Summary
Many natural products that show therapeutic activities are oftentimes difficult to synthesize or isolate and have unknown targets, hindering their development as drugs. Identifying druggable hotspots targeted by covalently-acting anti-cancer natural products can enable pharmacological interrogation of these sites with more synthetically tractable compounds. Here, we used chemoproteomic platforms to discover that the anti-cancer natural product withaferin A targets C377 on the regulatory subunit PPP2R1A of the tumor suppressor protein phosphatase 2A (PP2A) complex leading to activation of PP2A activity, inactivation of AKT, and impaired breast cancer cell proliferation. We developed a more synthetically tractable cysteine-reactive covalent ligand, JNS 1–40, that selectively targets C377 of PPP2R1A to impair breast cancer signaling, proliferation, and in vivo tumor growth. Our study highlights the utility of using chemoproteomics to map druggable hotspots targeted by complex natural products and subsequently interrogating these sites with more synthetically tractable covalent ligands for cancer therapy.
eTOC Blurb
Many natural products that exhibit therapeutic activity, but translating these molecules into drugs is hindered by difficulty in their synthesis and isolation. Grossman and Ward et al. show that chemoproteomic technologies can be used to discover simpler molecules that react with the same sites as those targeted by natural products.

Introduction
There are countless natural products that have been isolated from microbes, plants, and other living organisms that demonstrate diverse biological action, including antibiotic, anti-inflammatory, and anti-cancer activities (Drahl et al., 2005). Among these natural products are agents that contain potential reactive electrophilic centers that can covalently react with nucleophilic amino acid hotspots on proteins to modulate their biological action (Drahl et al., 2005). Examples include natural product scaffolds such as β-lactones and β-lactams, many of which show antibiotic activity through covalent modification and inhibition of the catalytic serines of transpeptidases. However, there are many more reactive natural products that bear other reactive moieties such as Michael acceptors, epoxides, and aldehydes which may react with other nucleophilic side chains such as cysteines and lysines (Drahl et al., 2005). However, the reactivity and direct targets of most of these natural products that bear reactive centers remain poorly understood. Traditional approaches for target identification of natural products entail synthesis or derivatization of the natural products so that they can be appended to enrichment handles or beads for subsequent isolation and chemoproteomic studies. However, many natural products are incredibly complex to synthesize or may not necessarily have appropriate sites for derivatization, hindering the ability to develop natural product analogs to allow traditional chemoproteomic target identification.
Isotopic tandem orthogonal proteolysis-enabled activity-based protein profiling (isoTOP-ABPP) has arisen as a complementary chemoproteomic approach for target discovery of covalently-acting small molecules. IsoTOP-ABPP uses reactivity-based chemical probes to map proteome-wide reactive, functional, and ligandable hotspots directly in complex proteomes. When used in a competitive manner, covalently-acting small molecules can be competed against the binding of reactivity-based probes directly in complex proteomes to map their proteome-wide reactivity and targets (Backus et al., 2016; Counihan et al., 2016; Roberts et al., 2016; Wang et al., 2014). While the disadvantages of this isoTOP-ABPP strategy include biasing oneself to profiling of sites targeted by reactivity-based probes and the indirect nature of the assay inherent to competing small molecules against probe binding, a major advantage of this method is that it enables identification of the specific amino acids targeted by covalently-acting small molecules. Furthermore, this type of competitive isoTOP-ABPP strategy can be performed with the original parent molecule without having to synthesize analogs or derivatize the molecule. Several recent studies have used ABPP strategies to identify direct targets of reactive natural products such as licochalcone A, celastrol, and curcumin (Abegg et al., 2015; Roberts et al., 2017b; Zhou et al., 2016).
The advantage of identifying the direct targets and druggable hotspots targeted by covalently-acting anti-cancer natural products is that these sites can subsequently be deconvoluted to identify the specific target(s) responsible for the bioactivity; the identified targets can then be further pharmacologically interrogated with other chemical scaffolds to advance drug discovery efforts. This method contrasts with having to perform medicinal chemistry efforts on natural product scaffolds that are oftentimes synthetically challenging, with readouts based on their bioactivity rather than affinity to specific protein targets. Furthermore, identifying the nucleophilic amino acid hotspots targeted by reactive natural products enables covalent ligand discovery against these sites toward developing more potent and selective covalent inhibitors; these targets may also be more synthetically accessible compared to the oftentimes more complex structures of natural products. Recent studies have shown that covalent ligand discovery can be used to identify selective lead ligands against unique nucleophilic druggable hotspots in proteins (Backus et al., 2016; Bateman et al., 2017a; Roberts et al., 2017a).
Here, we have used the isoTOP-ABPP platform to couple target identification of a covalently-acting anti-cancer natural product with covalent ligand screening to identify a lead ligand that selectively interacts with the same target of interest. For this study, we have chosen to investigate the proteome-wide reactivity and targets of the natural product withaferin A, a steroidal lactone from the Ayurvedic plant Withania somnifera, which has been shown to possess anti-inflammatory, anti-diabetic, and anti-cancer activity (Dar et al., 2015; Lee and Choi, 2016; Lee et al., 2016; M et al., 2016). Withaferin A bears a Michael acceptor that may react with cysteine nucleophilic hotspots in protein targets (Fig. 1A). Previous studies have shown that withaferin A binds to functional cysteines in targets such as vimentin and NF-κB which have been postulated to account for its cancer and anti-inflammatory activities, respectively (Bargagna-Mohan et al., 2007; Heyninck et al., 2014). However, these studies either used a derivatized form of withaferin A, which could have missed targets that did not interact with this derivatized form, or performed experiments with specific proteins. Thus, withaferin A may potentially interact with additional targets that may be responsible for its anti-cancer activity. We have determined that withaferin A targets a particular cysteine on a regulatory subunit of the tumor suppressor protein phosphatase 2A (PP2A) to activate PP2A activity and inactivate multiple oncogenic signaling pathways which contributes to impairments in breast cancer pathogenicity and metabolism. We have also identified a significantly more synthetically tractable covalent ligand that selectively targets this same site to recapitulate the effects observed with withaferin A.
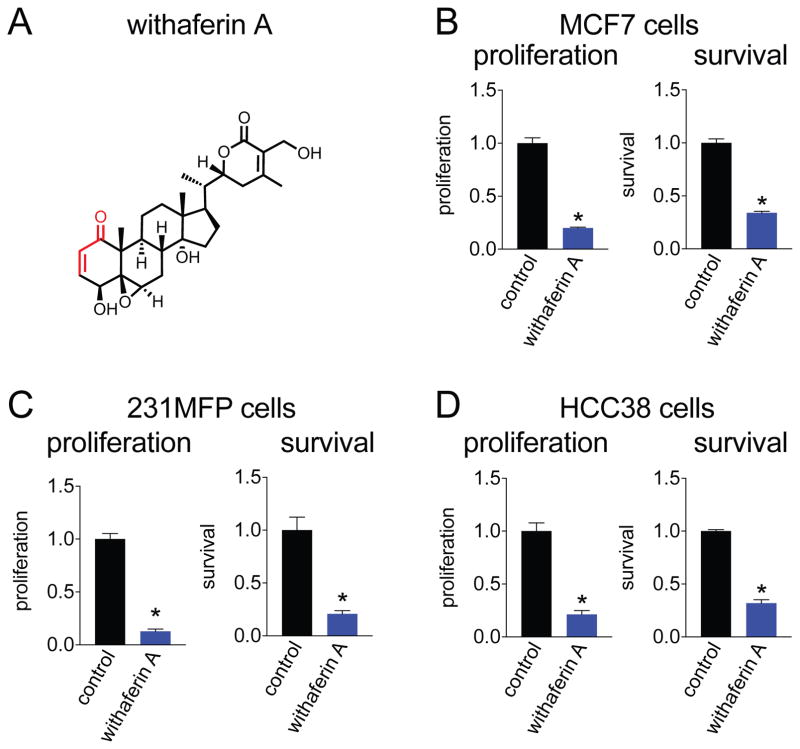
Figure 1. Withaferin A impairs breast cancer cell pathogenicity.
