Abstract
Allergy is defined as an inappropriate immune response to something normally considered harmless. The symptomatic immune response is driven by IgE antibodies directed against allergens. The study of allergens has contributed significantly to our understanding of allergic disease in three main areas. First, identifying allergens as the source of patient symptoms and developing allergen standards has led to many advances in exposure assessment and patient diagnostics. Second, a biochemical understanding of allergens has suggested a number of hypotheses related to the mechanisms of allergic sensitization. And finally, studies of allergen-antibody interactions have contributed to understanding the cross-reactivity of allergens, mapping patient epitopes, and the development of hypo-allergens. In this review, a few select cases are highlighted where structural biology in particular has contributed significantly to allergen research and provided new avenues to investigate.
Keywords: Allergens, Structural Biology, Sensitization, Standardization
Introduction
Structural Biology is a branch of science concerned with the atomic level description of biological macromolecules and how the three-dimensional arrangement of those atoms affects the function of the molecule. An allergic patient usually has other concerns, namely the relief of medical symptoms either temporarily or permanently. These symptoms are initiated by allergens binding to patient IgE antibodies. How can the study of the three-dimensional structure of allergens help the allergic patient? This review is intended to describe a number of vignettes where structural biology has contributed and suggested new directions for the study of allergens with the ultimate goal of benefiting the patient.
Standardization
Setting standards of measurement units is crucial to any enterprise from trade to science. In allergy research, several databases exist that help to define the protein sequences of reported allergens so that researchers can accurately communicate and compare results [1]. A common vocabulary is crucial for setting exposure recommendations, patient testing, and levels of allergens used in immunotherapies. The CREATE and follow up BSP-090 initiatives have made significant strides in standardizing allergen measurements and creating panels of allergens for patient diagnostics [2–6].
In the case of single domain protein systems, defining an allergen follows a typical routine. IgE binding proteins are identified, and subsequently fragments of the protein are sequenced by various technologies. With fragments of sequence information, the protein can typically be cloned, or identified from available genomic data. This allows for calculation of a molecular weight so the protein can be identified on a gel and the stoichiometry of binding to antibodies determined for accurate quantitative comparison. However, a more difficult case is when the allergen contains repeated genetic elements, like the group 1 cockroach allergens.
The group 1 cockroach allergens belong to a family of proteins restricted to insects [7]. Two different genetic duplication events prior to the emergence of Insecta appear to have created a family of proteins with an AB-AB-AB… pattern, where A and B are distantly but obviously related sequences [7,8]. The prevailing theory is that in the first genetic event A duplicated to form AA, which diverged over time to become AB, and then subsequently the AB unit was further duplicated. The number of duplications and number of proteins with AB units varies considerably from insect to insect [7,9]. This creates a major problem for standardization because the stoichiometry of antibody binding is unknown. An early prediction for one member of the allergen group, Per a 1, suggested these were membrane bound proteins based on the studious observation that the secondary structure was predicted to be highly helical, and the primary sequence contains a much higher proportion of hydrophobic residues than is typical for soluble proteins [10]. To extend this prediction further, one might then suggest that the helical and hydrophobic AB regions would be buried in a membrane and inaccessible to antibodies, and the connecting loops between AB would be exposed to solvent. These loops have the highest degree of evolutionary divergence, and would be expected to represent unique epitopes.
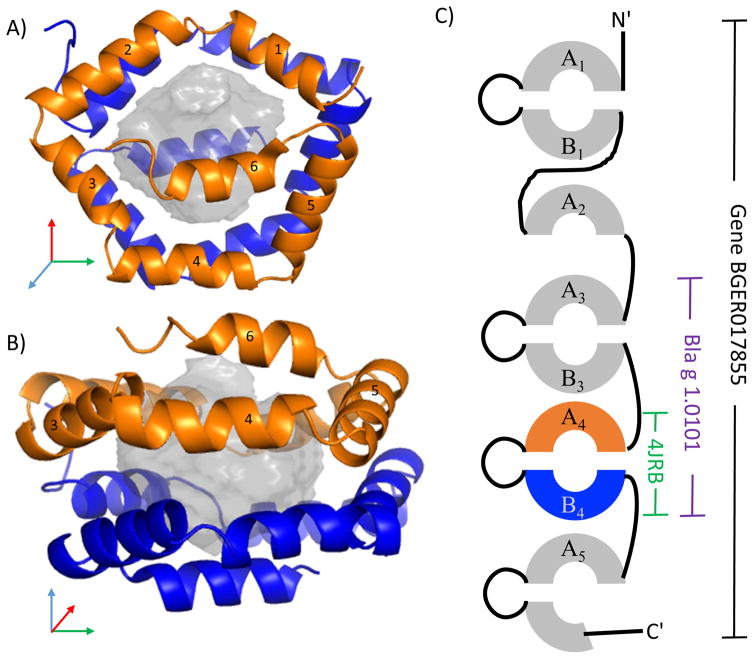
The structure of Bla g 1 resolved many, but not all, of these issues. The AB unit forms a soluble domain, which is a spherical ball of helices, Figure 1 [11]. Both the A and B hemispheres are nearly structurally identical (Cα RMSD of 1.4 Å) despite only 26% sequence identity. Each hemisphere is made up of a pentagon shaped ring of 5 helices, with a 6th helix capping and bisecting the pentagon. A large interior cavity appears to be a vehicle for lipids as it is lined exclusively with hydrophobic residues, explaining the excess abundance of these sidechains. Further, the structure suggested that the allergenic unit accessible to antibodies is the soluble, spherical AB unit. This was further confirmed with a Western Blot analysis of various sized Bla g 1 constructs [11]. Equivalent titration curves were found on a molar basis for constructs containing A3B3-A4B4 and only A4B4 (Figure 1). Therefore, the allergenic units can be standardized to a molecular weight and grams of proteins, as opposed to the arbitrary standard of units per gram of cockroach extract. This structure further qualitatively explained the typical multi-banded pattern of Bla g 1 staining on a gel. The flexible loops between A and B, and between AB units are variably susceptible to proteolysis. This creates a molecular ladder effect dependent on the extent of degradation in the sample [12].
Figure 1.
Bla g 1. Panels A and B show two 90° rotated views of the Bla g 1 structure (4JRB [11]) where helices from hemisphere A are colored orange and hemisphere B are colored blue. The interior cavity is rendered with a grey semitransparent sphere. Panel C is a schematic of the gene from which the 4JRB structure and the named allergen Bla g 1.0101 was derived. Sequence similarity of the repeated units is noted by the A or B nomenclature.
