Abstract
Growing evidence from both epidemiology and basic science suggest an inverse association between Alzheimer’s disease (AD) and cancer. We examined the genetic relationship between AD and various cancer types using GWAS summary statistics from the IGAP and GAME-ON consortia. Sample size ranged from 9,931 to 54,162; SNPs were imputed to the 1000 Genomes European panel. Our results based on cross-trait LD Score regression showed a significant positive genetic correlation between AD and five cancers combined (colon, breast, prostate, ovarian, lung; rg = 0.17, P = 0.04), and specifically with breast cancer (ER-negative and overall; rg = 0.21 and 0.18, P = 0.035 and 0.034) and lung cancer (adenocarcinoma, squamous cell carcinoma and overall; rg = 0.31, 0.38 and 0.30, P = 0.029, 0.016, and 0.006). Estimating the genetic correlation in specific functional categories revealed mixed positive and negative signals, notably stronger at annotations associated with increased enhancer activity. This suggests a role of gene expression regulators in the shared genetic etiology between AD and cancer, and that some shared variants modulate disease risk concordantly while others have effects in opposite directions. Due to power issues, we did not detect cross-phenotype associations at individual SNPs. This genetic overlap is not likely driven by a handful of major loci. Our study is the first to examine the co-heritability of AD and cancer leveraging large-scale GWAS results. The functional categories highlighted in this study need further investigation to illustrate the details of the genetic sharing and to bridge between different levels of associations.
Keywords: Alzheimer’s disease, cancer, GWAS, cross-trait genetic correlation, polygenic inheritance
Introduction
Alzheimer’s disease (AD) and cancer are complex diseases of aging that impose an enormous public health burden worldwide (Brookmeyer et al. 2007; Lopez et al. 2006; Thun et al. 2010). There is a growing understanding that these seemingly disparate conditions have substantial overlap. The pathophysiology of AD includes most if not all of the hallmarks of cancer, including abnormal cell cycle entry, metabolic deregulation, oxidative stress, DNA damage, inflammation, and angiogenesis (Hanahan and Weinberg 2011). All of these similarities suggest the diseases would be comorbid, but the weight of epidemiological evidence points to an unusual inverse association (Catala-Lopez et al.; Driver et al. 2012; Musicco et al. 2013; Realmuto et al. 2012; Roe et al. 2005; Roe et al. 2010).
While it is difficult to know for sure that this “inverse comorbidity” represents a true association and is not the result of survival bias, there is convincing biological evidence for it. A transcriptomic meta-analyses using gene-expression data from relevant tissues found a substantial number of shared genes and their corresponding pathways to be upregulated in AD but downregulated in lung, colorectal and prostate cancers, and vice versa (Ibanez et al. 2014). Differential expression of microRNAs between cancer and Alzheimer’s disease has also been demonstrated (Holohan et al. 2012). A number of shared proteins and pathways have been identified that are differentially regulated by cancer cells and degenerating neurons. This includes the enzyme Pin, which is overexpressed in most cancers but depleted in AD (Bao et al. 2004); tumor suppressor p53, which promotes apoptosis but protects against cancer (van Heemst et al. 2005); and the Wnt cell survival pathway, which is activated in cancer but downregulated in AD (Inestrosa and Toledo 2008). Genetics play an important role in these underlying biological pathways, and therefore is expected to contribute to the inverse relationship between the two disorders either additively or through interaction with external factors (Demetrius and Simon 2013; Ibanez et al. 2014; Tabares-Seisdedos and Rubenstein 2013).
However, beyond these three long-suspected but yet-to-be-confirmed candidates (Pin1, p53 and Wnt), very little is known about the genetic overlap between AD and cancer. Using genome-wide association study (GWAS) individual level data or summary statistics, one might be able to identify significant single nucleotide polymorphisms (SNPs) common to both disorders and estimate the cross-trait heritability. Existing methods based on genotype data, such as bivariate restricted maximum likelihood estimation (REML) as implemented in Genome-wide Complex Trait Analysis (GCTA) (Lee et al. 2012; Yang et al. 2011) and genetic risk score profiling (Cross-Disorder Group of the Psychiatric Genomics 2013; Purcell et al. 2009), have been applied to a number of traits for estimating genetic correlations. Another approach is LD Score regression of summary statistics, as was recently applied to 24 traits to assess their pairwise genetic correlations (Bulik-Sullivan et al. 2015a). Patterns of genetic overlap among 42 traits were also examined using a Bayesian approach (Pickrell et al. 2016). No study has yet reported the genetic correlation between cancer and AD.
In the present study, we investigated the genetic overlap between AD and a variety of cancer types using SNP-trait GWAS summary statistics. We first estimated the genome-wide genetic correlation between the two diseases, then evaluated sharing heritability in specific functional categories, and finally tested cross-disease associations at individual SNPs. We used AD GWAS meta-analysis summary-level data acquired from the International Genomics of Alzheimer’s Project (IGAP) and nine cancer GWAS meta-analysis results from the Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium. There were 54,162 individuals included in the IGAP dataset and a sample size ranging from 9,931 to 33,832 among the GAME-ON datasets. All were imputed with over 7 million SNPs from the 1000 Genomes Project. This to our knowledge is the first study to investigate the genetic overlap between AD and specific cancer types using large-scale GWAS summary results where no individual genotype data is required.