(A) Structure of withaferin A. Reactive Michael acceptor is indicated in red. (B) Withaferin A (10 μM) impairs cell proliferation and serum-free cell survival after 48 h in MCF7, 231MFP, and HCC38 cells compared to DMSO-treated controls. Data are presented as mean ± sem, n=5. Significance is shown as *p<0.05 compared to vehicle-treated controls.
Results and Discussion
Anti-Cancer Activity of Withaferin A in Breast Cancer Cells
We first tested the anti-cancer activity of withaferin A across several breast cancer cell lines including the receptor-positive MCF7 cells and triple-negative breast cancer (TNBC) cells 231MFP and HCC38 devoid in estrogen, progesterone, and HER2 receptors. TNBCs are highly aggressive breast cancers that show the worst prognosis with little to no targeted therapies (Bianchini et al., 2016). Identifying agents and new druggable hotspots in anti-cancer targets that are capable of impairing TNBC pathogenicity would contribute significantly towards combatting breast cancer. Consistent with previous studies, we show that withaferin A impairs serum-free cell survival and proliferation in MCF7, 231MFP, and HCC38 breast cancer cells (Fig. 1B–1D). We show that withaferin A impairs 231MFP cell proliferation in a dose-dependent manner with a 50 % effective concentration (EC50) of 7.5 μM (Fig. S1A).
Mapping Withaferin A Targets with IsoTOP-ABPP
We next used competitive isoTOP-ABPP platforms to map the proteome-wide cysteine-reactivity of withaferin A in 231MFP breast cancer cell proteomes. While previous studies have demonstrated biological effects of withaferin A at lower concentrations (Bargagna-Mohan et al., 2007), in this study, we used the minimum concentration of withaferin A (10 μM) that yielded the maximal anti-proliferative effect. To ascertain the direct targets of withaferin A without potential confounding effects from protein expression changes, we initially performed isoTOP-ABPP studies in vitro, in which we competed withaferin A against the reactivity of a broadly cysteine-reactive iodoacetamide-alkyne (IAyne) probe in 231MFP breast cancer cell proteomes. We subsequently appended isotopically light (for vehicle-treated) or heavy (for withaferin A-treated) enrichment handles by copper-catalyzed azide-alkyne cycloaddition (CuAAC), followed by combining vehicle and withaferin A-treated proteomes in a 1:1 ratio, and enrichment and isolation of probe-modified tryptic peptides for quantitative proteomic analysis (Fig. 2A). Out of the >3000 total probe-modified peptides identified, we only interpreted those peptides that were present in at least 2 out of 3 biological replicates. Through this analysis, we identified C377 of PPP2R1A, a regulatory subunit of PP2A, as the primary and only target that showed a light to heavy ratio >5 across all three biological replicates (Fig. 2B). We also confirmed that C377 of PPP2R1A was the primary in situ target of withaferin A in 231MFP cells showing an isotopically light to heavy ratio of 4.0. (Fig. S1B).
Figure 2. Using isoTOP-ABPP platforms to map proteome-wide targets of withaferin A in breast cancer cells.
(A) Competitive isoTOP-ABPP method. We mapped the cysteine-reactivity of withaferin A by pre-incubating withaferin A (10 μM) for 30 min in 231MFP breast cancer cell proteomes, prior to labeling with the cysteine-reactive iodoacetamide-alkyne (IAyne) probe (100 μM, 30 min). Probe labeled proteins were then tagged with an isotopically light (for control) or heavy (for withaferin A-treated) biotin-azide tag bearing a TEV protease recognition site by CuAAC. Control and treated proteomes were then mixed in a 1:1 ratio, probe labeled proteins were avidin-enriched and tryptically digested, probe-labeled tryptic peptides were avidin-enriched again, and released by TEV protease and analyzed by quantitative proteomic methods and light to heavy peptide ratios were quantified. (B) Competitive isoTOP-ABPP analysis of withaferin A cysteine-reactivity in 231MFP breast cancer cell proteomes in vitro. Light to heavy ratios of ~1 indicate peptides that were labeled by IAyne, but not bound by withaferin A. We designate light to heavy ratios of >5 as targets that were bound by withaferin A. Also shown are the peptide sequences and sites of modification of probe-modified peptides identified for PPP2R1A and the light to heavy ratios of C377 and C390 on PPP2R1A. (C) Validation of PPP2R1A as a target of withaferin A. Withaferin A was pre-incubated with pure human PPP2R1A protein followed by IAyne. Probe-labeled proteins conjugated to rhodamine-azide by CuAAC and analyzed by SDS/PAGE and in-gel fluorescence. Shown above is fluorescence detection of IAyne labeling and shown below is silver staining of the gel showing PPP2R1A protein expression. (D) Crystal structure of PP2A complex showing C377 of PPP2R1A (shown in white), the catalytic subunit shown in blue, and another regulatory subunit shown in green. PDB structure used is 2IAE. (E) PP2A activity assay with PP2A complex proteins PPP2R1A wild-type (WT) or C377A mutant and PPP2R2A and PPP2CA subunits measuring phosphate release from a PP2A substrate phosphopeptide. This PP2A complex was treated in vitro with DMSO or withaferin A (10 μM) for 30 min prior to initiation of the assay. (F) Withaferin A (10 μM, 4 h) treatment significantly reduces phospho-AKT levels in 231MFP breast cancer cells and this reduction is rescued by cotreatment with cantharidin (10 μM, 4 h). Data in (B) is average ratios from n=3. Gel in (C) is a representative gel from n=3. Data in (E–F) are presented as mean ± sem, n=3. Significance expressed as *p<0.05 compared to vehicle-treated controls and #p<0.05 compared to withaferin A-treated control. NS refers to not significant compared to the vehicle-treated C377A PPP2R1A group. Raw isoTOP-ABPP proteomic data can be found in Table S1. Withaferin A in situ isoTOP-ABPP analysis can be found in Fig. S1. Figure 1 is related to Figure S1 and Table S1.
