Abstract
Objective
Recent transcriptomic studies describe two subgroups of adults with sepsis differentiated by a sepsis response signature (SRS). The implied biology and related clinical associations are comparable to recently reported pediatric sepsis endotypes, labeled “A” and “B”. We classified adults with sepsis using the pediatric endotyping strategy and the SRS, and determined how endotype assignment, SRS membership, and age interact with respect to mortality.
Design
Retrospective analysis of publically available transcriptomic data representing critically ill adults with sepsis from which the SRS groups were derived and validated.
Setting
Multiple intensive care units.
Patients
Adults with sepsis
Interventions
None.
Measurements and Main Results
Transcriptomic data were co-normalized into a single data set yielding 549 unique cases with SRS assignments. Each subject was assigned to endotype A or B using the expression data for the 100 endotyping genes. There were 163 subjects (30%) assigned to endotype A and 386 to endotype B. There was a weak, positive correlation between endotype assignment and SRS membership. Mortality rates were similar between patients assigned endotype A and those assigned endotype B. A multivariable logistic regression model fit to endotype assignment, SRS membership, age, and the respective two-way interactions revealed that endotype A, SRS1 membership, older age, and the interactions between them were associated with mortality. Subjects co-assigned to endotype A and SRS1 had the highest mortality.
Conclusions
Combining the pediatric endotyping strategy with SRS membership might provide complementary, age-dependent, biological and prognostic information.
Keywords: sepsis, sub-classification, gene expression, immune suppression, endotypes
INTRODUCTION
Recent transcriptomic studies describe two clinically relevant subgroups of adults with sepsis, which are differentiated by a sepsis response signature (SRS) reflecting immune suppression [1, 2]. Membership in SRS1 is associated with higher mortality relative to membership in SRS2, therefore suggesting clinical utility for SRS assignment. The biology associated with the SRS and the differences in mortality are comparable to our previous reports of pediatric sepsis endotypes “A” and “B” [3–7]. The expression signature differentiating the pediatric sepsis endotypes consists of 100 genes corresponding to adaptive immunity and glucocorticoid receptor signaling. The majority of these genes are repressed among endotype A patients relative to endotype B patients, and allocation to endotype A is independently associated with poor outcomes. In addition, corticosteroid prescription is independently associated with increased risk of mortality among endotype A patients.
Using the publically available transcriptomic data from which the SRS groups were derived and validated [1, 2], we classified adults with sepsis using the endotyping strategy previously developed for pediatric sepsis. We assessed the overlap between the SRS and endotype groupings to determine whether the strategies provide equivalent or complementary information. We also determined if two-way interactions between endotype assignment, SRS membership, and age are associated with mortality among adults with sepsis.
METHODS
Since we used de-identified, publically available transcriptomic data, the study was exempt from Institutional Review Board approval. The 100 endotyping genes and the method for using them in endotyping were previously reported [6]. Gene expression data sets E-MTAB-4421, E-MTAB-4451, E-MTAB-5273, and E-MTAB-5274 were downloaded from ArrayExpress. These data from the GAinS study represent adults with sepsis used in the discovery and validation of the SRS groupings taken from the earliest available time point after enrollment [1, 2]. Since the two cohorts were from the same study and represent the same clinical circumstances, we used ComBat normalization [8, 9] to co-normalize the cohorts into a single dataset representing 549 unique cases with SRS assignments. From this dataset, we extracted expression data for the 100 endotyping genes. We generated individual gene expression mosaics for each study subject using the Gene Expression Dynamics Inspector [6]. These were compared to reference mosaics using computer-assisted image analysis to assign the study subjects into endotype A or B, as previously detailed [6]. The Phi coefficient was used to assess the correlation between SRS membership and endotype assignment. We used multivariable logistic regression to explain associations between endotype assignment, SRS membership, and mortality within 28 days. Since the endotyping strategy was initially derived in a pediatric cohort, we also considered whether age modified the association between endotype, SRS membership, and mortality.
RESULTS AND DISCUSSION
There were 163 subjects (30%) allocated to endotype A and 386 subjects allocated to endotype B. This distribution is similar to what was reported in children with sepsis [6]. Mortality was similar in the endotype A (23%) and endotype B (25%) subgroups. Among the 220 subjects in the SRS1 group, 34 were assigned to endotype A and 186 were assigned to endotype B. Among the 329 subjects in the SRS2 group, 129 were assigned to endotype A and 200 were assigned to endotype B. There was a weak, positive correlation between endotype assignment and SRS membership (Phi coefficient = 0.25, p < 0.001).
