Abstract
Loss-of-function mutations in the tyrosine kinase JAK3 cause autosomal recessive (AR) severe combined immunodeficiency (SCID). Defects in this form of SCID are restricted to the immune system, which led to the development of the immunosuppressive JAK inhibitors. We find that the B6.Cg-Nr1d1tm1Ven/LazJ mouse line purchased from Jackson Laboratories harbors a spontaneous mutation in Jak3 generating a SCID phenotype, with an inability to generate antigen-independent professional cytokine-producing innate lymphoid cells (ILCs). Mechanistically, Jak3 deficiency blocks ILC differentiation in the bone marrow at the ILC progenitor (ILCP) and the pre-NK cell progenitor (pre-NKP). We further demonstrate that the pan-JAK inhibitor tofacitinib and specific JAK3 inhibitor PF-06651600 impair the ability of human intraepithelial ILC1 (iILC1) to produce IFN-γ, without affecting ILC3 production of IL-22. Both inhibitors impaired the proliferation of iILC1 and ILC3 and differentiation of human ILC in vitro. Tofacitinib is currently approved for the treatment of moderate-to-severely active rheumatoid arthritis (RA) and is under clinical trial for several other immune-mediated conditions. Our data suggests that therapeutic inhibition of JAK may also impact ILCs and, to some extent, underlie clinical efficacy.
Keywords: ILC, innate immune, Jak3, development, immunodeficiency, human
INTRODUCTION
Primary immunodeficiencies (PIDs) are inborn errors that cause developmental and/or functional defects in the immune system1. Most frequently rare and monogenic, PIDs present clinically with a broad array of phenotypes including but not limited to increased susceptibility to infection, autoimmunity, and cancer. One of the most deadly categories of PID is SCID, which typically presents early in childhood with failure to thrive and severe recurrent infections, and is universally fatal if untreated2. SCID is invariably caused by severe developmental and/or functional defects of T lymphocytes, but may also present with variable defects of B and/or Natural Killer (NK) cells depending on the type of causative mutation1, 3.
The most common variant of SCID is X-linked (X-SCID) and is caused by inactivating mutations in IL2RG, which encodes the common gamma chain or γc3, 4. Although the γc is shared between many cytokines (IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21), defects in lymphocyte generation are due to loss of IL-7 and IL-15 signaling, which regulate thymocyte and NK cell differentiation and survival, respectively5. Therefore, γc mutations cause a severe defect of both T and NK lymphocytes. Furthermore, mice (but not humans) with Il2rg defects also present an additional defect in the development of B cells due to a species-difference in their requirement for IL-75. In both human and mouse, the γc signals through the tyrosine kinase JAK3; therefore, inactivating mutations in JAK3 phenocopy X-SCID, except for their AR inheritance pattern6, 7.
Patients with either X-SCID or AR JAK3 SCID are cured by hematopoietic cell transplant (HCT) and lack significant deficits in other organ systems2, 8. Based on the restricted immune defect in these SCID variants, the JAK inhibitor tofacitinib was developed to induce therapeutic immunosuppression, with the rationale that a more targeted approach may be clinically superior to calcineurin inhibitors and corticosteroids, which act on many different cell types and can have substantial off-target effects8. Reminiscent of T cell immunodeficiency in SCID, tofactinib has been shown to inhibit CD4+ T cell polarization9, and it is now an effective, approved treatment for moderate-to-severely active RA10. Although tofacitinib was originally developed as a JAK3 specific inhibitor, it is now recognized to have pan-JAK activity. Thus, a new selective JAK3 inhibitor, PF-06651600, was recently engineered from tofacitinib by targeting one of two unique residues in the JAK3 ATP-binding domain, Cyp-90911, 12.
Innate lymphoid cells (ILCs) are a recently discovered class of lymphocytes enriched at mucosal surfaces in both human and mice that are antigen-independent, cytokine-activated correlates of T cells13. These cells include NK cells, an innate counterpart to CD8+ killer T cells, as well as three additional groups of ILCs that mirror the polarized CD4+ helper T cell subsets Th1, Th2, and Th17, called ILC1, ILC2, and ILC3 respectively. Each ILC and CD4+ T cell polarization module shares core transcription factor (TF) regulators and the capacity to produce shared signature cytokines effectors13, 14. Thus, ILC1s and Th1s use the TF TBET and produce IFN-γ, ILC2s and Th2s utilize GATA3 and secrete IL-5 and IL-13, and ILC3s and Th17s express RORγt and generate IL-22 and/or IL-1714–17. In mice, ILC3s are an especially diverse lineage. These cells include: a fetal lymphoid tissue inducer (LTi) cell responsible for initiating lymphoid tissue development via lymphotoxin-β signaling to the stroma; an adult CCR6+ LTi-like ILC3 population; and two other ILC3 populations which are developmentally distinct from LTi and LTi-like cells but share core TFs and the ability to produce IL-22, called NKp46+ ILC3 and NKp46−CCR6− double negative (DN) ILC316. Recent data from single-cell analysis of human ILCs suggest there may also be similar diversity among human ILC318.