Scientists should be explicit about the isoform using the current nomenclature for consistent analysis of standards for the group 1 cockroach allergens. (Isoform sequences, names, and nomenclature guidelines can be found at the website, www.allergen.org, which is maintained by World Health Organization/International Union of Immunological Societies, WHO/IUIS.) A problem to be aware of is that the number of antibodies that can bind a particular isoform is variable. For example, Bla g 1.0101 and Bla g 1.0201 [8,12] contain two AB units, so the stoichiometry of antibody binding in an ELISA is the same, but not for Bla g 1.0102 [13,14]. In Periplaneta americana, the molecular weight and number of AB units differs by a factor of two for the various characterized Per a 1 iso-allergens [15]. Therefore, the methods of each paper should be carefully reviewed for strict comparisons.
Further complicating the study of these allergens is an examination of the Blatella germanica genome and RNAseq data suggests that there are five genes expressed with multiple AB units, and a few fragments of A or B. Figure 1 shows the gene product BGER017855 containing Bla g 1.0101. In data to be published, unique peptides from all 5 gene products were identified by mass spectrometry from cockroach frass. All of these genes have highly interrelated sequence identities ranging from 57% to 98% identical, so the amount of Bla g 1-like allergen in cockroach frass may be underestimated depending on the cross-reactivity of the detecting antibody, which is currently unknown. Sorting the details of potential cross-reactivity and the implications for standardization will require further studies.
The cockroach group 1 proteins are a really interesting case where structural data has informed the standardization of the measurement unit, and highlighted the need to be explicit with the nomenclature to accurately communicate the quantities of allergen content in exposure and extract measurements.
Mechanisms of Sensitization
One fascinating thing about allergens is that humans are exposed to thousands of foreign proteins every day, yet a few select proteins consistently become major allergens in Homo sapiens. This has led many researchers to look for common properties of allergens that might skew the immune response towards allergy. For example, the major mite allergen Der p 1 was identified as a cysteine protease from sequence information [16] and subsequently many studies have been conducted to examine if proteolytic activity could influence barrier function [17], innate immune signaling [18], and adaptive immunity [19,20]. Similar experiments were suggested for Bla g 2, given the high sequence identity with aspartyl proteases. However, a crystal structure of Bla g 2 revealed an incompetent active site [21]. This is a useful example where structural biology ruled out a series of further experiments.
Since the function of Der p 1 appeared to have important allergenic properties, the biological functions of allergens have been carefully catalogued to look for other patterns. While many allergen sources including mites, cockroaches [22], fungi [23], and plants [24,25] have proteases as prominent allergens, the most common functional property of allergens is the binding of hydrophobic ligands [26]. Prominent allergens like Bet v 1 and Der p 2 have hydrophobic cavities that can bind different lipids. Various lipids are known to skew the immune response towards allergy in a variety of relevant cases, and this has recently been well reviewed [27].
The group 2 mite allergens are an excellent case study where structural biology was extremely useful in suggesting a series of experiments regarding allergenicity and the affinity for lipids. When the structure of mite group 2 allergens was first described it was not obviously related to a unique protein function [28–30]. The secondary structure was loosely related to the traditional immunoglobulin greek-key motif, but proteins with a similar fold have a broad range of possible biological functions. In retrospect, there were some clues as to the function of the protein. For example, the NMR structure of Der f 2 was determined in the presence of the detergent N-octyl-β-D-glucoside [30] and the same compound qualitatively changed the NMR spectra of Der p 2 indicating a possible interaction [29]. In the crystal structure of Der p 2, a long aliphatic chain was identified in the central cavity of the protein, which did not resemble N-octyl-β-D-glucoside [31]. With the benefit of hindsight, these observations hinted at the lipid binding properties of the allergen. These structures predated the explosion of research on Toll-like receptors (TLR) and the structure of the accessory protein MD-2 [32]. MD-2 binds bacterial lipopolysaccharide (LPS) and binds to TLR4, which stimulates innate immune pathways. When the structure of MD-2 was determined, the researchers noticed that the structure aligned very closely with Der p 2. Karp and co-workers very elegantly described a series of experiments which demonstrated that Der p 2 could functionally substitute for MD-2 in murine models [33]. This suggested that Der p 2 could act as its own auto-adjuvant.
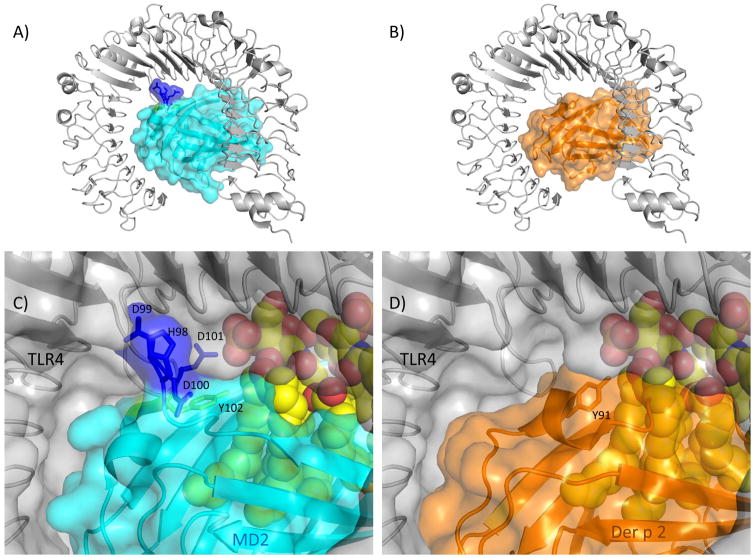
While the Der p 2/MD-2 mimicry paper has been widely cited, there have been few follow up experiments. Herein, an analysis of the structural biology suggests some questions about the affinity of Der p 2 for TLR4, which could test how Der p 2 might compete with endogenous MD-2. All of the experiments demonstrating that Der p 2 binds to TLR4 were immunoprecipitations that do not directly measure affinity [33]. MD-2 and TLR4 bind with a 0.8 nM dissociation constant [34]. An examination of the interactions from the crystal structures of both human and murine TLR4:MD-2 complexes [35–37] reveals that residues 98–104 (HDDDYSF) provide approximately 22% of the buried surface area for the interaction. In fact, a longer peptide including these specific residues is able to ablate binding of MD-2 to TLR4, implying these are functionally important residues for the interaction [38]. Similarly, a triple mutant of MD-2 where 99-DDD-101 was mutated to ANA showed reduced ability to respond to LPS [39]. Alignments of Der p 2 with human and murine MD-2 clearly indicate a five residue gap in Der p 2 resulting in the loss of the DDD motif in the mite. Using the protein interaction analysis program PISA [40] the DDD residues alone provide 5 potential hydrogen bonds, and 6 potential salt bridges. A further examination of conserved residues at the TLR4:MD-2 interface suggests that 15 of the 22 H-bonds, and 13 of the 16 salt bridges would not be possible based on the amino acid changes when aligning MD-2 with Der p 2. The lack of the DDD motif and other mutations casts some doubt on the ability of Der p 2 to bind human TLR4, Figure 2. This is clearly a testable hypothesis with the performance of binding assays that would directly measure the affinity of Der p 2 for TLR4. This would also facilitate estimating dosages of Der p 2 that would be likely to influence sensitization by knowing how strongly Der p 2 could compete with endogenous MD-2 for TLR4 interactions.