Previous studies have uncovered several targets of withaferin A, including C328 on vimentin as well as several cysteines on Keap1 (Bargagna-Mohan et al., 2007; Heyninck et al., 2016). In our study, we identified C328 on vimentin as a site of IAyne labeling, but this site was not a target of withaferin A, as evidenced by a light to heavy ratio of 1.0 from in vitro treatment of 231MFP breast cancer cell proteomes with withaferin A (10 μM) (Table S1) as well as a lack of competition observed between withaferin A and IAyne labeling of pure human vimentin by gel-based ABPP (Fig. S1C). While we did not observe IAyne labeled KEAP1 peptides in our isoTOP-ABPP studies, we also showed no competition between withaferin A and IAyne labeling of pure human KEAP1 by gel-based ABPP studies (Fig. S1C). These results do not negate the possibility that withaferin A may still interact with these targets under other conditions, but suggest that these proteins are likely not the primary targets of withaferin A in 231MFP breast cancer cells. We thus focused on further investigating the role of withaferin A interactions with PPP2R1A and its influence on PP2A activity and breast cancer pathogenicity.
Withaferin A Interactions with PPP2R1A
PP2A is a tumor suppressor that dephosphorylates and inactivates oncogenic signaling pathways such as AKT. There has been considerable interest in developing direct or indirect activators of PP2A for cancer therapy (Sangodkar et al., 2016). While our IAyne probe labeled both C377 and C390 on PPP2R1A, withaferin A specifically targets C377 but not C390 on PPP2R1A (Fig. 2B). We confirmed this interaction as demonstrated by competition of withaferin A against IAyne labeling of pure human PPP2R1A protein using gel-based ABPP methods (Fig. 2C). In these gel-based studies, we used a lower concentration of IAyne than our isoTOP-ABPP studies, which may explain why we observe full competition of withaferin A against IAyne labeling. C377 sits at an interface between three subunits of the core PP2A complex based on previously solved crystal structures of the PP2A heterotrimeric holoenzyme complex (Fig. 2D) (Cho and Xu, 2007). We postulated that withaferin A activates PP2A activity through targeting C377 on PPP2R1A to impair 231MFP breast cancer cell proliferation. Consistent with this premise, we showed that withaferin A activated PP2A activity in a reconstituted in vitro biochemical assay with purified human wild-type PPP2R1A protein and the regulatory and catalytic subunits PPP2R2A and PPP2CA, respectively, but not with the PPP2R1A C377A mutant protein (Fig. 2E). Treatment of 231MFP cells with withaferin A also reduced phosphorylated AKT levels and this effect was rescued by co-treatment with the PP2A-selective inhibitor cantharidin (Fig. 2F). Further confirming that targeting of PPP2R1A is involved in withaferin A effects, PPP2R1A knockdown with short interfering RNA (siPPP2R1A) significantly attenuated the anti-proliferative effects observed with withaferin A treatment in 231MFP breast cancer cells (Fig. S1D, S1E). The lack of complete attenuation of withaferin A-induced anti-proliferative effects in siPPP2R1A cells may be due to residual PPP2R1A protein expression in the knockdown cells or the contribution of additional withaferin A targets to the anti-proliferative effects. Nonetheless, our data indicate that withaferin A targeting of C377 of PPP2R1A and activation of PP2A activity is in-part involved in the observed anti-proliferative effects.
Screening Cysteine-Reactive Fragment Libraries to Reveal PPP2R1A Ligands
To identify more synthetically tractable covalent ligands against C377 of PPP2R1A, we next screened a library of cysteine-reactive small molecule ligands in 231MFP breast cancer cells to identify any compounds that recapitulated the phenotypes of withaferin A in impairing 231MFP cell proliferation (Fig. 3A, 3B; Table S2). The top hit that arose from this screen was the chloroacetamide DKM 2-90 (Fig. 3B–3C, 4A).
Figure 3. Screening of covalent ligand libraries in breast cancer cells.
(A) Coupled screening of a cysteine-reactive covalent ligand library in 231MFP breast cancer cells with competitive isoTOP-ABPP platforms to identify anti-cancer lead compounds, targets, and ligandable hotspots within these targets. (B) We screened a cysteine-reactive fragment library consisting of acrylamides and chloroacetamides in 231MFP breast cancer cells (100 μM) to identify any leads that significantly impaired 231MFP breast cancer cell proliferation. Cell viability was assessed 48 h after treatment by Hoescht staining. (C) Shown is the structure of the lead covalent ligand DKM 2-90. Reactive chloroacetamide warheads are designated in red. Data in (B) are presented as mean ± sem, n=3. Significance expressed as *p<0.05 compared to vehicle-treated controls. Figure 3 is related to Table S2.
Figure 4. Target identification of DKM 2-90 using competitive isoTOP-ABPP platforms.
(A) Dose-responsive effects of DKM 2-90 on cell proliferation in 231MFP breast cancer cells. 231MFP cells were treated with DMSO or DKM 2-90 and proliferation was assessed 48 h after treatment by Hoechst staining. (B) IsoTOP-ABPP analysis of DKM 2-90 in 231MFP cell proteomes in vitro. 231MFP proteomes were pre-treated with DMSO or DKM 2-90 (100 μM) for 30 min prior to labeling proteomes with IAyne (100 μM) and subjected to the isoTOP-ABPP method. A light to heavy ratio of 1 indicates that the probe-labeled cysteine-bearing peptide was not bound by the covalent ligand, whereas a ratio >5 indicates bound sites. (C) Competition of DKM 2-90 against IAyne labeling of pure human PPP2R1A protein. DKM 2-90 was pre-incubated with pure PPP2R1A protein for 30 min prior to labeling with IAyne (100 μM) for 30 min. Rhodamine-azide was appended on by copper-catalyzed azide-alkyne cycloaddition and proteins were separated by SDS/PAGE and analyzed by in-gel fluorescence. (D) Levels of total and phosphorylated AKT (p-AKT) and vinculin as a loading control in 231MFP breast cancer cells. 231MFP cells were treated with vehicle, DKM 2-90 (100 μM), or cantharidin (10 μM) and DKM 2-90 (100 μM) for 5 h. Proteins were blotted for p-AKT, total AKT, and vinculin loading control. All data shown represents n=3–5/group. Raw data for (B) can be found in Table S1. IsoTOP-ABPP analysis of DKM 2-90 in situ can be found in Figure S1. Figure 2 is associated with Figure S1 and Table S1.
We next performed competitive isoTOP-ABPP experiments to identify the targets of DKM 2-90 through competition of this lead fragment against IAyne labeling of 231MFP proteomes. While this ligand was not potent, only showing anti-proliferative effects at 100 μM, we found that DKM 2-90 showed considerable selectivity in targeting C377 of PPP2R1A in vitro (Fig. 4B; Table S1). We also confirmed this through gel-based ABPP methods, showing significant competition of DKM 2–90 against IAyne labeling of pure human PPP2R1A protein with a 50 % inhibitory concentration (IC50) of 10 μM. (Fig. 4C). We also show that other chloroacetamide ligands of similar structures do not bind to PPP2R1A, suggesting that DKM 2-90 interacts with PPP2R1A in vitro relatively specifically, despite its simple structure (Fig. S1F). IsoTOP-ABPP analysis of DKM 2-90 treatment in 231MFP cells in situ also showed targeting of C377 of PPP2R1A with an isotopically light to heavy ratio of 5.9. However, four additional targets were also evident that showed an isotopically light to heavy ratio >5, including TXNDC17 C43, CLIC4 C35, ACAT1 C196, and SCP2 C307 (Fig. S1G). Nonetheless, while DKM 2-90 was a very simple and non-potent covalent ligand, it showed remarkable overall selectivity with only 5 total sites showing >5 ratio out of >1000 cysteines profiled (Fig. S1G; Table S1). Despite additional targets of DKM 2-90, we still observed an attenuation of DKM 2-90-mediated anti-proliferative effects in siPPP2R1A 231MFP cells compared to DKM 2-90-treated siControl cells (Fig. S1H). We also recapitulated the reduced levels of phosphorylated AKT and cantharidin rescue with DKM 2-90 treatment in 231MFP cells (Fig. 4D).