The median age of the cohort was 68 years (range 18 to 92). Table 1 provides the results of the multivariable logistic regression model wherein endotype assignment, SRS membership, age, and the corresponding two-way interactions are considered as predictor variables for mortality. SRS1 membership, endotype A assignment, and older age were associated with increased risk of mortality. The interaction between endotype and age was statistically significant, showing increased risk of mortality with endotype A and younger age. There were no statistically significant interactions when considering the interactions between SRS1 membership and older age, and between SRS1 membership and endotype A assignment, respectively.
Table 1.
Results of multivariable logistic regression testing for associations between the listed variables and mortality.
| Variable | Odds Ratio | 95% C.I. | p value |
|---|---|---|---|
| SRS1 | 16.23 | 1.31 to 200.58 | 0.03 |
| Endotype A | 64.70 | 4.90 to 853.63 | 0.002 |
| Age | 1.07 | 1.04 to 1.10 | <0.001 |
| Endotype A x Age | 0.94 | 0.90 to 0.97 | <0.001 |
| SRS1 x Age | 0.97 | 0.94 to 1.00 | 0.060 |
| SRS1 x Endotype A | 2.57 | 0.96 to 6.91 | 0.062 |
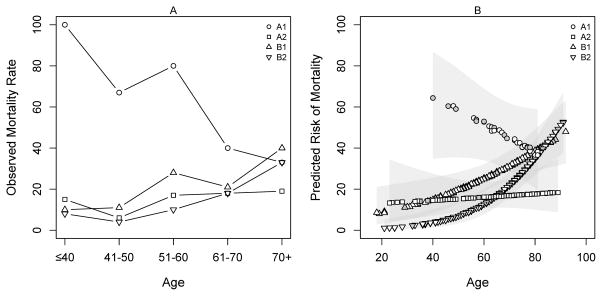
Figure 1A shows the observed mortality rate for subjects ≤ 40 years of age, and for those between 41 and 50, 51 and 60, 61 and 70, and > 70, cross classified by both endotype and SRS. Figure 1B shows the mortality predicted from the multivariable logistic regression model with 95% confidence intervals. Age was modeled as a continuous variable. The youngest patients co-assigned to endotype A and SRS1 had a >60% mortality risk, which decreased with older age. In contrast, mortality risk was <10% and increased with age among those co-assigned endotype B and SRS1 or SRS2. Mortality was relatively constant over the age spectrum among those co-assigned endotype A and SRS2. The overall mortality rates for the co-assignment groups were 47% (endotype A/SRS1), 16% (endotype A/SRS2), 28% (endotype B/SRS1), and 23% (endotype B/SRS2) (p = 0.001, Chi-square, 3 degrees of freedom).
Figure 1. Mortality associated with cross classification by both endotype and SRS, and age.
(A) The observed mortality rate based on cross classification and age groupings. A1 = subjects co-assigned endotype A and SRS1; A2 = subjects co-assigned endotype A and SRS2; B1 = subjects co-assigned endotype B and SRS1; and B2 = subjects co-assigned endotype B and SRS2. Among the A1 subjects, 1 subject was ≤ 40 years old, 3 were 41 to 50 years old, 5 were 51 to 60 years old, 10 were 61 to 70 years old, and 15 were > 70 years old. (B) The mortality predicted from the multivariable logistic regression model, where age is a continuous variable. The 95% confidence intervals are shown by grey shading. Grey filled symbols represent subjects who died by 28 days.
The interaction between age and endotype assignment indicates that the endotyping strategy developed among children with sepsis might be more directly applicable to younger adults than to older adults. The interactions between endotype assignment and SRS membership suggest that the gene expression patterns associated with both endotype A and with SRS1 are both involved in sepsis pathobiology. Accordingly, we compared how the 100 endotyping genes are differentially expressed between the A1 group and the other three endotype/SRS co-assignment groups. Using a Benjamini-Hochberg False Discovery Rate of 1%, we found 28 differentially regulated genes (Supplemental Table 1). Pathway analysis revealed that the 28 genes correspond to the T cell receptor signaling pathway. The majority of these genes had decreased expression in the A1 group, relative to the other three co-assignment groups.