Materials and Methods
Data: GWAS summary statistics for AD and each cancer type
Summary statistics of association analysis for late-onset AD were obtained from the International Genomics of Alzheimer’s Project (IGAP; (Lambert et al. 2013)), available upon request. International Genomics of Alzheimer’s Project (IGAP) is a large two-stage study based upon genome-wide association studies (GWAS) on individuals of European ancestry. In stage 1, IGAP used genotyped and imputed data on 7,055,881 single nucleotide polymorphisms (SNPs) to meta-analyse four previously-published GWAS datasets consisting of 17,008 Alzheimer’s disease cases and 37,154 controls (The European Alzheimer’s disease Initiative – EADI, the Alzheimer Disease Genetics Consortium – ADGC, The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium – CHARGE, The Genetic and Environmental Risk in AD consortium – GERAD; Table 1). European population reference haplotype data in the 1000 Genomes Project (2010 release) was used for genotype imputation, and genomic control correction was applied to each study before meta-analysis. In stage 2, 11,632 SNPs were genotyped and tested for association in an independent set of 8,572 Alzheimer’s disease cases and 11,312 controls. Finally, a meta-analysis was performed combining results from stages 1 & 2.
Table 1.
Summary of Cancer (from GAME-ON) & AD (from IGAP) GWAS meta-analysis data
| Dataset | #SNPs | #Studya | #Casea | #Controla | #Total samples | Imputation QC |
|---|---|---|---|---|---|---|
| GAME-ON GWAS | ||||||
| All Colon | 8,840,515 | 6 | 5,100 | 4,831 | 9,931 | info ≥ 0.7; certainty≥0.9; concordance≥0.9 |
| Breast ER-negative | 10,988,257 | 8 | 4,939 | 13,128 | 18,067 | r2 >0.3 |
| Breast (overall) | 11,099,926 | 11 | 15,748 | 18,084 | 33,832 | |
| Prostate aggressive | 9,671,146 | 6 | 4,450 | 12,724 | 17,174 | r2 >0.3 |
| Prostate (overall) | 9,760,825 | 6 | 14,160 | 12,724 | 26,884 | |
| Ovarian (overall) | 15,344,587 | 4 | 4,369 | 9,123 | 13,492 | r2 >0.25 |
| Lung adenocarcinoma | 8,897,683 | 6 | 3,718 | 15,871 | 19,589 | |
| Lung squamous cell carcinoma | 8,909,656 | 6 | 3,422 | 16,015 | 19,437 | r2 ≥ 0.3 or info ≥ 0.4 |
| Lung (overall) | 8,945,892 | 6 | 12,160 | 16,838 | 28,998 | |
|
| ||||||
| IGAP GWAS | ||||||
| AD (stage1) | 7,055,881 | 4 | 17,008 | 37,154 | 54,162 | r2 ≥ 0.3 or info ≥ 0.3 |
Max. number; may differ by SNP
Summary statistics for cancers were acquired from the Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium, which included meta-analysis results for nine cancer types (colon cancer, ER-negative breast cancer, overall breast cancer, aggressive prostate cancer, overall prostate cancer, ovarian cancer, lung adenocarcinoma, lung squamous cell carcinoma, and overall lung cancer (Table 1). Study designs included population-based, hospital-based, or family-based case-control studies. SNPs in individual studies were genotyped using Affymetrix or Illumina platform, and SNP imputation was performed using IMPUTE2, MiniMAC or MACH with 1000 Genomes Project (March 2012) data as reference. In each study, principle components were adjusted in association analysis to control for confounding by population stratification. Imputed SNPs were filtered according to imputation quality (r2) before meta-analysis, which was implemented using METAL (Willer et al. 2010). The final number of SNPs ranged from 9M to 15M across cancer types, the number of samples also varied to some extent, with colon cancer and ovarian cancers having a smaller number (~10K–13K) while prostate, breast, and lung cancers having a larger number of samples (~20K–30K).
Study subjects in IGAP and GAME-ON were all of European ancestry and originated from countries in Europe, Canada, the United States, or Australia. There is no sample sharing between AD and any of the cancer studies used in our analysis.
Estimation of genome-wide genetic correlation
Genetic correlation between AD and each cancer type was estimated by cross-trait LD Score regression (Bulik-Sullivan et al. 2015a). This is a recently developed method based on GWAS summary statistics that quantifies the genetic covariance (ρg; analogous to co-heritability) between two traits by regressing the product of the z-scores (Z1jZ2j) from two studies of traits against the LD Score (lj) for each SNP j, assuming both traits follow polygenic inheritance. Genetic correlations were obtained as by normalizing genetic covariance by SNP-heritability for each trait estimated from single-trait LD Score regression (Bulik-Sullivan et al. 2015b). AD and cancer as complex diseases likely possess a polygenic genetic architecture and therefore it is appropriate for using cross-trait LD score regression to estimate their genetic correlation. Empirical genetic correlations between AD and cancer were also calculated by taking Pearson’s correlation coefficients of AD z-score and cancer z-score from all SNP to get an initial sense of the direction and magnitude of the genetic parameter and to be compared with the rg estimates from LD Score regression.
The analysis was implemented using the LDSC v1.0.0 software package (Bulik-Sullivan et al. 2015b). First, LD scores of all SNPs from individuals of European descent in the 1000 Genomes Project were computed. Next, genetic correlation of each cancer type with AD was estimated via cross-trait LD Score regression. Only IGAP stage1 data was used in the following analysis, because LDSC is designed for genome-wide analysis with SNPs scattered across the genome and does not work well for a concentrated set of SNPs showing significant association with a trait of interest. Intercepts from cross-trait LD Score regression were constrained to zeros as there is no sample overlap. The number of overlapping SNPs between AD and each cancer dataset was around 5M to 6M. Before investigating the genome-wide relationship of AD with individual cancer types, we examined the genetic correlation between AD and “any cancer type” combined using five independent GAME-ON cancer data sets that do not share samples with one another, including that of colon cancer, overall breast cancer, overall prostate cancer, ovarian cancer, and overall lung cancer. The summary association statistics for “any cancer” were obtained by meta-analyzing GWAS summary statistics data from the five cancer types using METAL (Willer et al. 2010).