JNS 1-40: An Optimized Covalent Ligand Targeting C377 of PPP2R1A
While DKM 2-90 was not a potent ligand against PPP2R1A, we showed that much simpler covalent ligands could be identified that hit the same druggable hotspots targeted by complex natural products like withaferin A with decent selectivity and cell penetration. We next sought to optimize the potency of DKM 2-90. We found that replacing the benzodioxan ring with a tetralin with JNS 1-37 dramatically reduced potency with an IC50 value of 300 μM (Fig. S2A) compared to 10 μM with DKM 2-90. Adding an N-benzyl group to DKM 2-90 with JNS 1-40 improved potency towards PPP2R1A by 16-fold with an IC50 of 630 nM (Fig. 5A). We thus moved forward with further characterization of JNS 1-40. Both in vitro and in situ isoTOP-ABPP analysis showed that JNS 1-40 selectively targets C377 of PPP2R1A in both 231MFP complex proteome and cells, and is the only target exhibiting an isotopically light to heavy ratio >5 (Fig. 5B; Fig. S2B; Table S1). Much like withaferin A, JNS 1-40 activated PP2A activity in vitro with purified PP2A complex proteins with wild-type PPP2R1A, but not with the PPP2R1A C377A mutant protein (Fig. 5C). Similarly, JNS 1-40 treatment in 231MFP cells significantly reduced phosphorylated AKT levels and impaired proliferation and survival (Fig. 5D–5F). The anti-proliferative effects observed with JNS 1-40 were also attenuated in siPPP2R1A 231MFP cells compared to siControl cells (Fig. S2C).
Figure 5. Optimized covalent ligand JNS 1-40 selectively targets C377 of PPP2R1A to activate PP2A activity and impair breast cancer pathogenicity.
(A) Structure of JNS 1-40 and gel-based ABPP analysis of its potency against PPP2R1A. Reactive chloroacetamide is shown in red. Pure human PPP2R1A was pre-treated with DMSO or JNS 1-40 for 30 min at 37 °C prior to IAyne labeling for 30 min at room temperature. Probe-labeled proteins were appended to rhodamine-azide by CuAAC and analyzed by SDS/PAGE and in-gel fluorescence. Shown above is fluorescence detection of IAyne labeling and shown below is silver staining of the gel showing PPP2R1A protein expression. (B) IsoTOP-ABPP analysis of JNS 1-40 treatment in 231MFP cells. 231MFP proteomes were treated in vitro with DMSO or JNS 1-40 (100 μM) for 30 min prior to IAyne labeling for 1 h and subjected to the isoTOP-ABPP method. Light to heavy ratios of probe-modified peptides are shown. (C) PP2A activity assay with PP2A complex proteins PPP2R1A wild-type (WT) or C377A mutant and PPP2R2A and PPP2CA subunits measuring phosphate release from a PP2A substrate phosphopeptide. This PP2A complex was treated in vitro with DMSO or JNS 1-40 (100 μM) for 30 min prior to initiation of the assay. (D) Levels of total and phosphorylated AKT (p-AKT) and vinculin as a loading control in 231MFP breast cancer cells. 231MFP cells were treated with vehicle or JNS 1-40 (100 μM) for 75 min. (E, F) JS 1-40 (100 μM) impairs cell proliferation and serum-free cell survival after 48 h in 231MFP cells. (G) 231MFP tumor xenograft growth in immune-deficient SCID mice. 231MFP cells were subcutaneously injected into mice. Daily once per day treatment with vehicle or JNS 1-40 (50 mg/kg ip) was initiated 15 days after tumor implantation. (H) isoTOP-ABPP analysis of 231MFP tumor xenografts from JNS 1-40-treated mice in vivo. Tumors harvested from the end of the study shown in (G) were labeled with IAyne ex vivo and subjected to the isoTOP-ABPP methodology. Data in (C–G) are presented as mean ± sem, n=3–7/group. Data in (B, H) is average ratios from n=3. Significance is shown as *p<0.05 compared to vehicle-treated controls. NS indicates not significant (p>0.05) compared to the vehicle-treated C377A PPP2R1A group. Raw proteomic data can be found in Table S1. IsoTOP-ABPP analysis of JNS 1-40 in situ can be found in Figure S2. Figure 5 is associated with Figure S2 and Table S1.
With the improved potency and selectivity observed with JNS 1-40, we next sought to determine whether JNS 1-40 targeting of PPP2R1A could reduce breast cancer tumor growth in vivo. Daily treatment of mice with JNS 1-40 (50 mg/kg ip) in vivo began 15 days after 231MFP tumor xenograft initiation and significantly attenuated tumor growth (Fig. 5G). Daily treatment with JNS 1-40 for >30 days did not cause any overt toxicity or body weight loss, suggesting that this compound is well tolerated in vivo (data not shown). To confirm target engagement, we also performed ex vivo isoTOP-ABPP analysis of 231MFP tumor xenografts from in vivo JNS 1-40 treated mice. We demonstrated that JNS 1-40 selectively targeted C377 of PPP2R1A in vivo in 231MFP tumors showing an isotopically light to heavy ratio of 5.7 (Fig. 5H, Table S1). Collectively, our data indicated that the improved covalent ligand JNS 1-40 selectively targeted C377 of PPP2R1A to activate PP2A activity and impair breast cancer cell proliferation and in vivo tumor growth.
Conclusion
In conclusion, we showed here that isoTOP-ABPP platforms can be used to identify the druggable hotspots targeted by bioactive covalently-acting natural products, which can in-turn be pharmacologically interrogated with more synthetically tractable covalent ligands. In this study, we revealed that withaferin A targets C377 of PPP2R1A, a unique druggable hotspot within the PP2A tumor suppressor complex, to activate PP2A activity, inhibit AKT signaling, and impair breast cancer cell proliferation. Through screening a library of fragment-based covalent ligands, we identified a much simpler covalent ligand DKM 2-90 that also targeted the same site as withaferin A. We further optimized this hit to generate JNS 1-40, a lead covalent ligand with nanomolar potency that selectively targets C377 of PPP2R1A in vitro, in situ, and in vivo to recapitulate the effects observed with withaferin A: activating PP2A, inactivating AKT signaling, and impairing breast cancer pathogenicity. Previous studies have identified various additional targets of withaferin A, including vimentin, KEAP1, Hsp90, and IKKβ (Bargagna-Mohan et al., 2007; Heyninck et al., 2014, 2016; Yu et al., 2010). Withaferin A has also been shown to affect important processes such as proteostasis (Tao et al., 2015). Our studies do not rule out that these and other protein targets may also play important roles in the anti-proliferative and pathogenic impairments observed with withaferin A, but instead reveal C377 of PPP2R1A as an additional target of this natural product and a unique druggable hotspot that can be targeted to activate PP2A and impair breast cancer pathogenicity. While we and many other groups have used ABPP platforms to identify targets of covalently-acting natural products (Abegg et al., 2015; Bateman et al., 2017a; Blewett et al., 2016; Roberts et al., 2017b; Wang et al., 2014; Zhou et al., 2016), we show here that this process can be coupled with covalent ligand discovery approaches, yielding much more synthetically tractable small molecules that do not require significant synthetic or isolation efforts, which recapitulate the effects observed with complex natural products. Overall, our study demonstrates the utility of using isoTOP-ABPP platforms to map the reactivity and targets of covalently-acting natural products towards identifying their therapeutic mechanism of action and discovering druggable hotspots that can be interrogated by alternative pharmacological scaffolds for future drug discovery efforts.