Figure 2.
Murine TLR4:MD-2 interactions compared with possible Der p 2 interactions. Murine TLR4 (5IJD [37]) is shown in grey with a ribbon and/or semitransparent surface. Lipid A is rendered as spheres in panels C and D. In panels A and C MD2 is rendered in cyan with the exception of the labeled residues 98-HDDD-101 shown in blue and Y102 shown in chartreuse. In panels B and D, shown in orange, Der p 2 (1KTJ [31]) was aligned with MD-2. Note the HDDD interactions of MD-2 with TLR4 would be missing in a putative Der p 2:TLR4 complex.
Another incongruous point about Der p 2 mimicking MD-2 is that the Y91A mutant of Der p 2 was inactive compared to WT [33]. This mutant was designed based on two separate studies showing that the comparable Y102A mutant of MD-2 was inactive, and the observation that this tyrosine is strongly conserved in related species [39,41]. In the structure of TLR4:MD-2, the tyrosyl side chain points away from the TLR4 molecule, but appears to interact with a hydrophobic lipid chain of the LPS mimic suggesting it is involved in lipid recognition, Figure 2 [35,36]. The affinity of Der f 2 for LPS was originally proposed from modeling [42] and then directly measured to have nanomolar affinity [43]. The authors also astutely observed that in mites this residue varies; Phe (e.g. Der f 2), His, or Val are represented indicating that the tyrosyl sidechain is not required [43]. So the mutation to alanine, which is also hydrophobic, in an already large hydrophobic pocket seems unlikely to strongly affect LPS binding nor does it appear to interact with TLR4. Further experiments on the recognition of LPS by group 2 allergens would be informative. So in the same way that structural information motivated research into allergenic stimulation by Der p 2, it should continue to motivate more experimentation that may disprove, augment, or refine the existing hypothesis for auto-adjuvant activity of the group 2 allergens.
Immunotherapy
Allergen immunotherapy fundamentally involves treating a patient with increasing dosages of a compound to which the patient is hypersensitive. Clearly this involves some risk and to reduce this risk an improved treatment would reduce the symptomatic IgE response, while retaining enough molecular information about the allergen to modify the existing immune response towards an improvement in symptoms. Two strategies will be discussed that utilized structural information to design hypoallergens. One involves global disruption of allergen structure, while the other uses a targeted approach of mutation of key residues in immunodominant epitopes. Finally, the appearance of allergen specific IgG4 antibodies is generally considered a marker of successful immunotherapy [44]. A clever strategy to monitor patient IgG and IgE epitopes during therapy is discussed that was based on structural similarities of PR-10 proteins.
Because the polyclonal nature of the immune response could theoretically recognize the entire allergen surface, some of the techniques attempt to globally disrupt the structure to reduce IgE binding [45]. There are a few nice cases where the structure of the allergens was used to suggest specific mutations to ablate antibody binding by destabilizing the proteins. For example, the structure of the allergen Mus m 1 was used to design a destabilized allergen with reduced IgE binding while retaining T-cell reactivity [46]. In silico screening of point mutations of Bet v 1 and Phl p 5 demonstrated similar results and in addition immunization with the hypoallergenic mutants produced antibodies that blocked human IgE epitopes [47]. Another computer analysis suggested a chimera of Bet v 1 and Mal d 1 would disrupt the fold of Bet v 1, which did result in a shift of the immune polarization compared to the wild type Bet v 1 [48]. These destabilizing predictions appear promising and it is notable that all of these examples did not require specific information about the patient epitopes, just the allergen structure [45].
On the other hand, if one wants to design site directed mutants for immunotherapy, the major difficultly is determining which residues are the most important for patients and how many should be mutated. Thus this topic is inherently interrelated with strategies to map epitopes, which are discussed further in the section, Antibody-Allergen Interactions. Continuing the example of PR-10 proteins like Bet v 1 and Mal d 1 mentioned above, it was determined that 6 simultaneous site directed mutants of Bet v 1 were required to consistently reduce IgE reactivity in patients [49]. Using the structural similarity of Mal d 1 for Bet v 1, a 5-mutant version of Mal d 1 was designed and had similar success in reducing IgE binding [50]. Other strategies for theoretically designing hypoallergens from known B- and T-cell epitopes utilizing structures have also been discussed [51]. For more examples, Tscheppe and Breiteneder have reviewed recombinant allergens including a section on mutants designed for immunotherapy [52]. Despite many promising results, there are regulatory concerns and questions about the broad applicability that need to be addressed for designer hypoallergens to replace extract based immunotherapy [52].
The structural similarity of the PR-10 proteins can be exploited in a different way to monitor the change in patient IgG and IgE epitopes during immunotherapy using a technique call grafting. The idea is to create a structural scaffold which holds an epitope of interest, which can be used as a tool in various assays. As a demonstration, a conformational epitope of Bet v 1 was grafted onto Mal d 1, and it was shown that a monoclonal antibody, which didn’t previously interact with Mal d 1 would bind to the chimera [53]. This grafting required careful comparison of the two structures to selectively mutate a limited number of residues. Further, this demonstrated the capacity of the PR-10 protein fold to accept multiple point mutations from another species.
Extending the use of grafted chimeras, four different Bet v 1 epitopes were grafted on to the celery allergen, Api g 1 [54]. Then, by testing birch allergic patients that were not allergic to celery with all four chimeras, the relative importance of each Birch epitope could be determined. This revealed that the IgE response was highly patient specific and recognized epitopes spread across the entire surface of Bet v 1. As epitopes are generally conformational in nature, this is a very elegant alternative to the use of peptides for epitope mapping because the structural context of the epitopes is maintained. Finally, the grafted epitopes were used to monitor epitope recognition by different antibody isotypes during immunotherapy in eleven patients [55]. It was found that the IgE, IgG1, and IgG4 recognized different epitope profiles. In summary, not only have the structures help design hypoallergens for immunotherapy, this new chimeric epitope mapping technique is illuminating changes in the immune response during immunotherapy.
