201602064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrawal G, Fassas SN, Xia ZJ, Subramani S. Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 2016;212:335–348. doi: 10.1083/jcb.201506141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal G, Joshi S, Subramani S. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9113–9118. doi: 10.1073/pnas.1018749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam SK, Yoda N, Schekman RA. Vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21523–21528. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoops K, Manivannan S, Cepinska MN, Krikken AM, Kram AM, Veenhuis M, et al. Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 2014;204:659–668. doi: 10.1083/jcb.201310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellu AR, Salomons FA, Kiel JA, Veenhuis M, Van Der Klei IJ. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 2002;277:42875–42880. doi: 10.1074/jbc.M205437200. [DOI] [PubMed] [Google Scholar]
- 19.Williams C, van der Klei IJ. Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 2013;438:395–401. doi: 10.1016/j.bbrc.2013.07.086. [DOI] [PubMed] [Google Scholar]
- 20.Wroblewska JP, Cruz-Zaragoza LD, Wei Y, Schummer A, Chuartzman SG, de Boer R, et al. Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim. Biophys. Ada. 2017 doi: 10.1016/j.bbamcr.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 22.Chang J, Mast FD, Fagarasanu A, Rachubinski DA, Eitzen GA, Dacks JB, et al. Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J. Cell Biol. 2009;187:233–246. doi: 10.1083/jcb.200902117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haan GJ, Baerends RJ, Krikken AM, Otzen M, Veenhuis M, van der Kiel IJ. Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 2006;6:186–194. doi: 10.1111/j.1567-1364.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- 24.Stasyk OV, Ksheminskaya GP, Kulachkovsky AR, Demchyshyn V, Cregg JM, Sibirny AA. New mutants of methylotrophic yeast Pichia pastoris defective in peroxisome degradation and biogenesis. In: Scheffers WA, Van Dijken JP, editors. Beijerinck Centennial Microbial Physiology and Gene Regulation: Emerging Principles and Applications. The Netherlands Delft University Press; The Hague: 1995. pp. 426–427. [Google Scholar]
- 25.Law KB, Bronte-Tinkew D, Di Pietro E, Snowden A, Jones RO, Moser A, et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017;13:868–884. doi: 10.1080/15548627.2017.1291470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 2014;10:835–845. doi: 10.4161/auto.28259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J. Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Ghaedi K, Fujiki Y, et al. Identification of PEX3 as the gene mutated in a Zellweger syndrome patient lacking peroxisomal remnant structures. Hum. Mol. Genet. 2000;9:1995–1999. doi: 10.1093/hmg/9.13.1995. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, et al. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J, Klute MJ, Tower RJ, Mast FD, Dacks JB, Rachubinski RA. An ancestral role in peroxisome assembly is retained by the divisional peroxin Pex11 in the yeast Yarrowia lipolytica. J. Cell Sci. 2015;128:1327–1340. doi: 10.1242/jcs.157743. [DOI] [PubMed] [Google Scholar]
- 33.Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J. Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honsho M, Hiroshige T, Fujiki Y. The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J. Biol. Chem. 2002;277:44513–44524. doi: 10.1074/jbc.M206139200. [DOI] [PubMed] [Google Scholar]
- 35.Yahraus T, Braverman N, Dodt G, Kalish JE, Morrell JC, Moser HW, et al. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 1996;15:2914–2923. [PMC free article] [PubMed] [Google Scholar]
- 36.Motley AM, Galvin PC, Ekal L, Nuttall JM, Hettema EH. Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J. Cell Biol. 2015;211:1041–1056. doi: 10.1083/jcb.201412066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knoops K, de Boer R, Kram A, van der Klei IJ. Yeast pex1 cells contain peroxisomal ghosts that import matrix proteins upon reintroduction of Pex1. J. Cell Biol. 2015;211:955–962. doi: 10.1083/jcb.201506059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 39.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halbach A, Rucktaschel R, Rottensteiner H, Erdmann R. The N-domain of Pex22p can functionally replace the Pex3p N-domain in targeting and peroxisome formation. J. Biol. Chem. 2009;284:3906–3916. doi: 10.1074/jbc.M806950200. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal G, Shang HH, Xia ZJ, Subramani S. Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis. J. Biol. Chem. 2017 doi: 10.1074/jbc.M116.774067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stasyk OV, Nazarko TY, Sibirny AA. Methods of plate pexophagy monitoring and positive selection for ATG gene cloning in yeasts. Methods Enzymol. 2008;451:229–239. doi: 10.1016/S0076-6879(08)03216-3. [DOI] [PubMed] [Google Scholar]
- 43.Lin Cereghino GP, Lin Cereghino J, Sunga AJ, Johnson MA, Lim M, Gleeson MA, et al. New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene. 2001;263:159–169. doi: 10.1016/s0378-1119(00)00576-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.