Significance
While there are countless natural products that exhibit therapeutic activity, translating these natural products into drugs has been hindered by difficulty in synthesis and isolation of these compounds. Here, we show that isoTOP-ABPP based chemoproteomic platforms and covalent ligand discovery approaches can be coupled to discover synthetically tractable covalent ligands that target druggable hotspots targeted by more complex covalently-acting natural products. In this study, we discovered that withaferin A specifically targets C377 of PPP2R1A, a regulatory subunit within the tumor suppressor PP2A, to activate PP2A activity, inactivate AKT signaling, and impair breast cancer pathogenicity. Through covalent ligand screening and optimization, we identify a synthetically accessible small-molecule that targets the same site as withaferin A and produces identical effects. Overall, our study provides an alternative strategy for translating natural products into therapeutics, by developing simpler covalent ligands that hit the same druggable hotspots targeted by natural products using chemoproteomics-enabled covalent ligand discovery platforms.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel K. Nomura (dnomura@berkeley.edu).
Experimental Model and Subject Details
Chemicals
Withaferin A was obtained from Sigma. IAyne was obtained from CHESS Gmbh. Heavy and light TEV-biotin tags were synthesized per previously described methods (Weerapana et al., 2010). The synthesis for most of our cysteine-reactive ligand library has been described previously (Bateman et al., 2017b). New ligands screened in this paper that were not previously described are in Supplemental Methods. All compounds in our library were confirmed to be >95 % pure. The structures of the covalent ligand library screened in this paper are all listed in Table S2.
In Vivo Animal Studies
Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of California, Berkeley. Female immune-deficient SCID mice (6 weeks old) purchased from Taconic laboratories were used for tumor xenograft studies. Tumor xenograft and pharmacology studies were performed as previously described (Benjamin et al., 2013; Nomura et al., 2010).
Cell Culture
The 231MFP cells were obtained from Prof. Benjamin Cravatt and were generated from explanted tumor xenografts of MDA-MB-231 cells. These cells have been previously characterized as a more aggressive variant of the MDA-MB-231 cells (Jessani et al., 2004, 2005). HCC38 and MCF7 cells were obtained from the American Type Culture Collection. 231MFP cells were cultured in L15 medium containing 10% FBS, supplemented with 1% glutamine (200 mM stock), and maintained at 37°C with 0% CO2. HCC38 and MCF7 cells were cultured in RPMI medium containing 10% FBS, supplemented with 1% glutamine (200 mM stock), and maintained at 37°C with 5% CO2. Unless otherwise specified, all cell culture materials were purchased from Gibco. Withaferin A, DKM 2-90, JNS 1-40, and all other covalent ligands were dissolved in DMSO and final DMSO concentrations in cells were 0.1%.
Method Details
Cellular Phenotype Studies
Cell survival and proliferation assays were performed as previously described using Hoechst 33342 dye (Invitrogen) according to manufacturer’s protocol (Louie et al., 2016). Cells were seeded into 96-well plates (40,000 for survival and 20,000 for proliferation) in a volume of 150 μl and allowed to adhere overnight. Cells were treated with an additional 50 μL of media containing 1:250 dilution of 1000x compound stock in DMSO. Medium was removed from each well and 100 μl of staining solution containing 10% formalin and Hoechst 33342 dye was added to each well and incubated for 15 min in the dark at room temperature. After incubation, staining solution was removed and wells were washed with PBS before imaging. Studies with HCC38 cells were also performed as above but were seeded with 20,000 cells for survival and 10,000 cells for proliferation.
Western Blotting
Antibodies to vinculin, phospho-Akt (Ser473), and Akt were obtained from Cell Signaling Technology and proteomes were blotted per recommended manufacturer’s’ procedure. Cells were lysed in lysis buffer (containing the following: 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM pyrophosphate, 50 mM NaF, 5 mM β-glycero-phosphate, 1 mM Na3VO4, 50 nM calyculin A (EMD Millipore), and protease inhibitors (Roche)). Lysate was incubated on a rotator at 4°C for 30 min, and insoluble material was removed via centrifugation at max speed for 10 min. Proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes using the iBlot system (Invitrogen). Blots were blocked with 5% nonfat milk in Tris-buffered saline containing Tween 20 (TBST) solution for 1 hour at room temperature, washed in TBST, and probed with primary antibody diluted in recommended diluent per manufacturer overnight at 4°C. Following washes with TBST, the blots were incubated in the dark with secondary antibodies purchased from Rockland and used at 1:10 000 dilution in 5% nonfat milk in TBST at room temperature. Blots were visualized using an Odyssey Li-Cor scanner after additional washes.
Purification of PPP2R1A and PPP2R2A Subunits
Wild-type mammalian expression plasmids with C-terminal FLAG tag were purchased from Origene (PPP2R1A: RC200056; PPP2R2A, MR207137). The PPP2R1A C337A mutant was generated with Agilent QuickChange Lightning site-directed mutagenesis kit according to manufacturer’s instructions. HEK293T cells (ATCC CRL-11268) were grown to 60% confluency in DMEM (Corning) supplemented with 10% FBS (Corning) and 2mM L-glutamine (Life Technologies) and maintained at 37°C with 5% CO2. Immediately prior to transfection, media was replaced with DMEM + 5% FBS. Each plate was transfected with 20 μg of overexpression plasmid with 100 μg PEI (Sigma). After 48hrs cells were collected in TBS, lysed by sonication, and batch bound with anti-DYKDDDDK resin (GenScript) for 1hr. Lysate and resin was loaded onto a gravity flow column and washed, followed by elution with 250ng/uL 3xFLAG peptide (ApexBio A6001). Purity and concentration were verified by PAGE, UV/Spectroscopy, and BCA assay.
In Vitro PP2A Activity Assay
Recombinant PPP2CA (40 nM, Origene TP301334) was combined with pulled-down WT or mutant PPP2R1A (50 nM) as well as PPP2R2A (50 nM) and incubated with 10 μM withaferin A, JS 1-40, or vehicle for 30 min at RT in TBS. Activity was assayed by addition of 60 μM Thr phosphopeptide (KRpTIRR, Millipore, 12-219) at 37° C for 25 min, and free phosphate was detected colorimetrically by malachite green kit (Cayman 10009325) per manufacturer’s instructions.