Estimation of annotation-specific genetic correlation
To characterize the genetic overlap at the level of functional categories, for each cancer type that showed significant genetic sharing with AD, we estimated genetic correlation between AD and cancer in eight large annotations using cross-trait LD score regression. These annotations included repressed region, introns, transcribed region, super enhancers, DNase I hypersensitivity sites (DHSs), and histone marks H3K27ac, H3K4me1, and H3K4me3 (Finucane et al. 2015; Gusev et al. 2014). Each of them contained more than 600,000 overlapping SNPs between the AD and cancer datasets that appropriates the use of LD score regression. For each annotation, we re-calculated LD scores for SNPs assigned to that particular category and then used the annotation-specific LD scores for estimating the AD-cancer genetic correlation.
Detection of individual SNPs associated with AD and cancers
Tests for cross-phenotype effects were carried out at individual loci to detect SNPs that show cross-phenotype (CP) associations with both AD and cancer, for the cancer types that have a significant genetic correlation with AD.
For each cancer type, among the SNPs overlapping between AD and cancer summary statistics, we started by picking out SNPs with a less stringent p-value cutoff at SNP-AD p-value < 0.001, then selecting SNPs every 100kb apart to mimic LD pruning and to appropriately evaluate statistical significance based on number of independent tests; SNPs selected within each window were those with the smallest SNP-AD p-values. Next, we looked for any additional signal from cancer beyond the existing SNP-AD association. Bonferroni correction was used to correct for multiple testing.
To search for SNPs of a possible CP effect on AD and one or more cancer types, we also conducted individual SNP meta-analysis using Cross-Phenotype Meta-Analysis (CPMA; (Cotsapas et al. 2011)) to explore if there is any SNP associated with some of the cancer types in addition to its correlation with AD. The filtered SNPs with a SNP-AD p-value < 0.001 again underwent distance pruning based on a window of 100kb. CPMA was performed among the remaining SNPs, followed by FDR control to correct for multiple testing. SNPs were assigned to genes via PLINK with SNP attributes--dbSNP build 129 and gene range list--hg19 for inference of a potential common biological process between the two traits. eQTL function for each top SNP was checked upon at the Genotype-Tissue Expression (GTEx) portal.
Results
Genetic correlation estimates between AD and cancer
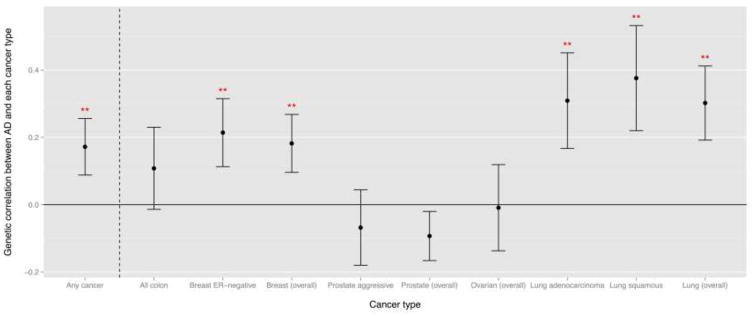
We observed an overall positive genetic correlation of 0.172 between AD and the five cancers combined (colon, breast cancer, prostate, ovarian, and lung cancers; p-value = 0.04) estimated via cross-trait LD Score regression from the 6 million SNPs included in both GWASs (Table 2; fig. 1).
Table 2.
Co-heritability and genetic correlation between AD and each cancer type, estimated by cross-trait LD score regressiona
| Trait 1 | Trait 2: Cancer | #SNPsc | co-h2 (SE) | rg | rg SE | p-value |
|---|---|---|---|---|---|---|
| Alzheimer’s Disease | Any cancerb | 4,799,343 | 0.007 (0.003) | 0.172 | 0.084 | 0.040 |
|
| ||||||
| All Colon | 4,772,982 | 0.008 (0.009) | 0.108 | 0.122 | 0.376 | |
| Breast ER-negative | 5,883,841 | 0.015 (0.007) | 0.214 | 0.101 | 0.035 | |
| Breast (overall) | 4,743,056 | 0.012 (0.005) | 0.182 | 0.086 | 0.034 | |
| Prostate aggressive | 5,405,868 | −0.005 (0.008) | −0.068 | 0.112 | 0.543 | |
| Prostate (overall) | 5,666,977 | −0.008 (0.006) | −0.093 | 0.073 | 0.204 | |
| Ovarian | 5,892,502 | −0.005 (0.008) | −0.009 | 0.128 | 0.947 | |
| Lung adenocarcinoma | 5,681,123 | 0.015 (0.006) | 0.309 | 0.142 | 0.029 | |
| Lung squamous | 5,681,315 | 0.018 (0.006) | 0.376 | 0.156 | 0.016 | |
| Lung (overall) | 5,681,066 | 0.019 (0.005) | 0.302 | 0.110 | 0.006 | |
All overlapping SNPs between AD-stage1 and cancer datasets were used. All cross-trait intercepts were constrained to 0, as there is no sample overlap.