ILCs are exquisitely interconnected with cytokines, depending on them for their development, activation, and function. In mice, the development of ILC2 and ILC3 predominantly requires IL-7 like T cells, while the development of ILC1 predominantly requires IL-15 like NK cells14, 16, 19. Furthermore, the polarized cytokines that mouse ILCs produce are now thought to substantially contribute to a variety of homeostatic and disease processes, which have been extensively reviewed20–23. However, the developmental requirements for human ILCs, their contribution to homeostasis and disease, and their degree of regulation by immunosuppressive drugs are substantially less clear24. Here, we identify a spontaneous mutation in Jak3 in the Jackson Nr1d1tm1Ven/LacZ mouse line causing a SCID phenotype with impaired lymphoid tissue development, and isolate the first Jak3 deficient line on the C56BL6/J background. We also identify a critical role for Jak3 in mouse ILC development and test the effect of acute pharmacologic JAK3 inhibition on human ILCs development, proliferation, and function.
RESULTS
AR phenotype in Nr1d1tm1Ven/LacZ line leads to impaired ILC development
We recently reported that Nr1d1, which encodes the transcriptional repressor REV-ERBα, is part of a core molecular signature that distinguishes ILC3 from other ILC subsets25. To test if REV-ERBα is important for ILC3, we purchased Nr1d1tm1Ven/LacZ mice (lacking REV-ERBα expression) from Jackson Laboratories. During the preliminary analysis of these mice, we noted a phenotype characterized by lack of lymphoid tissue development that emerged in the F1 from original Jackson breeders and that segregated independently from Nr1d1tm1Ven alleles, which we called Wolverine (WLVRN) (Figure 1A). Grossly, mice with the WLVRN phenotype lacked Peyer’s patches (PPs) and peripheral lymph nodes as measured by inguinal lymph node presence or absence, and also demonstrated thymic aplasia compared to littermates (Figure 1B and data not shown). Pedigree analysis demonstrated an AR pattern of inheritance (Figure 1A). We tested this interpretation by backcrossing an Nr1d1+/+ mouse with the WLVRN phenotype to C57BL6/J and intercrossing the F1. Supporting the conclusion that the phenotype was AR, F1 mice were phenotypically normal, while the WLVRN phenotype reemerged in the F2 (Figure 1C). Collectively, these data suggested that the Jackson-derived Nr1d1tm1Ven/LacZ mouse line most likely carried a second mutation unrelated to Nr1d1.
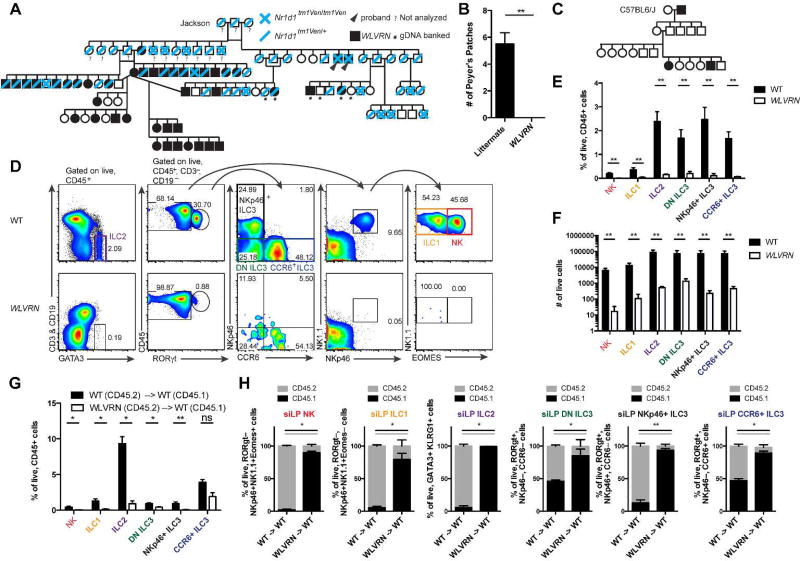
Figure 1. ILCs fail to develop in WLVRN mice.
(A) Pedigree analysis of intercrossed heterozygous B6.Cg-Nr1d1tm1Ven/LazJ mice beginning with original breeding trio purchased from Jackson Laboratories. WLVRN probands emerge in F1. (B) Number of PPs in WLVRN mice compared to littermate controls. (C) Pedigree analysis of an Nr1d1+/+ WLVRN mouse backcrossed to C57BL6/J. (D–F) Evaluation of ILC populations from the siLP of Nr1d1+/+ WLVRN or unrelated WT control mice. (D) Gating strategy and representative flow plots. (E) Frequency and (F) total number of ILC subsets. (G–H) CD45.2 WT or WLVRN donors were transplanted into CD45.1 WT lethally irradiated recipients. (G) Frequency of recovered siLP ILCs from bone marrow chimeras and (H) the degree of chimerism per subset 8 weeks post-irradiation.. (B, D–H) Data represent (B, D–F) n=5 mice per group or (G–H) n=4–5 mice per group from 2–3 independent experiments. *p<.05, ** p<.01, two-tailed Mann-Whitley test.