PPP2R1A Knockdown Studies
PPP2R1A was transiently knocked down with siRNA using previously described methods (Benjamin et al., 2014). siRNA for a scrambled RNA oligonucleotide control and pooled RNA oligonucleotides targeting PPP2R1A were purchased from Dharmacon.
IsoTOP-ABPP
IsoTOP-ABPP studies were done as previously reported (Backus et al., 2016; Bateman et al., 2017a; Weerapana et al., 2010). Cells were lysed by probe sonication in PBS and protein concentrations were measured by BCA assay. Proteome samples diluted in PBS (4 mg of proteome per biological replicate) were treated with a covalently-acting small molecule or vehicle for 30 min at 37°C. Then, IAyne labeling (100 μM) was performed for 1 h at room temperature. CuAAC was used by sequential addition of tris(2-carboxyethyl)phosphine (1 mM, Sigma), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (34 μM, Sigma), copper (II) sulfate (1 mM, Sigma), and biotin-linker-azide, the linker functionalized with a TEV protease recognition sequence along with an isotopically light or heavy valine for treatment of control or treated proteome, respectively. After click reactions, proteomes were precipitated by centrifugation at 6500 x g, washed in ice-cold methanol, combined in a 1:1 control/treated ratio, washed again, then denatured and resolubilized by heating in 1.2% SDS/PBS to 80°C for 5 minutes. Insoluble components were precipitated by centrifugation at 6500 x g and soluble proteome was diluted in 5 ml 0.2% SDS/PBS. Labeled proteins were bound to avidin-agarose beads (170 μl resuspended beads/sample, Thermo Pierce) while rotating overnight at 4°C. Bead-linked proteins were enriched by washing three times each in PBS and water, then resuspended in 6 M urea/PBS (Sigma) and reduced in TCEP (1 mM, Sigma), alkylated with iodoacetamide (18 mM, Sigma), then washed and resuspended in 2 M urea and trypsinized overnight with 0.5 μg/μl sequencing grade trypsin (Promega). Tryptic peptides were eluted off. Beads were washed three times each in PBS and water, washed in TEV buffer solution (water, TEV buffer, 100 μM dithiothreitol) and resuspended in buffer with Ac-TEV protease and incubated overnight. Peptides were diluted in water and acidified with formic acid (1.2 M, Spectrum) and prepared for analysis.
MS Analysis
Peptides from all proteomic experiments were pressure-loaded onto a 250 μm inner diameter fused silica capillary tubing packed with 4 cm of Aqua C18 reverse-phase resin (Phenomenex # 04A-4299) which was previously equilibrated on an Agilent 600 series HPLC using gradient from 100% buffer A to 100% buffer B over 10 min, followed by a 5 min wash with 100% buffer B and a 5 min wash with 100% buffer A. The samples were then attached using a MicroTee PEEK 360 μm fitting (Thermo Fisher Scientific #p-888) to a 13 cm laser pulled column packed with 10 cm Aqua C18 reverse-phase resin and 3 cm of strong-cation exchange resin for isoTOP-ABPP studies. Samples were analyzed using an Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) using a 5-step Multidimensional Protein Identification Technology (MudPIT) program, using 0 %, 25 %, 50 %, 80 %, and 100 % salt bumps of 500 mM aqueous ammonium acetate and using a gradient of 5–55 % buffer B in buffer A (buffer A: 95:5 water:acetonitrile, 0.1 % formic acid; buffer B 80:20 acetonitrile:water, 0.1 % formic acid). Data was collected in data-dependent acquisition mode with dynamic exclusion enabled (60 s). One full MS (MS1) scan (400–1800 m/z) was followed by 15 MS2 scans (ITMS) of the nth most abundant ions. Heated capillary temperature was set to 200° C and the nanospray voltage was set to 2.75 kV.
Data was extracted in the form of MS1 and MS2 files using Raw Extractor 1.9.9.2 (Scripps Research Institute) and searched against the Uniprot mouse database using ProLuCID search methodology in IP2 v.3 (Integrated Proteomics Applications, Inc) (Xu et al., 2015). Cysteine residues were searched with a static modification for carboxyaminomethylation (+57.02146) and up to two differential modifications for methionine oxidation and either the light or heavy TEV tags (+464.28596 or +470.29977, respectively). Peptides were required to have at least one tryptic end and to contain the TEV modification. ProLUCID data was filtered through DTASelect to achieve a peptide false-positive rate below 1%. Only those probe-modified peptides that were evident in two out of three biological replicates were interpreted for their isotopic light to heavy ratios. MS1 peak shapes were confirmed to be of good quality for interpreted peptides. Targets of covalently-acting molecules are defined here as targets that showed >4 light to heavy ratios across all three biological replicates.
Gel-Based ABPP
Gel-based ABPP methods were performed as previously described (Bateman et al., 2017b; Medina-Cleghorn et al., 2015). Recombinant pure human proteins were purchased from Origene. Pure proteins (0.2 μg) were pre-treated with DMSO or covalently-acting small molecules for 30 min at 37°C in an incubation volume of 50 μL PBS, and were subsequently treated with IAyne (10 μM final concentration) for 30 min at room temperature. CuAAC was performed to append rhodamine-azide (1 μM final concentration) onto IAyne probe-labeled proteins. The samples were separated by SDS/PAGE and scanned using a ChemiDoc MP (Bio-Rad Laboratories, Inc). Inhibition of target labeling was assessed by densitometry using ImageJ.
General synthetic methods
Chemicals and reagents were purchased from major commercial suppliers and used without further purification. Reactions were performed under a nitrogen atmosphere unless otherwise noted. Silica gel flash column chromatography was performed using EMD or Sigma Aldrich silica gel 60 (230–400 mesh). Proton and carbon nuclear magnetic resonance (1H NMR and 13C NMR) data was acquired on a Bruker AVB 400, AVQ 400, or AV 600 spectrometer at the University of California, Berkeley. High resolution mass spectrum were obtained from the QB3 mass spectrometry facility at the University of California, Berkeley using positive or negative electrospray ionization (+ESI or −ESI). Yields are reported as a single run.
General Procedure A
The amine (1 eq.) was dissolved in DCM (5 mL/mmol) and cooled to 0°C. To the solution was added acryloyl chloride (1.2 eq.) followed by triethylamine (1.2 eq.). The solution was warmed to room temperature and stirred overnight. The solution was then washed with brine and the crude product was purified by silica gel chromatography (and recrystallization if necessary) to afford the corresponding acrylamide.
General Procedure B
The amine (1 eq.) was dissolved in DCM (5 mL/mmol) and cooled to 0°C. To the solution was added chloroacetyl chloride (1.2 eq.) followed by triethylamine (1.2 eq.). The solution was warmed to room temperature and stirred overnight. The solution was then washed with brine and the crude product was purified by silica gel chromatography (and recrystallization if necessary) to afford the corresponding chloroacetamide.
Quantification and Statistical Analysis
Microsoft Excel and Graphpad Prism software were used for statistical analysis. Statistical and quantification details of experiments can be found in the figure legends. Significance was defined a p<0.05 between comparison groups.