Any of the 5 overall cancer types: colon, breast, prostate, ovarian, and lung
The number of overlapping SNPs between AD-stage1 and cancer datasets merged to the EUR LD score reference panel
Fig. 1. Genetic correlation between AD and each cancer type, estimated by cross-trait LD score regression.
Error bars are displayed as point estimate ± SE; “**” denotes p-value for genetic correlation < 0.05; “Any cancer” category includes all colon cancer, breast cancer (overall), prostate cancer (overall), ovarian cancer (overall), and lung cancer (overall).
Stratifying by cancer type, ER-negative and overall breast cancer showed significant positive genetic correlations with AD at rg = 0.21 (p-value = 0.04) and rg = 0.18 (p-value = 0.03), respectively. Lung adenocarcinoma, lung squamous cell carcinoma, and overall lung cancer also had a prominent positive genetic correlation with AD at rg = 0.31 (p-value = 0.03), 0.38 (p-value = 0.02), and 0.30 (p-value = 0.01). This implied that the two traits—AD and breast cancer, or AD and lung cancer—may share some common genetic background across the genome and the shared gene variants modulate the diseases risk in the same direction. On the contrary, the genetic correlation between AD and aggressive and overall prostate cancer were negative but not statistically significant (rg = −0.07 and −0.09; p-value = 0.54 and 0.20, respectively). The genetic correlation was around 0.1 between AD and all colon cancer and was slightly below zero between AD and ovarian cancer, and both estimates were not significant. These two cancer types also had the smallest sample size and might be underpowered for detecting genetic correlation.
The genetic correlation estimates from cross-trait LD Score regression were consistent in terms of direction, relative magnitude, and statistical significance with our initial inspection of empirical correlation estimates between AD and each cancer type calculated as the Pearson’s correlation coefficients between z-scores for all SNPs from the two traits (Online Resource Table S1), when LD between SNPs were not taken into account.
After learning the genome-wide relationship between AD and a variety of cancer types, we attempted to characterize the genetic sharing at regional and at individual SNP level between AD and the 2 cancer types that have a significant signal of genetic correlation, i.e. breast and lung cancers (overall).
Genetic correlation between AD and cancer by functional category
The first approach was evaluating the genetic correlation between AD and cancers by functional annotations to pin down specific regions on the genome that may explain more of the genetic sharing than others. This analysis additionally evaluated the annotation-specific relationship between AD and “any cancer type” where a notable positive rg was also observed.
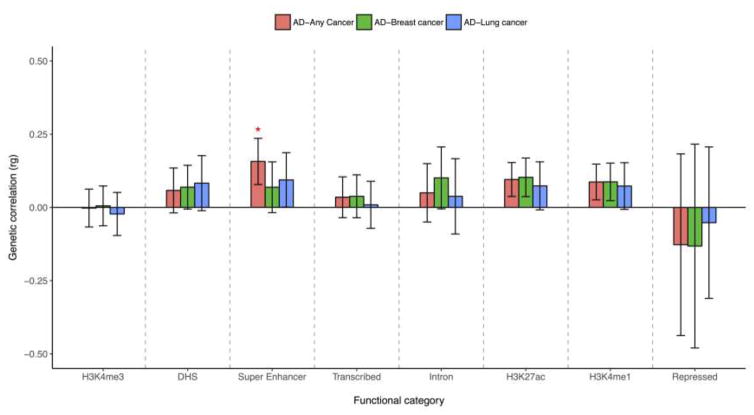
Our results showed that annotation-specific genetic correlations were comprised of a mixture of positive and negative signals (Figure 2; Table S2). Effect sizes of genetic variants in the repressed and the H3K4me3 annotations were negatively correlated between AD and breast cancer, lung cancer or the “any cancer” category, whereas positive genetic correlations were observed in the other six annotations. The only significant relationship between AD and the five cancer types combined appeared at super enhancers. Examining across all functional categories, three regions that represent active enhancer marks on the genome, including super enhancers, H3K27ac, and H3K4me1, all showed stronger and positive AD-cancer genetic correlations. This indicated a possible role of gene expression regulation with respect to enhancer activity in the shared genetic etiology of AD and cancer.
Figure 2. Annotation-specific genetic correlations (± SE) between AD and each cancer type.
“*” denotes p-value for genetic correlation < 0.05; functional categories on the x-axis were ordered based on its size, from the smallest (left) to the largest (right)
Cross-phenotype associations between AD and cancer
In order to investigate if cross-trait genetic correlation could be explained by major genetic loci, we went down to individual locus level to find pleiotropic SNPs associated with both AD and cancer, the existence of which may implicate common genetic pathways shared by the two diseases.
For each cancer type, we searched for any AD-related SNPs that also have an effect on cancer. A total of 11,788 out of the 4,743,056 SNPs common in both AD and breast cancer summary statistics and 14,655 out of the 5,681,315 AD-lung cancer overlapping SNPs remained after the filtering procedure (SNP-AD p-value < 0.001). There were 1507 SNPs present in both AD and breast cancer and 1648 SNPs present in both AD and lung cancer datasets after the 100kb window based pruning of SNPs. Among them, no SNP was significant after Bonferroni correction for breast cancer (top SNP rs59776273; chr4:47,792,047, SNP-breast cancer p-value = 9.9*10−5). While for lung cancer there were two candidate SNPs that survived the correction: rs56117933 at chr15:78,832,349 (unadjusted SNP-lung cancer p-value = 4.1*10−20, corrected p-value = 6.7*10−17; AD z-score = −3.34, lung cancer z-score = 9.19), in close proximity to the PSMA4 gene encoding for proteasome subunit alpha 4, whose polymorphisms have been related to lung cancer susceptibility from published GWAS (Hung et al. 2008), and rs11249708 at chr5: 179821728 (unadjusted SNP-lung cancer p-value = 1.5*10−5, corrected p-value = 0.025; AD z-score = −3.43, lung cancer z-score = 4.33), for which no previous genome-wise associations have been reported.