Cross-reactivity
In the clinic, the cross-reactivity of various allergens from different sources complicates clinical diagnosis of the sensitizing allergen. The cross-reactivity or co-recognition is due to antibodies recognizing similar structures on the different allergens. And indeed, many retrospective analyses of structural similarities correlate well with measures of cross-reactivity. Studies of Bet v 1 in complex with an Fab fragment identified a dominant epitope, which is also the site of cross-reactivity for homologous allergens [56,57]. An in depth analysis of the structure of a murine monoclonal in complex with Der p 1 and Der f 1 showed how this antibody could interact with both proteins at a site known to be involved in IgE binding [58]. Models of homologous proteins can also aid in understanding cross-reactivity when the actual allergen structures are not available [59]. Allergen structural models can be found at the Structural Database of Proteins [60], or using available homology modeling software as was done in the study of Fel d 7 and Can f 1 cross-reactivity [61]. In the other extreme, the lack of cross-reactivity could be explained from the differences in surface residues of various GST-allergens based on their crystal structures[62].
However, predicting cross-reactivity from structure is difficult and time consuming. Fortunately, measures of sequential homology are often adequate [45,63]. As a general rule sequential identity greater than 70% suggests cross-reactivity is likely and is rare at less than 50% sequential identity [64]. However, there are some exceptions where the sequence identity was very limited such as recently described cross-reactivity between the peanut allergens Ara h 2, and Ara h 1 and Ara h 3 [65]. The kiwi fruit allergen Act d 11 showed very low sequence identity to Bet v 1, but was demonstrated to be recognized by IgE of birch pollen allergic patients [66]. The sequence identity between these proteins is not greater that 21% over any sliding window of 80 residues, which is below the usual cutoff of 35% for potential allergens in food products [66]. From an examination of the limited homology and the structure of Bet v 1, the authors noticed that residues reported to belong to IgE epitopes are conserved in Act d 11 suggesting a structural reason for the co-recognition of Bet v 1 and Act d 11. Thus structural biology helped explain how a very distant sequence identity could generate cross-reactivity.
Antibody-Allergen Interactions
Antibodies have the potential to interact with any surface exposed element of a protein [67]. However, Aalberse and Crameri have presented a number of lines of evidence suggesting that IgE epitopes may be different from the epitopes of other isotypes [45]. Evidence for this includes limited analysis of the few known IgE epitopes that appear more planar than other known epitopes, e.g. [68]. The spatial clustering of the Phl p 2 epitopes was demonstrated by a single mAb which can block 80% of IgE epitopes [69]. This also suggested that IgE epitopes may preferentially recognize certain features of protein structures. Another study suggested that IgE antibodies are more cross-reactive than IgG4 antibodies [70]. Clearly structural biology can contribute to a better understanding of IgE epitopes and what differentiates them from other isotypes, but many technical hurdles exist.
Obtaining high resolution, three-dimensional structural information about patient epitopes has proven to be rather challenging. For evidence of this, there are approximately 100 allergen structures in the protein data bank, but there are less than 10 structures of different allergens in complex with antibodies [71]. The review of Dall’Antonia et al nicely presents high resolution techniques utilizing crystallography and NMR for mapping antigen-antibody epitopes, with particular attention to allergen complexes [72]. More complex structures will be needed to generalize about the repertoire of IgE epitopes. However, it seems rather implausible that high-resolution structures can be obtained for all the antibodies and all the allergens. Dealing with the heterogeneous nature or polyclonal response of the patient(s) is the key problem. However, by combining some high throughput low to medium resolution techniques with high resolution structures it may be possible to make strides on the questions mentioned above. Some concepts and ideas are outlined below.
A few attempts have been made to use polyclonal antibodies and high resolution NMR techniques to map patient epitopes. Two examples are the studies using Art v 1 [73] and Bet v 1 [74]. In the study of Art v 1, IgE specific for Art v 1 was purified from a patient pool, while in the study of Bet v 1, IgE from 3 different birch allergic patients were studied. Both studies looked for chemical shift changes in the amide resonances of the allergen upon addition of the antibodies, and both observed a rather unconvincing and small effect. However, in both experiments, the design of the study was likely at fault. The allergen was added in roughly 10:1 excess, possibly due the quantity of IgE available. This poses two sensitivity problems for NMR. First, the signal will be dominated by the free allergen, which is in excess. The second problem is that the amide signals from the backbone become weaker and broader with increasing molecular weight, or more specifically, with the slower overall tumbling rate of the molecule. So attaching an IgE molecule of 190 kDa to a small allergen would make the amide signals for the bound allergen nearly impossible to observe without special techniques. The two problems likely both contributed to the minor spectral changes observed.
However, the concept has significant merit. While backbone amides can provide perhaps the best molecular coverage in terms of information content, methyl groups are much less sensitive to the loss in sensitivity at high molecular weights, and have been studied in very large molecular complexes [75,76]. Using 13C labeled methyl groups in a background of 2H labeling [77] should overcome the sensitivity problems of the previous study and the allergen should be added in equal proportions to the antibodies. This could provide atom specific information for epitope mapping. In light of the polyclonal nature of patient antibodies, care must be taken to interpret the results. In a simple example of one antibody interacting with an allergen, any shift in the NMR signal from atoms in the allergen compared to the apo state can be cautiously interpreted as reflecting a conformational change in the vicinity of that atom due to complex formation. The disappearance of the old signal and the emergence of the new signal would provide epitope information. If many antibodies bind at once, the situation could become highly complex with many new atomic frequencies possible, and many at low population, i.e. low intensity. But most likely, if the atomic frequencies do not change upon complex formation those atoms can be ruled out as likely epitope locations. By comparing the signals remaining versus the signals that disappear it may be possible to globally map the relative importance of epitope locations in a complex mixture of antibodies interacting with an allergen.
Another emerging structural technique that could contribute to IgE epitope mapping is cryo-electron microscopy (Cryo-EM) that is experiencing a major renaissance due largely to the improvement in detectors [78]. The resolution of some structures rivals that of traditional X-ray crystallography, although it is not clear if this will generally be the case [79]. Much of the current work utilizes a hybrid approach where atomic resolution structures are modeled into the Cryo-EM data. Cryo-EM has the advantage over crystallography that proteins do not need to be crystallized; they are instead flash frozen into vitrified ice. Another advantage of Cryo-EM is the ability to work with much larger biomolecular complexes, like the ribosome [80]. In addition, much less sample is required and at lower concentrations than is typically required for crystallography. However, the lower limit of complexes that can be studied routinely at high resolution with this technique is about 200 kDa and the biomolecules must be ‘well-ordered’ [81]. There is, therefore, an opportunity to directly examine the structure of an IgE molecule of 190 kDa in complex with an allergen.