As lymphoid tissue inducer (LTi) cells are critically important for lymphoid tissue development and are a fetal member of the ILC3 lineage16, we assessed ILCs in Nr1d1+/+ WLVRN mice compared to unrelated WT controls. In adult mice, we found that NK cells, ILC1, ILC2, and ILC3 subsets excluding CCR6+ ILC3 were significantly reduced in frequency in the small intestine lamina propria (siLP) of mice with the WLVRN phenotype. All ILCs were significantly reduced in total number, with each subset amounting to less than 1500 cells in WLVRN mice, a reduction of over 100-fold compared to WT mice for all subsets except DN ILC3, which were reduced by 36-fold (Figure 1D–F). This phenotype was intrinsic to the immune system, as significantly fewer ILCs could be recovered from the siLP of irradiated CD45.1 mice that received CD45.2 WLVRN marrow compared to those that received CD45.2 WT marrow after 8 weeks of reconstitution (Figure 1G). Moreover, among the ILCs that could be recovered, there was significantly less CD45.2+ donor reconstitution by transplanted WLVRN marrow than by WT among all of the ILC subsets. In fact, most ILCs from recipients of WLVRN marrow remained CD45.1+ host-derived at 8 weeks, while in recipients of WT marrow, only radioresistant DN and CCR6+ ILC326 subsets remained substantially CD45.1+ host-derived (Figure 1H). Thus, the putative mutation causing the WLVRN phenotype led to a cell-intrinsic block in ILC development, and confounded the interpretation of ILC biology in the Nr1d1 tm1Ven/LacZ mouse line.
WLVRN phenotype impairs lymphoid and increases myeloid development
Given the immune-intrinsic deficiency in all ILC subsets, we next asked if the development of other lymphoid cells was similarly impaired in Nr1d1+/+ WLVRN mice. Evaluation of splenic lymphocytes demonstrated a dramatic reduction in both innate and adaptive cells (Figure 2A), with significantly reduced frequency of B cells, CD8+ T cells, and NK cells among CD45+ immune cells (Figure 2B). All lymphocytes were significantly reduced in total number (Figure 2C). Similar to siLP ILCs, the most dramatic decrease was found among splenic NK cells with an over 100-fold reduction in their total number between WT and WLVRN mice (341-fold), followed by B cells (150-fold), CD8+ T cells (91-fold) and CD4+ T cells (6-fold) (Figure 2C). Consistent with the relative preservation of CD4+ T cells by total number, these cells were not significantly reduced in frequency compared to WT mice (Figure 2B), and were the major population of lymphocytes found in mice with the WLVRN phenotype (Figure 2D). As CD4+ T cells appeared to be selectively expanded, we next asked if T cell subsets in WLVRN mice were activated or naive (Figure 2D). The majority of T cells in both the CD4+ and CD8+ lineages from WLVRN mice were CD62L−CD44+ effector memory cells, which were significantly greater in frequency compared to WT mice (Figure 2E). We also found a significant and concomitant decrease in naive CD62L+CD44− cells in both T cell subsets, and a significant decrease in CD62L+CD44+ central memory T cells among CD4+ T cells from WLVRN mice compared to WT. This phenotype was reminiscent of Il2rg−/− SCID mice, which have dramatically impaired lymphoid development and expansion of residual T cells3, 4.
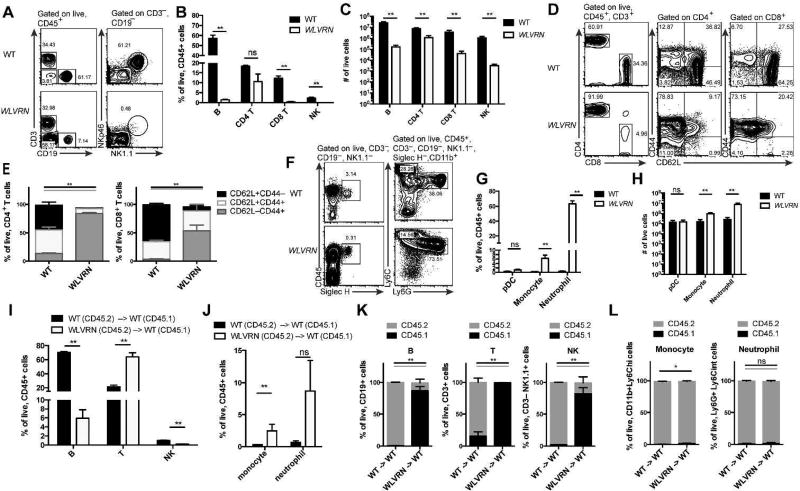
Figure 2. Wolverine mice have decreased lymphoid development.