Both the pediatric and adult stratification strategies implicate immune suppression as a biological feature, albeit via different gene expression signatures. While this is well aligned with current paradigms of sepsis pathobiology [10] and reflects the large number of genes involved in immune function, neither strategy has measured immune suppression directly. However, since the two signatures appear to be measuring different components of immune dysregulation [2], the finding that patients exhibiting immune suppression signatures by both measures are at the greatest risk of mortality suggests the combination of endotype A with SRS1 might identify a population with a heightened degree of immune suppression. Functional studies are required to directly assess the specific biology reflected by these two signatures and how they interact in the context of sepsis pathobiology. The counter-intuitive effect of age in the endotype A/SRS1 group requires replication, particularly given the very small sample size of the group, which may lead to an imprecise effect estimate. For the remaining groups, age effects may be related to comorbidity burden; older adults are likely to have a greater comorbidity burden and decreased resiliency to critical illness.
The general purpose of both the SRS and endotyping strategies is not to provide a directly prognostic tool, but rather to uncover novel molecular subtypes of sepsis whose clinical relevance is reinforced by the finding of different mortality rates. The lack of redundancy between SRS and the endotypes does not necessarily question their relative robustness. Since they were discovered using different approaches, they were fit to independent patient cohorts and considered different variables. It appears that combining the two signatures might provide complementary, age-dependent biological and prognostic information, suggesting utility in considering both sets of variables in future analyses. However, it should be noted that the pediatric studies used RNA samples obtained during the first 24 hours of admission, whereas the GAinS study allowed for sample procurement up five days from the time of enrollment.
In conclusion, recent studies and our current results show that clinically relevant sepsis sub-classification is possible using gene expression-based strategies. Future studies should carefully consider the influence of age on the association between immune function and sepsis outcomes, and should directly link gene expression patterns with biological function.
Supplementary Material
Acknowledgments
FUNDING SOURCE
Supported by National Institutes of Health Grants RO1GM099773 and R01GM108025.
Footnotes
Copyright form disclosure: Drs. Wong, Hart, and Lindsell’s institutions received funding from the National Institutes of Health (NIH), and they received support for article research from the NIH. Drs. Wong and Lindsell are named as co-inventors for a provisional U.S. patent application based on the endotyping strategy reported in the manuscript. Dr. Sweeney received funding from Inflammatix. Dr. Khatri disclosed stock ownership in Inflammatix, and he received support for article research from the NIH and Bill & Melinda Gates Foundation. Dr. Lindsell received funding from Digital Bioscapes (stock options for participating on a scientific advisory board; no relevance to current work).
References
- 1.Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4(4):259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnham KL, Davenport EE, Radhakrishnan J, et al. Shared and Distinct Aspects of the Sepsis Transcriptomic Response to Fecal Peritonitis and Pneumonia. Am J Respir Crit Care Med. 2017;196(3):328–339. doi: 10.1164/rccm.201608-1685OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong HR, Cvijanovich N, Lin R, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong HR, Wheeler DS, Tegtmeyer K, et al. Toward a clinically feasible gene expression-based subclassification strategy for septic shock: proof of concept. Crit Care Med. 2010;38(10):1955–1961. doi: 10.1097/CCM.0b013e3181eb924f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong HR, Cvijanovich NZ, Allen GL, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011;39(11):2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong HR, Cvijanovich NZ, Anas N, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. 2015;191(3):309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong HR, Sweeney TE, Lindsell CJ. Simplification of a Septic Shock Endotyping Strategy for Clinical Application. Am J Respir Crit Care Med. 2017;195(2):263–265. doi: 10.1164/rccm.201607-1535LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney TE, Perumal TM, Henao R, et al. Mortality prediction in sepsis via gene expression analysis: a community approach. bioRxiv. 2016 doi: 10.1038/s41467-018-03078-2. http://dx.doi.org/10.1101/095489. [DOI] [PMC free article] [PubMed]
- 10.Hotchkiss RS, Sherwood ER. Immunology. Getting sepsis therapy right. Science. 2015;347(6227):1201–1202. doi: 10.1126/science.aaa8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.