The conceptual framework would be to isolate IgE from either a patient or mix of many patients and complex it with an allergen. A number of technical challenges need to be addressed. Just to name two: First, is the IgE structure ‘well-ordered’ enough for high resolution work or will it need to be restrained in some way? Second, how well can Cryo-EM handle different orientations of allergen binding to IgE? Presumably in this mixture and at Cryo-EM resolution the IgE molecules will all be similar, but the allergen will be turned in different directions in the images. Previously, 8 different substructures of the ribosome in its catalytic cycle could be resolved during Cryo-EM image processing [80]. These substructures could ultimately be assigned a population value. In the same way, the orientations of the allergen bound to the IgE could be sorted to evaluate which epitopes were more highly populated. Most likely, high resolution structures of the allergens (either NMR or X-ray) will be needed to model the orientation of the allergens into the Cryo-EM maps of the complex.
The methods presented above reflect opportunities to address the hypotheses that IgE epitopes cluster, or are directed to certain features of allergens [45]. Both NMR and/or Cryo-EM methodologies appear to have the potential to address the heterogeneous nature of the patient response. This could lead to a better characterization of the nature of IgE response and new treatment modalities.
Conclusions
The goals of allergy research are to improve the health of allergic patients and to recommend prophylactic interventions for future generations. The study of allergens has been crucial for setting standards for exposure measurements, diagnostic assays, and treatment formulations, which all contribute to these goals. The various techniques of structural biology have contributed in various ways to setting standards, informing hypotheses for sensitization mechanisms, and the interaction of IgE with allergens that also contributed to these goals. In the future there are still many ways for structural biology to contribute, using a variety of different techniques.
Acknowledgments
The author wishes to thank Dr. Thomas Randall for probing the Blatella germanica genome to help make Figure 1. The author thanks Dr. Lars Pedersen and Dr. Jessica Wojtaszek for a critical review of the manuscript. This study was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences (Z01- ES102906-01).
Footnotes
Conflicts of Interest
The author affirms there are no conflicts of interest to report.
References
- 1.Radauer C. Navigating through the Jungle of Allergens: Features and Applications of Allergen Databases. Int Arch Allergy Immunol. 2017;173:1–11. doi: 10.1159/000471806. [DOI] [PubMed] [Google Scholar]
- 2.Filep S, Tsay A, Vailes LD, Gadermaier G, Ferreira F, Matsui E, King EM, Chapman MD. Specific allergen concentration of WHO and FDA reference preparations measured using a multiple allergen standard. J Allergy Clin Immun. 2012;129:1408–1410. doi: 10.1016/j.jaci.2011.12.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filep S, Tsay A, Vailes L, Gadermaier G, Ferreira F, Matsui E, King EM, Chapman MD. A multi-allergen standard for the calibration of immunoassays: CREATE principles applied to eight purified allergens. Allergy. 2012;67:235–241. doi: 10.1111/j.1398-9995.2011.02750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Ree R, Chapman MD, Ferreira F, Vieths S, Bryan D, Cromwell O, Villalba M, Durham SR, Becker WM, Aalbers M, Andre C, Barber D, Bahima AC, Custovic A, Didierlaurent A, Dolman C, Dorpema JW, Di Felice G, Eberhardt F, Fernandez Caldas E, Fernandez Rivas M, Fiebig H, Focke M, Fotisch K, Gadermaier G, Das RG, Gonzalez Mancebo E, Himly M, Kinaciyan T, Knulst AC, Kroon AM, Lepp U, Marco FM, Mari A, Moingeon P, Monsalve R, Neubauer A, Notten S, Heer PO, Pauli G, Pini C, Purohit A, Quiralte J, Rak S, Raulf-Heimsoth M, SanMiguel Moncin MM, Simpson B, Tsay A, Vailes L, Wallner M, Weber B. The CREATE Project: development of certified reference materials for allergenic products and validation of methods for their quantification. Allergy. 2008;63:310–326. doi: 10.1111/j.1398-9995.2007.01612.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaul S, Zimmer J, Dehus O, Costanzo A, Daas A, Buchheit KH, Asturias JA, Barber D, Carnes J, Chapman M, Dayan-Kenigsberg J, Doring S, Fuhrer F, Hanschmann KM, Holzhauser T, Ledesma A, Moingeon P, Nony E, Pini C, Plunkett G, Reese G, Sandberg E, Sander I, Strecker D, Valerio C, van Ree R, Vieths S. Standardization of allergen products: 3. Validation of candidate European Pharmacopoeia standard methods for quantification of major birch allergen Bet v 1. Allergy. 2016;71:1414–1424. doi: 10.1111/all.12898. [DOI] [PubMed] [Google Scholar]
- 6.Himly M, Nandy A, Kahlert H, Thilker M, Steiner M, Briza P, Neubauer A, Klysner S, van Ree R, Buchheit KH, Vieths S, Ferreira F. Standardization of allergen products: 2. Detailed characterization of GMP-produced recombinant Phl p 5. 0109 as European Pharmacopoeia reference standard. Allergy. 2016;71:495–504. doi: 10.1111/all.12824. [DOI] [PubMed] [Google Scholar]
- 7.Randall TA, Perera L, London RE, Mueller GA. Genomic, RNAseq, and molecular modeling evidence suggests that the Major Allergen domain in Insects evolved from a homodimeric origin. Genome Biol Evol. 2013 doi: 10.1093/gbe/evt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomes A, Melen E, Vailes LD, Retief JD, Arruda LK, Chapman MD. Novel allergen structures with tandem amino acid repeats derived from German and American cockroach. Journal of Biological Chemistry. 1998;273:30801–30807. doi: 10.1074/jbc.273.46.30801. [DOI] [PubMed] [Google Scholar]
- 9.Fischer HM, Wheat CW, Heckel DG, Vogel H. Evolutionary origins of a novel host plant detoxification gene in butterflies. Mol Biol Evol. 2008;25:809–820. doi: 10.1093/molbev/msn014. [DOI] [PubMed] [Google Scholar]