(A–H) Spleen was isolated from Nr1d1+/+ WLVRN or unrelated WT control mice. Lymphoid (A–E) and myeloid (F–H) populations were assessed. (A) Gating strategy and representative flow plots for lymphocyte analysis. (B) Frequency and (C) total number of lymphocyte subsets. (D) Gating strategy, representative flow plots, and (E) quantification of T cell populations. (F) Gating strategy and representative flow plots for myeloid analysis. (G) Frequency and (H) total number of selected myeloid subsets. (I–L) CD45.2 WT or WLVRN donors were transplanted into CD45.1 WT lethally irradiated recipients. Frequency of recovered (I) spleen lymphocytes and (J) spleen myeloid cells from bone marrow chimeras and (K–L) the degree of chimerism per subset 8 weeks post-irradiation. (A–L) Data represent n= 5 mice per group from 2–3 independent experiments. *p<.05, ** p<.01, two-tailed Mann-Whitley test.
Because myeloid development is intact in SCID mice, we next asked if myeloid development was normal in Nr1d1+/+ WLVRN mice. Indeed, development of pDC, monocytes, and neutrophils was intact (Figure 2F), with a significantly higher frequency and total number of monocytes and neutrophils in the spleen of WLVRN mice compared to WT controls (Figure 2G–H). Lymphoid and myeloid phenotypes were intrinsic to the immune system, because bone marrow chimeras generated with CD45.2 WLVRN marrow or CD45.2 WT marrow into CD45.1 recipient mice demonstrated a significant reduction in recovered B and NK cells in mice that received WLVRN marrow compared to those that received WT marrow (Figure 2I), and a significant increase in recovered monocytes with a trend for recovered neutrophils (Figure 2J). Moreover, among the cells that were recovered, splenic T, B, and NK cells from mice that received WLVRN marrow were predominantly host-derived after 8-weeks reconstitution compared to those that received WT marrow (Figure 1K), while splenic monocytes and neutrophils were almost entirely donor-derived from both WLVRN and WT donors (Figure 1L). From these data we concluded that WLVRN mice have a SCID phenotype with selective and profound inability to generate all lymphocytes. The developmental block in lymphocyte generation was greatest for ILCs, which may lack an intrinsic program to undergo significant and rapid clonal expansion. Given that this phenotype originated at Jackson, this observation is additionally notable because most studies of REV-ERBα deficient mice have thus far studied macrophages or non-immune populations and could potentially miss this confounding immune defect.
WLVRN phenotype is caused by a spontaneous frameshift mutation in Jak3, abrogating C-terminal kinase activity
To determine if there was a spontaneous mutation in the Nr1d1tm1Ven/LacZ line responsible for the WLVRN phenotype, we performed whole exome sequencing (WES) on genomic DNA banked from a male mouse with the WLVRN phenotype lacking Nr1d1tm1Ven alleles (Figure 1A). Analysis of sequencing results revealed 2423 SNPs and 571 insertions or deletions compared to the reference C57BL6/J genome, of which 126 SNPs and 136 insertions/deletions were homozygous (data not shown). Among these were 5 frameshift variants (Table 1), which included Jak3. Published mouse studies from Jak3 deficient mice describe a phenotype similar to WLVRN mice; Jak3tm1Ljb/tm1Ljb mice have diminished lymphoid and increased myeloid development, with residual lymphocytes skewed to activated, oligoclonally expanded CD4+ T cells27, 28. Furthermore, ILC development is known to be critically dependent upon γc15, 16, which signals through Jak35. Thus, we hypothesized that the WLVRN phenotype was caused by a Jak3 mutation.
Table 1.
| Function | Gene | Type | Chr | Start | End | Reference | Observed |
|---|---|---|---|---|---|---|---|
| frameshift variant | Tctn2 | Indel | chr5 | 124624334 | 124624334 | T | TAC |
| frameshift variant | Auts2 | Indel | chr5 | 131476702 | 131476702 | A | AG |
| frameshift variant | Jak3 | Indel | chr8 | 71684243 | 71684243 | G | GC |
| frameshift variant | Bean1 | Indel | chr8 | 104182033 | 104182033 | TC | T |
| frameshift variant+stop lost | Mmp1a | Indel | chr9 | 7465083 | 7465083 | T | TG |
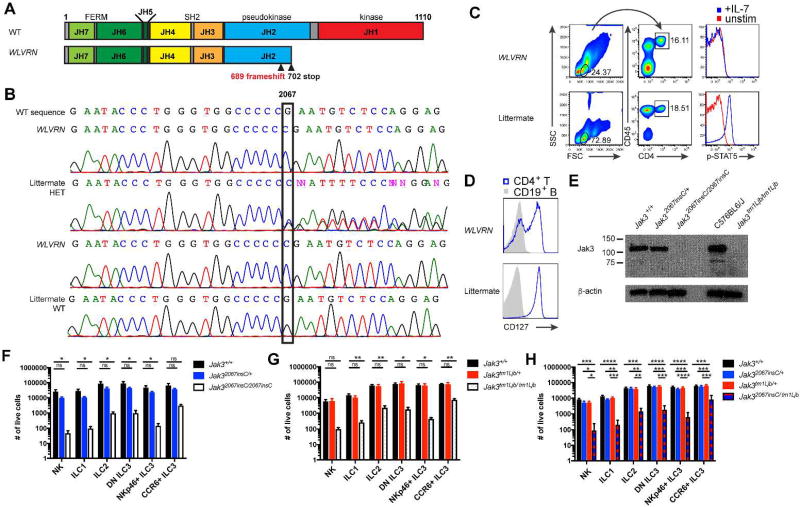