Data and Software Availability
N/A
Additional Resources
N/A
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Vinculin | Abcam | ab129002 |
| Rabbit anti-p-Akt (S473) | CST | D9E |
| Mouse anti-total Akt | CST | 40D4 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Withaferin A | Sigma | W4394 |
| N-Hex-5-ynyl-2-iodo-acetamide (IAyne) | Chess Gmbh | 3187 |
| Sequencing grade trypsin (porcine) | Promega | V5111 |
| AcTEV protease | Invitrogen | 12575015 |
| 1,4-benzodioxan- 6-amine | Sigma | 193232 |
| Chloroacetyl chloride | Sigma | 104493 |
| Triethylamine | Sigma | T0886 |
| Sodium Hydride | Sigma | 452912 |
| Benzyl Bromide | Sigma | B17905 |
| 5,6,7,8-Tetrahydro-2-napthylamine | Sigma | 678864 |
| Biotin-TEV-azide | (Weerapana et al., 2010) | |
| Critical Commercial Assays | ||
| Malachite Green Phosphate Assay Kit | Cayman | 10009325 |
| Hoechst 33342 dye | Invitrogen | H3570 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| 231MFP | (Jessani et al., 2004) | |
| HCC38 | ATCC | CRL-2314 |
| MCF7 | ATCC | HTB-22 |
| MCF10A | ATCC | CRL-10317 |
| HEK293T/17 | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| C.B17 SCID mice | Taconic | CB17SC-F |
| Oligonucleotides | ||
| PPP2R1A SMARTPOOL siRNA | Dharmacon | L-010259-00-0005 |
| Recombinant DNA | ||
| PPP2R1A (Myc-DDK-tagged) expression plasmid | Origene | RC200056 |
| PPP2R2A (Myc-DDK-tagged) expression plasmid | Origene | MR207137 |
| Software and Algorithms | ||
| IP2 proteomics pipeline 4.1.1 | Integrated Proteomics Applications | |
| Other | ||
| Streptavidin agarose | ThermoFisher | 20347 |
Supplementary Material
We mapped the cysteine-reactivity of withaferin A and DKM 2-90 by pre-incubating withaferin A (10 μM) or DKM 2-90 (100 μM) for 30 min in 231MFP breast cancer cell proteomes, prior to labeling with the cysteine-reactive iodoacetamide-alkyne (IAyne) probe (100 μM, 30 min). Probe labeled proteins were then tagged with an isotopically light (for control) or heavy (for withaferin A- or DKM 2-90-treated) biotin-azide tag bearing a TEV protease recognition site by copper-catalyzed azide-alkyne cycloaddition. Control and treated proteomes were then mixed in a 1:1 ratio, probe labeled proteins were avidin-enriched and tryptically digested, probe-labeled tryptic peptides were avidin-enriched again, and released by TEV protease and analyzed by quantitative proteomic methods and light to heavy peptide ratios were quantified. Competitive isoTOP-ABPP analysis of withaferin A and DKM 2-90 cysteine-reactivity in 231MFP breast cancer cell proteomes. Light to heavy ratios of ~1 indicate peptides that were labeled by IAyne, but not bound by withaferin A or DKM 2-90. We designate light to heavy ratios of >10 as targets that were bound by withaferin A or DKM 2-90. Tabs 1 and 3 shows average light versus heavy ratios from those probe-modified peptides found in at least 2 out of 3 biological replicates.
Tabs 1–7. IsoTOP-ABPP analysis of withaferin A (10 μM), DKM 2-90 (100 μM), JNS 1-40 (100 μM) treatment in vitro in 231MFP proteomes or in situ in 231MFP cells, or 231MFP tumors from JNS 1-40 daily in vivo treatment in mice. For in vitro experiments, DMSO or compounds were pre-treated for 30 min prior to labeling with IAyne. For in situ experiments, cells were treated with DMSO or compounds for 4 hours and cell proteomes were subsequently labeled with IAyne. For in vivo study in Tab 7, tumors were taken from the study shown in Fig. 5G.
Tabs 8–14. All peptides identified in isoTOP-ABPP experiments from Tabs 1–7.
Table S2. Structures of cysteine-reactive covalent ligand library screened in this study; Table S2 is related to Figure 3.
Highlights.
We used chemoproteomics to discover druggable hotspots targeted by withaferin A
Withaferin A targets C377 of PPP2R1A, a regulatory subunit of the PP2A complex
Withaferin A activates PP2A, inhibits AKT, and impairs breast cancer pathogenicity
We developed a more synthetically tractable covalent ligand that targets PPP2R1A
Acknowledgments
We thank the members of the Nomura Research Group for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (R01CA172667 and T32 GM066698 Training Grant for CCW and EAG), American Cancer Society Research Scholar Award (RSG14-242-01-TBE), and Department of Defense Breast Cancer Research Program Breakthroughs Award (CDMRP W81XWH-15-1-0050).
Footnotes
Author Contributions. EAG and CCW are co-first authors and contributed equally to this paper; EAG, CCW, JNS, JIK, and DKN performed experiments, analyzed and interpreted data, and wrote the paper. JNS, LAB, TRH, DKM synthesized compounds and chemical reagents for this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abegg D, Frei R, Cerato L, Prasad Hari D, Wang C, Waser J, Adibekian A. Proteome-Wide Profiling of Targets of Cysteine reactive Small Molecules by Using Ethynyl Benziodoxolone Reagents. Angew Chem Int Ed Engl. 2015;54:10852–10857. doi: 10.1002/anie.201505641. [DOI] [PubMed] [Google Scholar]
- Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, et al. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargagna-Mohan P, Hamza A, Kim Y, Khuan Abby Ho Y, Mor-Vaknin N, Wendschlag N, Liu J, Evans RM, Markovitz DM, Zhan CG, et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem Biol. 2007;14:623–634. doi: 10.1016/j.chembiol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman LA, Nguyen TB, Roberts AM, Miyamoto DK, Ku W-M, Huffman TR, Petri Y, Heslin MJ, Contreras CM, Skibola CF, et al. Chemoproteomics-enabled covalent ligand screen reveals a cysteine hotspot in reticulon 4 that impairs ER morphology and cancer pathogenicity. Chem Commun Camb Engl. 2017a doi: 10.1039/c7cc01480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman LA, Nguyen TB, Miyamoto DK, Ku W-M, Huffman TR, Heslin MJ, Contreras CM, Skibola CF, Olzmann JA, Nomura DK. Covalent Ligand Screening Reveals Cysteine Hotspot within RTN4 that Impairs ER Morphology and Colorectal Cancer Pathogenicity. Submitt ACS Chem Biol. 2017b doi: 10.1039/c7cc01480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci U S A. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Louie SM, Mulvihill MM, Kohnz RA, Li DS, Chan LG, Sorrentino A, Bandyopadhyay S, Cozzo A, Ohiri A, et al. Inositol phosphate recycling regulates glycolytic and lipid metabolism that drives cancer aggressiveness. ACS Chem Biol. 2014;9:1340–1350. doi: 10.1021/cb5001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewett MM, Xie J, Zaro BW, Backus KM, Altman A, Teijaro JR, Cravatt BF. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci Signal. 2016;9:rs10. doi: 10.1126/scisignal.aaf7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- Counihan JL, Ford B, Nomura DK. Mapping proteome-wide interactions of reactive chemicals using chemoproteomic platforms. Curr Opin Chem Biol. 2016;30:68–76. doi: 10.1016/j.cbpa.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar NJ, Hamid A, Ahmad M. Pharmacologic overview of Withania somnifera, the Indian Ginseng. Cell Mol Life Sci CMLS. 2015;72:4445–4460. doi: 10.1007/s00018-015-2012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahl C, Cravatt BF, Sorensen EJ. Protein-reactive natural products. Angew Chem Int Ed Engl. 2005;44:5788–5809. doi: 10.1002/anie.200500900. [DOI] [PubMed] [Google Scholar]