- 10.Diraphat P, Sookrung N, Chaicumpa W, Pumhirun P, Vichyanond P, Tapchaisri P, Kalambaheti T, Mahakunkijchareon Y, Sakolvaree Y, Bunnag C. Recombinant American cockroach component, per a 1, reactive to IgE of allergic Thai patients. Asian Pac J Allergy. 2003;21:11–20. [PubMed] [Google Scholar]
- 11.Mueller GA, Pedersen LC, Lih FB, Glesner J, Moon AF, Chapman MD, Tomer KB, London RE, Pomes A. Novel structure of cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. J Allergy Clin Immun. 2013;132:1420–1426. doi: 10.1016/j.jaci.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomes A, Vailes LD, Helm RM, Chapman MD. IgE reactivity of tandem repeats derived from cockroach allergen, Bla g 1. European Journal of Biochemistry. 2002;269:3086–3092. doi: 10.1046/j.1432-1033.2002.02990.x. [DOI] [PubMed] [Google Scholar]
- 13.Helm R, Cockrell G, Stanley JS, Brenner RJ, Burks W, Bannon GA. Isolation and characterization of a clone encoding a major allergen (Bla g Bd90K) involved in IgE-mediated cockroach hypersensitivity. J Allergy Clin Immun. 1996;98:172–180. doi: 10.1016/s0091-6749(96)70240-3. [DOI] [PubMed] [Google Scholar]
- 14.Helm RM, Bandele EO, Swanson MC, Campbell AR, Wynn SR. Identification of a German Cockroach-Specific Allergen by Human Ige and Rabbit Igg. Int Arch Aller a Imm. 1988;87:230–238. doi: 10.1159/000234678. [DOI] [PubMed] [Google Scholar]
- 15.Wang NM, Lee MF, Wu CH. Immunologic characterization of a recombinant American cockroach (Periplaneta americana) Per a 1 (Cr-PII) allergen. Allergy. 1999;54:119–127. doi: 10.1034/j.1398-9995.1999.00902.x. [DOI] [PubMed] [Google Scholar]
- 16.Chua KY, Stewart GA, Thomas WR, Simpson RJ, Dilworth RJ, Plozza TM, Turner KJ. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. J Exp Med. 1988;167:175–182. doi: 10.1084/jem.167.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deb R, Shakib F, Reid K, Clark H. Major house dust mite allergens Dermatophagoides pteronyssinus 1 and Dermatophagoides farinae 1 degrade and inactivate lung surfactant proteins A and D. Journal of Biological Chemistry. 2007;282:36808–36819. doi: 10.1074/jbc.M702336200. [DOI] [PubMed] [Google Scholar]
- 18.Lewkowich IP, Day SB, Ledford JR, Zhou P, Dienger K, Wills-Karp M, Page K. Protease-activated receptor 2 activation of myeloid dendritic cells regulates allergic airway inflammation. Resp Res. 2011:12. doi: 10.1186/1465-9921-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gough L, Schulz O, Sewell HF, Shakib F. The cysteine protease activity of the major dust mite allergen Der p 1 selectively enhances the immunoglobulin E antibody response. Journal of Experimental Medicine. 1999;190:1897–1901. doi: 10.1084/jem.190.12.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghaemmaghami AM, Shakib F. Human T cells that have been conditioned by the proteolytic activity of the major dust mite allergen Der p 1 trigger enhanced immunoglobulin E synthesis by B cells. Clinical and Experimental Allergy. 2002;32:728–732. doi: 10.1046/j.1365-2222.2002.01374.x. [DOI] [PubMed] [Google Scholar]
- 21.Pomes A, Chapman MD, Vailes LD, Blundell TL, Dhanaraj V. Cockroach allergen Bla g 2 - Structure, function, and implications for allergic sensitization. Am J Resp Crit Care. 2002;165:391–397. doi: 10.1164/ajrccm.165.3.2104027. [DOI] [PubMed] [Google Scholar]
- 22.Sudha VT, Arora N, Gaur SN, Pasha S, Singh BP. Identification of a serine protease as a major allergen (Per a 10) of Periplaneta americana. Allergy. 2008;63:768–776. doi: 10.1111/j.1398-9995.2007.01602.x. [DOI] [PubMed] [Google Scholar]
- 23.Shen HD, Tam MF, Chou H, Han SH. The importance of serine proteinases as aeroallergens associated with asthma. Int Arch Allergy Immunol. 1999;119:259–264. doi: 10.1159/000024202. [DOI] [PubMed] [Google Scholar]
- 24.Bouley J, Groeme R, Le Mignon M, Jain K, Chabre H, Bordas-Le Floch V, Couret MN, Bussieres L, Lautrette A, Naveau M, Baron-Bodo V, Lombardi V, Mascarell L, Batard T, Nony E, Moingeon P. Identification of the cysteine protease Amb a 11 as a novel major allergen from short ragweed. J Allergy Clin Immunol. 2015;136:1055–1064. doi: 10.1016/j.jaci.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Pastorello EA, Conti A, Pravettoni V, Farioli L, Rivolta F, Ansaloni R, Ispano M, Incorvaia C, Giuffrida MG, Ortolani C. Identification of actinidin as the major allergen of kiwi fruit. J Allergy Clin Immunol. 1998;101:531–537. doi: 10.1016/S0091-6749(98)70360-4. [DOI] [PubMed] [Google Scholar]
- 26.Thomas WR, Hales BJ, Smith WA. Structural biology of allergens. Curr Allergy Asthm R. 2005;5:388–393. doi: 10.1007/s11882-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 27.Bublin M, Eiwegger T, Breiteneder H. Do lipids influence the allergic sensitization process? J Allergy Clin Immun. 2014;134:521–529. doi: 10.1016/j.jaci.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller GA, Smith AM, Williams DC, Hakkaart GAJ, Aalberse RC, Chapman MD, Rule GS, Benjamin DC. Expression and secondary structure determination by NMR methods of the major house dust mite allergen Der p 2. Journal of Biological Chemistry. 1997;272:26893–26898. doi: 10.1074/jbc.272.43.26893. [DOI] [PubMed] [Google Scholar]
- 29.Mueller GA, Benjamin DC, Rule GS. Tertiary structure of the major house dust mite allergen Der p 2: Sequential and structural homologies. Biochemistry. 1998;37:12707–12714. doi: 10.1021/bi980578+. [DOI] [PubMed] [Google Scholar]
- 30.Ichikawa S, Hatanaka H, Yuuki T, Iwamoto N, Kojima S, Nishiyama C, Ogura K, Okumura Y, Inagaki F. Solution structure of Der f 2, the major mite allergen for atopic diseases. Journal of Biological Chemistry. 1998;273:356–360. doi: 10.1074/jbc.273.1.356. [DOI] [PubMed] [Google Scholar]
- 31.Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller GA, Rule GS, Benjamin DC. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. Journal of Molecular Biology. 2002;318:189–197. doi: 10.1016/S0022-2836(02)00027-X. [DOI] [PubMed] [Google Scholar]