The mutation identified in Jak3 corresponds to the insertion of an additional cytosine residue 3’ to a stretch of five cytosines, at position 2067 of the coding mRNA, in exon 14 of the gene (Table 1). This leads to a frameshift at position 689 within the pseudokinase domain of Jak3 and the introduction of a stop codon at position 702, thereby resulting in the expression of a truncated Jak3 molecule lacking the C-terminal kinase domain responsible for transmitting γc–dependent signals (Figure 3A). PCR amplification around the predicted mutation in exon 14 from genomic DNA of littermates discordant for the WLVRN phenotype in the original pedigree (Figure 1A), followed by Sanger sequencing, confirmed homozygosity for the c.2067insC mutation in phenotypically affected mice, whereas phenotypically normal littermates were either homozygous for the wild-type sequence or showed two traces diverging at position 2067 (Figure 3B). We next assessed if Nr1d1+/+ WLVRN mice were unable to signal through γc cytokines. As predicted, splenic CD4+ T cells from WLVRN mice were unable to phosphorylate STAT5 in response to 15 minutes of IL-7 stimulation, while littermate CD4+ T cells could robustly do so (Figure 3C). This defect was not simply due to lack of the receptor expression, as CD4+ T cells from both WLVRN mice and littermates expressed the receptor for IL-7, CD127 (Figure 3D), albeit at lower levels in WLVRN mice. To test if Jak3 protein was present in WLVRN mice, we next performed an immunoblot, using an N-terminal anti-Jak3 antibody to bone marrow derived macrophage (BMDM) lysates generated from Sanger-sequenced Nr1d1+/+ littermates. In Jak3+/+, Jak32067insC/+, and unrelated WT mice, expression of the Jak3 protein was clearly detected, but it was absent in Jak32067insC/2067insC mice and Jak3tm1Ljb/tm1Ljb negative controls. Thus, WLVRN mice failed to express full-length Jak3 with an intact C-terminus kinase domain. Although we did not detect a truncated protein in mice with Jak32067insC alleles, we hypothesize that the frameshifted variant may be unstable or improperly folded and therefore undergo degradation.
Figure 3. Jak32067insC causes the WLVRN phenotype.
(A) Diagram of mouse Jak3 with location of predicted frameshift mutation and introduced stop codon based on WES. (B–H) Analysis of littermates discordant for the WLVRN phenotype, either from (B) original Nr1d1tm1Ven/LazJ pedigree or (C–H) isolated Nr1d1+/+ WLVRN line. (B) Sanger sequencing of Jak3 exon 14 from banked genomic DNA (1A). Representative flow plots of (C) p-STAT5 from CD4+ T cells after 15 minutes of IL-7 stimulation or (D) CD127 expression from resting CD4+ T cell and CD19+ B cells. (E) Immunoblot for Jak3 from BMDMs. (F–H) Total number of ILCs in littermates from (F) Jak32067insC/+ intercross, (G) Jak3tm1Ljb/+ intercross, and (H) Jak32067insC/+ × Jak3tm1Ljb/+ intercross as gated in 1D, but without the use of NK1.1 for Jak3tm1Ljb mice which are on a mixed 129S4 and C57BL6/J background. Data represent (F) n=4 (G) n=5 or (H) n= 6 mice per genotype and (C–D, F–G) 3 (E) 1, or (H) 4 independent experiments. *p<.05, **p<.01, ***p<.001, ****p<.0001 one-way ANOVA with Tukey’s multiple comparisons test.
To determine if the Jak32067insC mutation caused the WLVRN phenotypic lack of siLP ILCs and loss of lymphoid tissue development, we first assessed littermates genotyped by Sanger sequencing. We found that there was no significant difference between WT and heterozygous mice, but significant decreases in all ILC subsets in homozygous mice (Figure 3F). Homozygous mice also lacked lymphoid tissue development (data not shown). This defect phenocopied littermates from intercrossed Jak3tm1Ljb/+ mice, which are on a mixed 129S4 and C57BL6/J background (Figure 3G and data not shown). Finally, to test directly if Jak3 deficiency caused these WLVRN phenotypes, we intercrossed Jak3tm1Ljb/+ and Jak32067insC/+ and found that Jak32067insC/ tm1Ljb mice had reductions in ILCs and lacked lymphoid tissue development, while there was no difference between WT and either heterozygous allele (Figure 3H and data not shown). From these data, we conclude that the WLVRN phenotype is caused by a spontaneous loss-of-function mutation in Jak3, leading to the first C57BL6/J Jak3 deficient mouse line. We also conclude that Jak3 is critically important for the development and/or maintenance of all ILCs.