We next carried out CPMA tests to find SNPs showing residual association with one or both cancer types, given its initial association with AD at SNP-AD p-value < 0.001. The results showed that, 11 out of 1458 SNPs after distance pruning had a CPMA p-value < 0.01, but only one of them passed the FDR 5% threshold (Table 3). This top SNP rs56117933 (CPMA p-value < 2.2*10−16; AD z-score = −3.34, breast cancer z-score = −0.59, lung cancer z-score = 9.19) was the same as discovered just previously, which had a highly significant association with lung cancer (p-value = 4.1*10−20) but a much larger p-value with breast cancer (>0.05). The significant AD SNP showing additional association with cancers was likely driven by one cancer type, similarly for the other 10 SNPs. This might reflect the heterogeneous nature of different cancer types and suggested to look for CP effects on AD and cancer independently by cancer type. The results also showed that cross-trait genetic relationships observed at the genome-wide level was not likely explained by several major variants, consistent with the polygenic architecture. Significant positive genetic correlations were found for between AD-breast cancer and AD-lung cancer, but as expected the SNP-AD z-score and the SNP-cancer z-score were not necessarily in the same direction. For example, SNP rs17466060 appeared to increase AD risk (z-score = 4.60) but decrease the risk of both breast cancer (z-score = −2.39) and lung cancer (z-score = −3.45). No significant eQTLs were found for the 11 SNPs in the most relevant tissues (brain or tumor) from the GTEx project, nor did they correspond to genetic variants on the previously reported candidate genes encoding p53, Pin1, or those involving in the Wnt pathway. The CP results on individual SNPs suggest that it would need a much larger sample size to obtain the same power as the cross-trait heritability estimate which aggregated information from all available SNPs on the genome or a particular functional category, and allow us to study the sharing genetic architecture of two diseases.
Table 3.
SNPs with potential cross-phenotype effect on AD and two cancer types (overall breast and overall lung cancers) detected by Cross Phenotype Meta-Analysis (CPMA)
| SNP | CHR | Position | Eff allele |
Ref allele |
AD | Breast cancer (overall) |
Lung cancer (overall) |
CPMA stat |
CPMA p-value |
FDR | Gene | Nearest gene |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| z-score | p-value | z-score | p-value | z-score | p-value | ||||||||||
| rs56117933 | 15 | 78832349 | C | T | −3.34 | 8.3E-04 | −0.59 | 5.6E-01 | 9.19 | 4.1E-20 | 73.98 | <2.2E-16 | <1.0E-15 | - | PSMA4 |
| rs11249708 | 5 | 179821728 | A | G | −3.43 | 6.0E-04 | 1.53 | 1.3E-01 | 4.33 | 1.5E-05 | 14.79 | 1.2E-04 | 0.087 | - | GFPT2 |
| rs59776273 | 4 | 47792297 | T | C | −3.52 | 4.4E-04 | −3.89 | 9.9E-05 | −2.14 | 3.2E-02 | 13.92 | 1.9E-04 | 0.093 | CORIN (intron) | |
| rs17466060 | 8 | 27422740 | A | G | 4.60 | 4.3E-06 | −2.39 | 1.7E-02 | −3.45 | 5.7E-04 | 12.12 | 5.0E-04 | 0.182 | - | EPHX2 |
| rs3843702 | 15 | 80639403 | A | G | −3.33 | 8.7E-04 | −0.78 | 4.4E-01 | 3.82 | 1.3E-04 | 9.16 | 2.5E-03 | 0.575 | - | LINC00927 |
| rs3204635 | 12 | 57637593 | A | G | −3.33 | 8.6E-04 | −3.80 | 1.5E-04 | −0.82 | 4.1E-01 | 9.10 | 2.5E-03 | 0.575 | STAC3 (exon) | |
| rs7725218 | 5 | 1282414 | A | G | −3.35 | 8.1E-04 | −0.10 | 9.2E-01 | 3.97 | 7.2E-05 | 8.96 | 2.8E-03 | 0.575 | TERT (intron) | |
| rs10896445 | 11 | 68967641 | T | C | 3.61 | 3.1E-04 | −2.69 | 7.1E-03 | 2.55 | 1.1E-02 | 8.71 | 3.2E-03 | 0.577 | - | MYEOV |
| rs77597338 | 2 | 53267773 | G | A | 4.39 | 1.1E-05 | 2.24 | 2.5E-02 | 2.84 | 4.5E-03 | 8.13 | 4.4E-03 | 0.705 | - | ASB3 |
| rs74766959 | 11 | 72065209 | G | A | 3.60 | 3.2E-04 | 1.39 | 1.7E-01 | 3.35 | 8.0E-04 | 7.88 | 5.0E-03 | 0.729 | CLPB (intron) | |
| rs1568485 | 1 | 151736876 | C | T | −3.30 | 9.7E-04 | −3.53 | 4.2E-04 | 0.89 | 3.7E-01 | 7.63 | 5.7E-03 | 0.762 | OAZ3 (intron) | |
Discussion
In this study using data from two large GWAS consortia, we found a significant positive genetic correlation between AD and cancer overall, and specifically with breast and lung cancer. We also observed a suspected negative genetic correlation between AD and prostate cancer. These results establish that there is shared genetic variation between AD and cancer, but suggests that the direction of the genome-wide association may differ by cancer type. Examining the genetic correlation between AD and each cancer type in specific functional categories revealed that annotations linked to enhancer activity could play a role in the genetic sharing between the two diseases. These annotations may harbor important genetic variants involved in common pathophysiological pathways. Although we did not find pleotropic or cross-phenotype SNPs, our study might not be well-powered enough to detect the associations at individual loci.