- 32.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 33.Trompette A, Divanovic S, Visintin A, Blanchard C, Hegde RS, Madan R, Thorne PS, Wills-Karp M, Gioannini TL, Weiss JP, Karp CL. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 2009;457:585–U591. doi: 10.1038/nature07548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J, Kim HJ, Lee SC, Hong SP, Park KW, Jeon YH, Kim DS, Cheong HK, Kim HS. Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein. Plos One. 2012:7. doi: 10.1371/journal.pone.0030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohto U, Yamakawa N, Akashi-Takamura S, Miyake K, Shimizu T. Structural Analyses of Human Toll-like Receptor 4 Polymorphisms D299G and T399I. Journal of Biological Chemistry. 2012;287:40611–40617. doi: 10.1074/jbc.M112.404608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Su L, Morin MD, Jones BT, Whitby LR, Surakattula MM, Huang H, Shi H, Choi JH, Wang KW, Moresco EM, Berger M, Zhan X, Zhang H, Boger DL, Beutler B. TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS. Proc Natl Acad Sci U S A. 2016;113:E884–893. doi: 10.1073/pnas.1525639113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slivka PF, Shridhar M, Lee GI, Sammond DW, Hutchinson MR, Martinko AJ, Buchanan MM, Sholar PW, Kearney JJ, Harrison JA, Watkins LR, Yin H. A Peptide Antagonist of the TLR4-MD2 Interaction. Chembiochem. 2009;10:645–649. doi: 10.1002/cbic.200800769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Re F, Strominger JL. Separate functional domains of human MD-2 mediate toll-like receptor 4-binding and lipopolysaccharide responsiveness. J Immunol. 2003;171:5272–5276. doi: 10.4049/jimmunol.171.10.5272. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki K, Nogawa H, Nishijima M. Identification of mouse MD-2 residues important for forming the cell surface TLR4-MD-2 complex recognized by anti-TLR4-MD-2 antibodies, and for conferring LPS and taxol responsiveness on mouse TLR4 by alanine-scanning mutagenesis. J Immunol. 2003;170:413–420. doi: 10.4049/jimmunol.170.1.413. [DOI] [PubMed] [Google Scholar]
- 42.Ichikawa S, Takai T, Inoue T, Yuuki T, Okumura Y, Ogura K, Inagaki F, Hatanaka H. NMR study on the major mite allergen Der f 2: Its refined tertiary structure, epitopes for monoclonal antibodies and characteristics shared by ML protein group members. Journal of biochemistry. 2005;137:255–263. doi: 10.1093/jb/mvi039. [DOI] [PubMed] [Google Scholar]
- 43.Ichikawa S, Takai T, Yashiki T, Takahashi S, Okumura K, Ogawa H, Kohda D, Hatanaka H. Lipopolysaccharide binding of the mite allergen Der f 2. Genes Cells. 2009;14:1055–1065. doi: 10.1111/j.1365-2443.2009.01334.x. [DOI] [PubMed] [Google Scholar]
- 44.Kouser L, Kappen J, Walton RP, Shamji MH. Update on Biomarkers to Monitor Clinical Efficacy Response During and Post Treatment in Allergen Immunotherapy. Curr Treat Options Allergy. 2017;4:43–53. doi: 10.1007/s40521-017-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aalberse RC, Crameri R. IgE-binding epitopes: a reappraisal. Allergy. 2011;66:1261–1274. doi: 10.1111/j.1398-9995.2011.02656.x. [DOI] [PubMed] [Google Scholar]
- 46.Ferrari E, Breda D, Longhi R, Vangelista L, Nakaie CR, Elviri L, Casali E, Pertinhez TA, Spisni A, Burastero SE. In Search of a Vaccine for Mouse Allergy: Significant Reduction of Mus m 1 Allergenicity by Structure-Guided Single-Point Mutations. Int Arch Allergy Imm. 2012;157:226–237. doi: 10.1159/000327551. [DOI] [PubMed] [Google Scholar]
- 47.Thalhamer T, Dobias H, Stepanoska T, Proll M, Stutz H, Dissertori O, Lackner P, Ferreira F, Wallner M, Thalhamer J, Hartl A. Designing hypoallergenic derivatives for allergy treatment by means of in silico mutation and screening. J Allergy Clin Immun. 2010;125:926–934. doi: 10.1016/j.jaci.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 48.Wallner M, Hauser M, Himly M, Zaborsky N, Mutschlechner S, Harrer A, Asam C, Pichler U, van Ree R, Briza P, Thalhamer J, Bohle B, Achatz G, Ferreira F. Reshaping the Bet v 1 fold modulates T-H polarization. J Allergy Clin Immun. 2011;127:1571–U1392. doi: 10.1016/j.jaci.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferreira F, Ebner C, Kramer B, Casari G, Briza P, Kungl AJ, Grimm R, Jahn-Schmid B, Breiteneder H, Kraft D, Breitenbach M, Rheinberger HJ, Scheiner O. Modulation of IgE reactivity of allergens by site-directed mutagenesis: potential use of hypoallergenic variants for immunotherapy. Faseb J. 1998;12:231–242. doi: 10.1096/fasebj.12.2.231. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y, Gadermaier G, Bohle B, Bolhaar S, Knulst A, Markovic-Housley Z, Breiteneder H, Briza P, Hoffmann-Sommergruber K, Ferreira F. Mutational analysis of amino acid positions crucial for IgE-binding epitopes of the major apple (Malus domestica) allergen, Mal d 1. Int Arch Allergy Immunol. 2006;139:53–62. doi: 10.1159/000089756. [DOI] [PubMed] [Google Scholar]
- 51.Vaughan K, Peters B, Larche M, Pomes A, Broide D, Sette A. Strategies to Query and Display Allergy-Derived Epitope Data from the Immune Epitope Database. Int Arch Allergy Imm. 2013;160:334–345. doi: 10.1159/000343880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tscheppe A, Breiteneder H. Recombinant Allergens in Structural Biology, Diagnosis, and Immunotherapy. Int Arch Allergy Imm. 2017;172:187–202. doi: 10.1159/000464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm J, Ferreras M, Ipsen H, Wurtzen PA, Gajhede M, Larsen JN, Lund K, Spangfort MD. Epitope grafting, re-creating a conformational Bet v 1 antibody epitope on the surface of the homologous apple allergen Mal d 1. J Biol Chem. 2011;286:17569–17578. doi: 10.1074/jbc.M110.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gepp B, Lengger N, Bublin M, Hemmer W, Breiteneder H, Radauer C. Chimeras of Bet v 1 and Api g 1 reveal heterogeneous IgE responses in patients with birch pollen allergy. J Allergy Clin Immun. 2014;134:188–194. doi: 10.1016/j.jaci.2013.12.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gepp B, Lengger N, Mobs C, Pfutzner W, Radauer C, Bohle B, Breiteneder H. Monitoring the epitope recognition profiles of IgE, IgG(1), and IgG(4) during birch pollen immunotherapy. J Allergy Clin Immun. 2016;137:1600–1603. doi: 10.1016/j.jaci.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 56.Spangfort MD, Mirza O, Ipsen H, van Neerven RJJ, Gajhede M, Larsen JN. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J Immunol. 2003;171:3084–3090. doi: 10.4049/jimmunol.171.6.3084. [DOI] [PubMed] [Google Scholar]