Jak3 deficiency blocks development at the ILCP and pre-NKP stages
The stage at which Jak3 is required for ILC development is unknown. Therefore, we next tested the developmental requirements for Jak3 within progenitor populations in the bone marrow. ILCs develop along a sequence of cells with progressively restricted fates15, 16: Downstream of the common lymphoid progenitor (CLP) that generates all lymphoid cells, early innate lymphoid progenitors (EILPs)29 and α–lymphoid precursors (αLP)30 give rise to all ILCs, a common helper ILC precursor (CHILP) to ILCs excluding NK cells31, an ILC precursor (ILCP) to ILC1, ILC2, and ILC3 cells excluding the CCR6+ ILC3 subset32, 33, and an ILC2 precursor (ILC2P) to ILC231, 34. Meanwhile, NK cells develop from the sequence of pre-NK precursors (pre-NKP), refined NK precursors (rNKP), and immature NK cells (iNK), with ILC1 phenotypic overlap most marked in iNK35, 36.
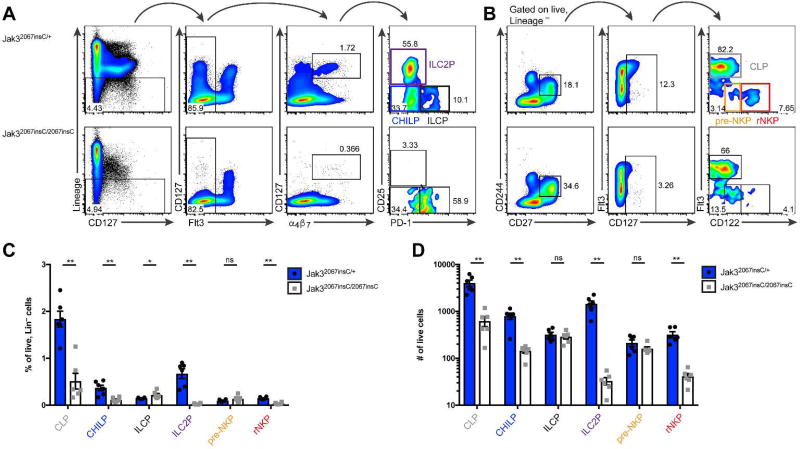
We assessed CLPs, CHILPs, ILCP, ILC2Ps, pre-NKP, and rNKP in C56BL6/J Jak32067insC /+ and Jak32067insC/2067insC littermates (Figure 4A–B), excluding EILP which require genetic means of identification. We found that CLP, CHILP, ILC2P, and rNKP were reduced in Jak3-deficient mice compared to sufficient littermates (Figure 4C–D), both as the frequency of lineage negative cells (Figure 4C) and the total number within two tibias (Figure 4D). In contrast, the frequency of ILCP was increased in Jak3-deficient mice than Jak3-sufficient littermates (Figure 4C). The frequency of pre-NKP and total numbers of ILCP and pre-NKPs were unchanged between genotypes (Figure 4C–D). We conclude that Jak3 deficiency blocks differentiation at the ILCP and pre-NKP stage to mature ILCs and NK cells, respectively.
Figure 4. Jak3 deficiency blocks ILC development at the ILCP and pre-NKP.
(A–D) Analysis of lymphoid progenitors from Jak32067insC/+ and sex-matched Jak32067insC/2067insC littermates. (A–B) Representative flow plots of bone marrow progenitors among lineage (CD3ε, CD4, CD8a, CD11b, CD11c, CD19, NK1.1, GR1, Ter-119, B220) negative cells electronically gated by FMO controls. (A) CHILP: CD127+Flt3−α4β7+CD25−PD-1−; ILCP: CD127+Flt3−α4β7+CD25−PD-1+; ILC2P: CD127+Flt3−α4β7+CD25+ PD-1−. (B) CLP: CD127+CD244+CD27+Flt3+; pre-NKP: CD127+CD244+CD27+Flt3−CD122lo; rNKP: CD127+CD244+CD27+Flt3−CD122+. (C) Frequency of progenitors among lineage negative cells. (D) Total number of progenitors in two tibias. Data represent n = 6 mice per genotype from 2 independent experiments. *p<.05 ** p<.01, two-tailed Mann-Whitley test.
JAK3-inhibition diminishes human ILC proliferation, function, and development
Based on our finding that Jak3 was critically important for mouse ILCs and prior reports that NK cells are reduced in patients treated with the JAK inhibitor tofacitinib37, we hypothesized that this inhibitor may also affect human ILCs. To test this, we sorted iILC1 and ILC3 from pediatric tonsil specimens and either expanded them in vitro or tested ILC3 directly ex-vivo, as previously described38. We first incubated cells acutely with low (20 nmol) or high doses (100 nmol) of tofactinib, corresponding to a selective but partial inhibition of JAK3 at low dose and dual JAK3/JAK1 inhibition with minimal JAK2 inhibition at high dose9. We found that both expanded iILC1 (Figure 5A–B) and ILC3 (Figure 5C–D) were substantially less proliferative in the presence of tofacitinib. Both iILC1 and ILC3 incorporated significantly less BrdU in a dose-dependent manner, and their proliferation almost completely blocked at high dose (Figure 5A–D). To test if the proliferative defect caused by tofactinib was truly JAK3 mediated, we next tested the effect of the JAK3-specific inhibitor PF-06651600 on iILC1 (Figure 5E) and ILC3 (Figure 5F) proliferation compared to tofactinib. Mirroring tofacitinib, we found that PF-06651600 abrogated ILC proliferation. Thus, human ILC proliferation is JAK3 dependent, at least in culture conditions.