- Heyninck K, Lahtela-Kakkonen M, Van der Veken P, Haegeman G, Vanden Berghe W. Withaferin A inhibits NF-kappaB activation by targeting cysteine 179 in IKKβ. Biochem Pharmacol. 2014;91:501–509. doi: 10.1016/j.bcp.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Heyninck K, Sabbe L, Chirumamilla CS, Szarc Vel Szic K, Vander Veken P, Lemmens KJA, Lahtela-Kakkonen M, Naulaerts S, Op de Beeck K, Laukens K, et al. Withaferin A induces heme oxygenase (HO-1) expression in endothelial cells via activation of the Keap1/Nrf2 pathway. Biochem Pharmacol. 2016;109:48–61. doi: 10.1016/j.bcp.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Mueller BM, Cravatt BF. Breast cancer cell lines grown in vivo: what goes in isn’t always the same as what comes out. Cell Cycle Georget Tex. 2005;4:253–255. [PubMed] [Google Scholar]
- Lee IC, Choi BY. Withaferin-A--A Natural Anticancer Agent with Pleitropic Mechanisms of Action. Int J Mol Sci. 2016;17:290. doi: 10.3390/ijms17030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Liu J, Feng X, Salazar Hernández MA, Mucka P, Ibi D, Choi JW, Ozcan U. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat Med. 2016;22:1023–1032. doi: 10.1038/nm.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie SM, Grossman EA, Crawford LA, Ding L, Camarda R, Huffman TR, Miyamoto DK, Goga A, Weerapana E, Nomura DK. GSTP1 Is a Driver of Triple-Negative Breast Cancer Cell Metabolism and Pathogenicity. Cell Chem Biol. 2016;23:567–578. doi: 10.1016/j.chembiol.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MM, Zameer F, Naidu A, MNNP, Dhananjaya BL, Hegdekatte R. Evaluating the inhibitory potential of Withania somnifera on platelet aggregation and inflammation enzymes: An in vitro and in silico study. Pharm Biol. 2016;54:1936–1941. doi: 10.3109/13880209.2015.1123729. [DOI] [PubMed] [Google Scholar]
- Medina-Cleghorn D, Bateman LA, Ford B, Heslin A, Fisher KJ, Dalvie ED, Nomura DK. Mapping Proteome-Wide Targets of Environmental Chemicals Using Reactivity-Based Chemoproteomic Platforms. Chem Biol. 2015;22:1394–1405. doi: 10.1016/j.chembiol.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AM, Ward CC, Nomura DK. Activity-based protein profiling for mapping and pharmacologically interrogating proteome-wide ligandable hotspots. Curr Opin Biotechnol. 2016;43:25–33. doi: 10.1016/j.copbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AM, Miyamoto DK, Huffman TR, Bateman LA, Ives AN, Akopian D, Heslin MJ, Contreras CM, Rape M, Skibola CF, et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem Biol. 2017a;12:899–904. doi: 10.1021/acschembio.7b00020. [DOI] [PubMed] [Google Scholar]
- Roberts LS, Yan P, Bateman LA, Nomura DK. Mapping Novel Metabolic Nodes Targeted by Anti-Cancer Drugs that Impair Triple-Negative Breast Cancer Pathogenicity. ACS Chem Biol. 2017b;12:1133–1140. doi: 10.1021/acschembio.6b01159. [DOI] [PubMed] [Google Scholar]
- Sangodkar J, Farrington CC, McClinch K, Galsky MD, Kastrinsky DB, Narla G. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J. 2016;283:1004–1024. doi: 10.1111/febs.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Tillotson J, Wijeratne EMK, Xu Y, Kang M, Wu T, Lau EC, Mesa C, Mason DJ, Brown RV, et al. Withaferin A Analogs That Target the AAA+ Chaperone p97. ACS Chem Biol. 2015;10:1916–1924. doi: 10.1021/acschembio.5b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Hamza A, Zhang T, Gu M, Zou P, Newman B, Li Y, Gunatilaka AAL, Zhan CG, Sun D. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem Pharmacol. 2010;79:542–551. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li W, Wang M, Zhang X, Zhang H, Tong X, Xiao Y. Competitive profiling of celastrol targets in human cervical cancer HeLa cells via quantitative chemical proteomics. Mol Biosyst. 2016;13:83–91. doi: 10.1039/c6mb00691d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We mapped the cysteine-reactivity of withaferin A and DKM 2-90 by pre-incubating withaferin A (10 μM) or DKM 2-90 (100 μM) for 30 min in 231MFP breast cancer cell proteomes, prior to labeling with the cysteine-reactive iodoacetamide-alkyne (IAyne) probe (100 μM, 30 min). Probe labeled proteins were then tagged with an isotopically light (for control) or heavy (for withaferin A- or DKM 2-90-treated) biotin-azide tag bearing a TEV protease recognition site by copper-catalyzed azide-alkyne cycloaddition. Control and treated proteomes were then mixed in a 1:1 ratio, probe labeled proteins were avidin-enriched and tryptically digested, probe-labeled tryptic peptides were avidin-enriched again, and released by TEV protease and analyzed by quantitative proteomic methods and light to heavy peptide ratios were quantified. Competitive isoTOP-ABPP analysis of withaferin A and DKM 2-90 cysteine-reactivity in 231MFP breast cancer cell proteomes. Light to heavy ratios of ~1 indicate peptides that were labeled by IAyne, but not bound by withaferin A or DKM 2-90. We designate light to heavy ratios of >10 as targets that were bound by withaferin A or DKM 2-90. Tabs 1 and 3 shows average light versus heavy ratios from those probe-modified peptides found in at least 2 out of 3 biological replicates.
Tabs 1–7. IsoTOP-ABPP analysis of withaferin A (10 μM), DKM 2-90 (100 μM), JNS 1-40 (100 μM) treatment in vitro in 231MFP proteomes or in situ in 231MFP cells, or 231MFP tumors from JNS 1-40 daily in vivo treatment in mice. For in vitro experiments, DMSO or compounds were pre-treated for 30 min prior to labeling with IAyne. For in situ experiments, cells were treated with DMSO or compounds for 4 hours and cell proteomes were subsequently labeled with IAyne. For in vivo study in Tab 7, tumors were taken from the study shown in Fig. 5G.
Tabs 8–14. All peptides identified in isoTOP-ABPP experiments from Tabs 1–7.
Table S2. Structures of cysteine-reactive covalent ligand library screened in this study; Table S2 is related to Figure 3.