- 57.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, Gajhede M. Dominant epitopes and allergic cross-reactivity: Complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol. 2000;165:331–338. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 58.Chruszcz M, Pomes A, Glesner J, Vailes LD, Osinski T, Porebski PJ, Majorek KA, Heymann PW, Platts-Mills TAE, Minor W, Chapman MD. Molecular Determinants for Antibody Binding on Group 1 House Dust Mite Allergens. Journal of Biological Chemistry. 2012;287:7388–7398. doi: 10.1074/jbc.M111.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Power TD, Ivanciuc O, Schein CH, Braun W. Assessment of 3D models for allergen research. Proteins-Structure Function and Bioinformatics. 2013;81:545–554. doi: 10.1002/prot.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivanciuc O, Schein CH, Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Research. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apostolovic D, Sanchez-Vidaurre S, Waden K, Curin M, Grundstrom J, Gafvelin G, Velickovic TC, Gronlund H, Thomas WR, Valenta R, Hamsten C, van Hage M. The cat lipocalin Fel d 7 and its cross-reactivity with the dog lipocalin Can f 1. Allergy. 2016;71:1490–1495. doi: 10.1111/all.12955. [DOI] [PubMed] [Google Scholar]
- 62.Mueller GA, Pedersen LC, Glesner J, Edwards LL, Zakzuk J, London RE, Arruda LK, Chapman MD, Caraballo L, Pomes A. Analysis of glutathione S-transferase allergen cross-reactivity in a North American population: Relevance for molecular diagnosis. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aalberse RC, Stadler BM. In silico predictability of allergenicity: From amino acid sequence via 3-D structure to allergenicity. Mol Nutr Food Res. 2006;50:625–627. doi: 10.1002/mnfr.200500270. [DOI] [PubMed] [Google Scholar]
- 64.Aalberse RC. Structural features of allergenic molecules. Chem Immunol Allergy. 2006;91:134–146. doi: 10.1159/000090277. [DOI] [PubMed] [Google Scholar]
- 65.Bublin M, Kostadinova M, Radauer C, Hafner C, Szepfalusi Z, Varga EM, Maleki SJ, Hoffmann-Sommergruber K, Breiteneder H. IgE cross-reactivity between the major peanut allergen Ara h 2 and the nonhomologous allergens Ara h 1 and Ara h 3. J Allergy Clin Immun. 2013;132:118–U212. doi: 10.1016/j.jaci.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 66.D’Avino R, Bernardi ML, Wallner M, Palazzo P, Camardella L, Tuppo L, Alessandri C, Breiteneder H, Ferreira F, Ciardiello MA, Mari A. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy. 2011;66:870–877. doi: 10.1111/j.1398-9995.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- 67.Benjamin DC, Berzofsky JA, East IJ, Gurd FR, Hannum C, Leach SJ, Margoliash E, Michael JG, Miller A, Prager EM, et al. The antigenic structure of proteins: a reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- 68.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM, Hakulinen N, Haahtela T, Takkinen K, Rouvinen J. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–1421. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 69.Flicker S, Steinberger P, Norderhaug L, Sperr WR, Majlesi Y, Valent P, Kraft D, Valenta R. Conversion of grass pollen allergen-specific human IgE into a protective IgG(1) antibody. Eur J Immunol. 2002;32:2156–2162. doi: 10.1002/1521-4141(200208)32:8<2156::AID-IMMU2156>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 70.Lin J, Bardina L, Shreffler WG, Andreae DA, Ge YC, Wang JL, Bruni FM, Fu ZY, Han YS, Sampson HA. Development of a novel peptide microarray for large-scale epitope mapping of food allergens. J Allergy Clin Immun. 2009;124:315–322. doi: 10.1016/j.jaci.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pomes A, Chruszcz M, Gustchina A, Minor W, Mueller GA, Pedersen LC, Wlodawer A, Chapman MD. 100 Years later: Celebrating the contributions of x-ray crystallography to allergy and clinical immunology. J Allergy Clin Immun. 2015;136:29–U87. doi: 10.1016/j.jaci.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dall’Antonia F, Pavkov-Keller T, Zangger K, Keller W. Structure of allergens and structure based epitope predictions. Methods. 2014;66:3–21. doi: 10.1016/j.ymeth.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Razzera G, Gadermaier G, de Paula V, Almeida MS, Egger M, Jahn-Schmid B, Almeida FCL, Ferreira F, Valente AP. Mapping the Interactions between a Major Pollen Allergen and Human IgE Antibodies. Structure. 2010;18:1011–1021. doi: 10.1016/j.str.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 74.Asam C, Batista AL, Moraes AH, de Paula VS, Almeida FCL, Aglas L, Kitzmuller C, Bohle B, Ebner C, Ferreira F, Wallner M, Valente AP. Bet v 1-a Trojan horse for small ligands boosting allergic sensitization? Clinical and Experimental Allergy. 2014;44:1083–1093. doi: 10.1111/cea.12361. [DOI] [PubMed] [Google Scholar]
- 75.Sprangers R, Velyvis A, Kay LE. Solution NMR of supramolecular complexes: providing new insights into function. Nat Methods. 2007;4:697–703. doi: 10.1038/nmeth1080. [DOI] [PubMed] [Google Scholar]
- 76.Zheng XH, Perera L, Mueller GA, DeRose EF, London RE. Asymmetric conformational maturation of HIV-1 reverse transcriptase. Elife. 2015:4. doi: 10.7554/eLife.06359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. Journal of Biomolecular Nmr. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 78.Egelman EH. The Current Revolution in Cryo-EM. Biophys J. 2016;110:1008–1012. doi: 10.1016/j.bpj.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramaniam S, Earl LA, Falconieri V, Milne JLS, Egelman EH. Resolution advances in cryo-EM enable application to drug discovery. Current Opinion in Structural Biology. 2016;41:194–202. doi: 10.1016/j.sbi.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Burger J, Mielke T, Scheerer P, Spahn CMT. Structural Snapshots of Actively Translating Human Ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, Subramaniam S. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]