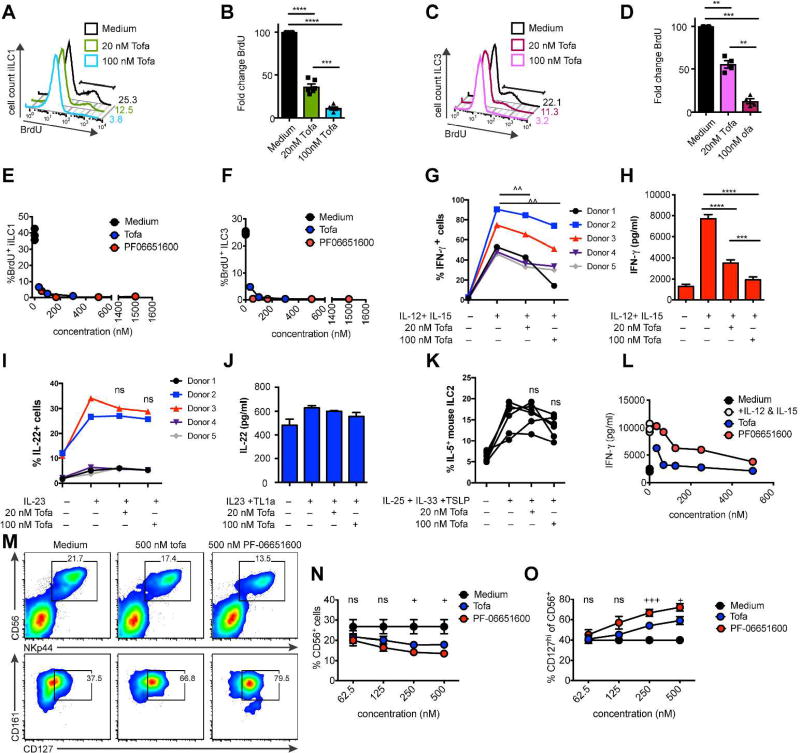
Figure 5. JAK3 inhibition impairs human ILC proliferation and differentiation.
(A–D) Effect of 48-hour tofacitinib and (E–F) PF-06651600 culture on BrdU incorporation in expanded (A–B, E) iILC1 and (C–D, F) ILC3. Data show (A,C) representative flow plots, (B,D) quantification, and (E–G) representative dose-response curves. (G–H) Effect of 48-hour tofacitinib culture on IFN-γ in expanded human tonsillar iILC1. Data show (G) percentage of IFN-γ+ cells and (H) representative IFN-γ protein in supernatant of Donor 3 by CBA. (I) Percentage of IL-22+ ILC3 cells after IL-23 stimulation ex-vivo in the presence of the indicated dose of tofacitinib. (J) Representative IL-22 protein in supernatant of expanded human tonsillar ILC3 after 48-hours of tofacitinib. (K) Percentage of IL-5+ mouse ILC2 after stimulation ex-vivo in the in the presence of the indicated dose of tofacitinib. (L) Representative dose-response curve of IFN-γ protein in supernatant of cells treated for 24 hours ex-vivo with tofacitinib and PF06651600. (M–O) Purified peripheral blood CD34+ cells were cultured in ILC-polarizing conditions for 14 days then incubated with the indicated concentration of tofacitinib and PF06651600. (M) Representative flow plots of differentiated human ILC. (N) Quantification of percent CD56+NKp44+ cells among lineage negative lymphocytes. (O) Quantification of percent CD127+ cells among CD56+NKp44+ cells. Data represent (A–D, G–J) n = 5 donors, (E–F, L) n= 2 donors, (K) n = 6 mice, or (M–O) n = 3 donors. ^p<.05, paired t-test. *p<.05, **p<.01, ***p<.001, ****p<.0001, one-way ANOVA with Tukey’s multiple comparison test. +p<.05, +++p<.001, ordinary oneway ANOVA.