As we went from genome-wide investigation to a more local analysis of genetic relationship, we observed mixed signals of positive and negative directions of shared genetic effect within specific annotations. We also noted a discordance in effect size and direction across AD and cancers at the level of individual SNPs. This is expected, and confirmed that the overall aggregated genetic correlation is a sum of positive and negative genetic correlations due to different functional regions or individual variants. The power to detect shared genetic architecture at whole-genome, whole-functional category would be dependent on the consistency of effect direction of genetic variants in those categories or even the whole genome.
Before interpreting the results of genetic correlation, it is important to acknowledge the possibility of confounding due to an imbalanced proportion of cancer survivors in AD cases vs. controls. If there were more cancer survivors among the AD patients than controls, genetic correlation analysis might pick up some cancer associations from cancer-related SNPs, resulting in a larger genetic overlap between AD and cancer than by chance, and contributing to an inflated positive genetic correlation. Unfortunately, we could not obtain information on cancer history from each study in order to directly examine the impact of cancer survivorship on our estimation. However, a study based on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort, one of the studies included in the ADGC of IGAP, suggested that the proportion of subjects with a history of cancer incidence in the study was around 31% (sample size = 1609), and those with AD at baseline were less likely to be cancer survivors than participants with mild cognitive impairment, subjective memory complaints and normal cognition (Nudelman et al. 2014).
Another possible confounder is the ratio of different cancer types in individual AD studies, as the genetic association between AD and cancer likely differs by cancer type. Non-lethal cancer (such as non-melamona skin cancer) and cancers that are screened for and cured (such as breast, prostate and colon) would be the most prevalent in an older population. In contrast, highly lethal cancers such as lung and pancreas, would have a lower prevalence and thus be less likely to act as a confounder. This indicates that our findings for lung cancer and aggressive subgroups of breast and prostate cancer might be more credible than the other analyses. Another situation when this bias would be less of a concern is when cases and controls are matched based on cancer history. Although in the present study we could not rule out the possibility of the potential confounding discussed herein, we suggest that future work report genetic correlations considering the effects of differential cancer survivor rates among AD cases and controls whenever this information is attainable. For example, one can conduct a sensitivity analysis excluding studies in which AD patients and controls have a dissimilar proportion of cancer survivors to evaluate the change in the estimates of genetic correlation.
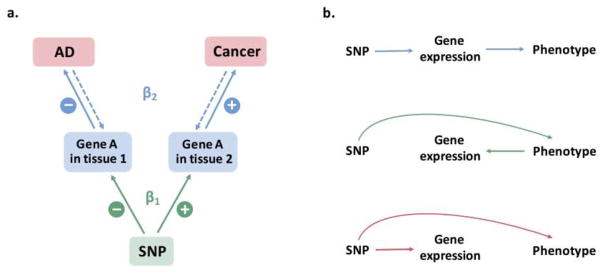
Our study found overall positive genetic correlations between AD and breast cancer and lung cancer, while epidemiological studies (Catala-Lopez et al.; Driver et al. 2012; Musicco et al. 2013) and a transcriptome meta-analysis (Ibanez et al. 2014) suggest an overall inverse association of AD with many cancer types. This might be due to the fact that phenotypic comorbidity, correlation of expression effect and correlation of genetic effect are different levels of association that should not be expected to be the same. The inverse comorbidity of two diseases could be due to the joint effect of genetics and environment, where the non-genetic effect could be negatively correlated and have a larger magnitude than the positively correlated genetic effect. In the context of survival bias, particularly for lung cancer, despite an inverse association at the phenotypic level, there might be a subset of individuals who are susceptible to both AD and lung cancer that is the driving force for the observed positive genetic correlation. A possible scenario in which a negative AD-cancer association based on differential gene expression in relevant tissues (Ibanez et al. 2014) can co-exist with a positive genetic correlation among SNPs is depicted in Fig. 3a; we note that this is only one of the numerous possibilities. In this case, the risk allele of a given SNP is associated with a decrease in expression of gene A in tissue 1 (eg. brain tissue) but an increase in its expression in tissue 2 (eg. tumor tissue). An increased expression of gene A in tissue 1 is associated with a reduced risk of AD, while its higher expression in tissue 2 is associated with an elevated risk of cancer, resulting in inverse molecular comorbidity. This level of association can in fact be bi-directional. The net SNP effects on the two diseases would be positive (βSNP=β1β2), and lead to a positive rg if the same is true for many more SNPs. In the analysis of partitioned co-heritability by functional categories, we observed both positive and negative genetic correlation in different categories. The functional annotation related to the negatively correlated category might explain the negatively correlated expression-AD association reported in previous studies and warrant further functional experiment.
Fig. 3. Relationship between SNP, gene expression, and observed phenotype(s).
(a) A possible scenario where an inverse correlation of gene expression effects (Ibanez et al. 2014) and a positive correlation of SNP effects between AD and cancer can be observed
(b) Possible causal pathways for the relationship between the three components if correlation exists between either two components. From up to down: causal effect of SNP on phenotype mediated through gene expression; gene expression reacts to phenotypic change due to SNP effect; pleiotropic effect of SNP on both gene expression and phenotype
Given the significant genome-wide associations of AD with some cancer types we have identified, we would need to gather gene expression data from brain and tumor tissues to establish a causal relationship linking SNP, gene expression, and both phenotypes together. Some possible scenarios for this are shown in Fig. 3b. This would ideally be accomplished in eQTL studies that can evaluate which SNPs have a direct effect on the phenotypes, which SNPs have an indirect effect mediated by gene expression, how those SNPs affect or regulate gene expression levels to exert their influences on the phenotypes, and what genes are being regulated. eQTLs might also have different effect directions in tissues relevant to AD and tissues relevant to cancers. Integration of these results with other –omics data (e.g. Methylation QTL) would help to better understand the underlying molecular mechanism of shared genetics and how that could lead to the suggested AD-cancer comorbidity, thereby allowing definition of a more accurate link between the phenotypic association and the genetic association of AD with cancer.
In addition, we noticed that most of our genetic correlation estimates were of small magnitude and have a relatively large standard error (SE). This is likely due to sample size and from using summary level GWAS data instead of genotype data. It has been shown that genetic correlation in bivariate analysis (rg) as a genetic parameter has a much larger sampling variance compared to proportion of phenotypic variance explained (hg2) by all SNPs in univariate analysis, which is true for both individual genotype data and in a pedigree design (Visscher et al. 2014). Simulation also showed that, when analyzing two case-controls studies of independent samples with an equal hg2 = 0.2 using genotype data, the power to detect an rg = 0.4 with a sample size of 10,000 for each study is 0.9 and is only 0.4 when rg = 0.2 (Visscher et al. 2014). Moreover, LD Score regression based on summary statistics generally yield bigger SEs than that from REML (GCTA) based on individual genotypes (Bulik-Sullivan et al. 2015a). Together these suggest that an even larger sample size is required for LD Score regression as compared to REML (GCTA) to achieve comparable power when estimating rg. The impact of sample size is evident in our results. We saw a larger SE around its rg estimate for cancer subtypes of smaller number of cases (ER-negative breast cancer, aggressive prostate cancer, lung adenocarcinoma, and lung squamous cell carcinoma) relative to their overall cancer type counterparts (Table 1&2). The two cancer types that have the smallest sample size in our datasets—colon cancer GWAS with less than 10,000 and ovarian cancer GWAS with less than 15,000 individuals—were found to have a non-significant rg surrounded by a wide confidence interval, but the effect size of rg between colon cancer and AD is in fact not negligible. Increasing sample size would likely reduce SEs and increase statistical power to detect a true genetic correlation.
In conclusion, we identified significant genetic correlations between AD and certain types of cancer that indicate the presence of shared genetic variants and disease mechanisms between the two diseases. To the best of our knowledge, this is the first investigation of genome-wide association between AD and cancer using GWAS summary statistics coming from large scale studies. Our functional category analysis suggests that regulation of gene expression in relation to enhancer activity might play an important role in this shared heritability. Integration with gene expression data or eQTL studies in specific tissues is needed to better define the overlapping biological pathways, find genes and regions on the genome to be targeted for functional studies, and connect the missing dots from genetic comorbidity discovered using SNP data to the association observed at the levels of gene expression and phenotype. We anticipate incorporating our current findings of a quantified and characterized genetic relationship between AD and a range of cancer types into functional studies that can generate a better understanding of the pathophysiology of AD and cancer and provide insights into novel therapeutic possibilities for both diseases.
Supplementary Material
Acknowledgments
We thank the International Genomics of Alzheimer’s Project (IGAP) and the GAME-ON network for providing summary results data for these analyses.
The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
We also thank members of individual studies from the GAME-ON network whose tremendous efforts altogether made this work possible:
Members of CORECT
CORECT acknowledges the following investigators: Kendra Blalock, Peter T. Campbell, Graham Casey, David V. Conti, Christopher K. Edlund, Jane Figueiredo, W. James Gauderman, Jian Gong, Roger C. Green, Stephen B. Gruber, John F. Harju, Tabitha A. Harrison, Eric J. Jacobs, Mark A. Jenkins, Shuo Jiao, Li Li, Yi Lin, Frank J. Manion, Victor Moreno, Bhramar Mukherjee, Ulrike Peters, Leon Raskin, Fredrick R. Schumacher, Daniela Seminara, Gianluca Severi, Stephanie L. Stenzel, and Duncan C. Thomas.
Members of DRIVE
DRIVE acknowledges the following GWASs and investigators that shared genome-wide summary data as part of the breast-cancer GWAS meta-analysis: the Australian Breast Cancer Family Study (ABCFS) (John L. Hopper, Melissa C. Southey, Enes Makalic, Daniel F. Schmidt), the British Breast Cancer Study (BBCS) (Olivia Fletcher, Julian Peto, Lorna Gibson, Isabel dos Santos Silva), the Breast and Prostate Cancer Cohort Consortium (BPC3) (David J. Hunter, Sara Lindström, Peter Kraft), the Breast Cancer Family Registries (BCFR) (Habib Ahsan, Alice Whittemore), the Dutch Familial Bilateral Breast Cancer Study (DFBBCS) (Quinten Waisfisz, Hanne Meijers-Heijboer, Muriel Adank, Rob B. van der Luijt, Andre G. Uitterlinden, Albert Hofman), German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) (Alfons Meindl, Rita K. Schmutzler, Bertram Müller-Myhsok, Peter Lichtner), the Helsinki Breast Cancer Study (HEBCS) (Heli Nevanlinna, Taru A. Muranen, Kristiina Aittomäki, Carl Blomqvist), the Mammary Carcinoma Risk Factor Investigation (MARIE) (Jenny Chang-Claude, Rebecca Hein, Norbert Dahmen, Lars Beckman), SardiNIA (Laura Crisponi), the Singapore and Sweden Breast Cancer Study (SASBAC) (Per Hall, Kamila Czene, Astrid Irwanto, Jianjun Liu), and the UK2 (Douglas F. Easton, Clare Turnbull, Nazneen Rahman).
Members of ELLIPSE
ELLIPSE acknowledges the following GWASs and investigators that shared genome-wide summary data as part of the prostate cancer GWAS meta-analysis: CRUK (Rosalind Eeles, Douglas F. Easton, Zsofia Kote-Jarai, Kenneth Muir, Graham Giles, Gianluca Severi, David Neal, Jenny L. Donovan, Freddie C. Hamdy), CAPS1 and CAPS2 (Fredrik Wiklund, Henrik Gronberg), BPC3-MEC (Christopher Haiman, Fred Schumacher), BPC3-EPIC (Ruth Travis, Elio Riboli), BPC3-Harvard (Peter Kraft, David Hunter), BPC3-ACS (Susan Gapstur), PEGASUS (Sonja Berndt, Stephen Chanock).
Members of TRICL
TRICL acknowledges the following investigators: Younghun Han, Li Su, Yongyue Wei, Rayjean J. Hung, Yonathan Brhane, John McLaughlin, Paul Brennan, James D. McKay, Heike Bickeböller, Albert Rosenberger, Richard S. Houlston, Neil Caporaso, Maria Teresa Landi, Joachim Heinrich, Angela Risch, Xifeng Wu, Yuanqing Ye, David C. Christiani, Christopher I. Amos.
Funding
This research was funded by the US Department of Veterans Affairs Merit Award Grant Clinical Science R&D [I01CX000934-01A1] (PI:Driver). The Genetic Association and Mechanisms in Oncology (GAME-ON) network was supported by the National Institutes of Health [U19CA148065 (DRIVE), U19CA148107 (CORECT), U19CA148127 (TRICL), and U19CA148537 (ELLIPSE)].
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015a;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015b;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala-Lopez F, Crespo-Facorro B, Vieta E, Valderas JM, Valencia A, Tabares-Seisdedos R. Alzheimer’s disease and cancer: current epidemiological evidence for a mutual protection. Neuroepidemiology. 2014;42(2):121–2. doi: 10.1159/000355899. Epub 2014 Jan 3. [DOI] [PubMed] [Google Scholar]
- Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetrius LA, Simon DK. The inverse association of cancer and Alzheimer’s: a bioenergetic mechanism. J R Soc Interface. 2013;10:20130006. doi: 10.1098/rsif.2013.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver JA, et al. Inverse association between cancer and Alzheimer’s disease: results from the Framingham Heart Study. Bmj. 2012;344:e1442. doi: 10.1136/bmj.e1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Holohan KN, Lahiri DK, Schneider BP, Foroud T, Saykin AJ. Functional microRNAs in Alzheimer’s disease and cancer: differential regulation of common mechanisms and pathways. Front Genet. 2012;3:323. doi: 10.3389/fgene.2012.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Ibanez K, Boullosa C, Tabares-Seisdedos R, Baudot A, Valencia A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014;10:e1004173. doi: 10.1371/journal.pgen.1004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa NC, Toledo EM. The role of Wnt signaling in neuronal dysfunction in Alzheimer’s Disease. Mol Neurodegener. 2008;3:9. doi: 10.1186/1750-1326-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001 systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- Musicco M, et al. Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology. 2013;81:322–328. doi: 10.1212/WNL.0b013e31829c5ec1. [DOI] [PubMed] [Google Scholar]
- Nudelman KN, Risacher SL, West JD, McDonald BC, Gao S, Saykin AJ Alzheimer’s Disease Neuroimaging I. Association of cancer history with Alzheimer’s disease onset and structural brain changes. Front Physiol. 2014;5:423. doi: 10.3389/fphys.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realmuto S, et al. Tumor diagnosis preceding Alzheimer’s disease onset: is there a link between cancer and Alzheimer’s disease? J Alzheimers Dis. 2012;31:177–182. doi: 10.3233/JAD-2012-120184. [DOI] [PubMed] [Google Scholar]
- Roe CM, Behrens MI, Xiong C, Miller JP, Morris JC. Alzheimer disease and cancer. Neurology. 2005;64:895–898. doi: 10.1212/01.WNL.0000152889.94785.51. [DOI] [PubMed] [Google Scholar]
- Roe CM, et al. Cancer linked to Alzheimer disease but not vascular dementia. Neurology. 2010;74:106–112. doi: 10.1212/WNL.0b013e3181c91873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabares-Seisdedos R, Rubenstein JL. Inverse cancer comorbidity: a serendipitous opportunity to gain insight into CNS disorders. Nat Rev Neurosci. 2013;14:293–304. doi: 10.1038/nrn3464. [DOI] [PubMed] [Google Scholar]
- Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31:100–110. doi: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, et al. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Visscher PM, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.