We next tested the role of JAK3 in ILC function. In response to stimulation with IL-12 and IL-15, significantly fewer expanded iILC1 cultured with tofacitinib produced IFN-γ at low dose compared to control, an effect that was even greater at high dose (Figure 5G). This corresponded to a highly significant, dose-dependent decrease in IFN-γ protein in the supernatant (Figure 5H). In contrast, in response to IL-23 stimulation ex-vivo, ILC3 stimulated in the presence of tofacitinib produced IL-22 equally well compared to control; there was no significant difference in the percent of ILC3 that produced IL-22 (Figure 5I). Similarly, we found no difference in IL-22 protein levels in the supernatant between expanded ILC3 stimulated in the presence of tofacitinib and control (Figure 5J), although expanded ILC3 also responded less to stimulation with IL-23 than primary cells as we have previously reported38. CRTH2+ ILC2 are rare populations within the human tonsil and could not be tested39, therefore we tested Jak inhibition in mouse ILC2. Like human ILC3, mouse ILC2 stimulated ex-vivo with IL-25, IL-33, and TSLP in the presence of tofacitinib also demonstrated no difference in IL-5 production (Figure 5K). We next asked if the reduction in IFN-γ by iILC1 was JAK3-dependent. Unlike proliferation data, JAK3-specific inhibition by PF-06651600 did not phenocopy tofacitinib, instead exhibiting a lesser effect (Figure 5L). We conclude that tofacitinib impairs cytokine production by iILC1 but not ILC3, but this effect is only partially JAK3-dependent.
Finally, we tested the effect of tofactinib or PF-06651600 inhibition on ILC differentiation from human peripheral blood CD34+ hematopoietic progenitor cells (HPC) in vitro. After 72 hours, we noted a significant dose-dependent reduction in the percentage of CD56+NKp44+ ILCs (Figure 5M–N), which have previously been shown to include NK cells and ILC340. Furthermore, the phenotype of cells from tofactinib or PF-06651600 cultures was altered, with a dose-dependent significant increase in expression of CD127 (Figure M, O). Both effects were equal or greater in PF-06651600 conditions compared to tofactinib (Figure M–O), suggesting that they are predominantly JAK3-dependent.
DISCUSSION
Collectively, we demonstrate that the Nr1d1tm1Ven/LacZ line harbors a spontaneous loss-of-function mutation in Jak3 that originated in Jackson Laboratories and is responsible for an AR SCID phenotype marked by loss of lymphoid tissue development and a lack of ILCs. Given that this mutation arose at Jackson and profoundly effects mice, these results may be particularly notable to investigators who have purchased this mouse line and can easily be tested through methods we describe. Isolated from Nr1d1 tm1Ven alleles, the Jak32067insC mutation is the first Jak3-deficient mouse model of SCID on the C57BL6/J background. To our knowledge, this is also the first time Jak3 has been shown to be critical to the development of adult ILCs in mouse.
How does loss of Jak3 impact ILC generation? Through analysis of bone marrow, we show that Jak3 deficiency causes a block in restricted ILC progenitors as they differentiate toward mature ILCs at the ILCP and pre-NKP stages. Surprisingly, progenitors both proximal and distal to these stages were reduced in frequency and total number, though the magnitude of this effect was greater in distal ILC2P and rNKP. These data suggest that that all lymphocyte progenitors require Jak3 for optimal fitness, but that Jak3 signaling is required for the normal differentiation of ILCP and pre-NKP and that these cells accumulate in its absence. It also remains possible that the phenotype of progenitors may be different in the absence of Jak3, which we could not directly test due to the inability of any progenitors to effectively generate mature ILCs. Similar to our data in mouse, acute JAK3 inhibition in vitro also reduces the generation of human CD56+NKp44+ ILCs from CD34+ HSCs. Interestingly, human ILCs are skewed to a CD127+ phenotype upon JAK3 blockade, which is a marker of human ILC3 but not NK cells38. Whether JAK3 inhibition skews progenitor polarization to ILC3, more acutely inhibits NK cell proliferation or survival than ILC3, or simply causes increased CD127 expression will require further investigation.
After development, Jak3 deficiency likely also impacts ILC survival in the periphery. Here, we demonstrate that tofacitinib dramatically reduced proliferation of mature human iILC1 and ILC3 in vitro, which was completely JAK3-dependent. These data are consistent with our recent report showing partial functional redundancy between IL-7 and IL-15 in ILC development in vivo and survival in vitro.19 In vitro, mouse ILC3 are also the most resistant to extended γc cytokine depletion19, 41.
In most human PID, ILCs have yet to be evaluated and may differ from mouse, similar to the divergent requirements for IL-7 in B cell development between species5. Recently, ILCs were shown to be absent in the peripheral blood of human subjects with JAK3 SCID. These populations did not recover after non-myeloblative bone marrow transplants, with no significant adverse affects over 7–39 years of follow-up.42 These data highlight the partial redundancy that we recently reported between human ILC and T cell transcriptional and epigenetic programs39. However, from this small sample of patients without thorough assessment of tissues where ILC predominantly reside, one cannot exclude that ILC may have important roles in tissue homeostasis, early infection control, or the development of disease, which may be context dependent.
The degree to which human ILC are affected by immunosuppressive therapies remains mostly unknown. As ILC are primarily a tissue-resident population in both human and mouse, we were unable to assess ILC function from patients treated with JAK3 inhibitors directly ex-vivo. However, we were able to test the role of acute-JAK3 inhibition in vitro on prevalent human tonsillar ILC1 and ILC3 populations using tofacitinib and PF-06651600. Beyond the most notable inhibition of ILC proliferation, tofacitinib substantially also reduced iILC1 production of IFN-γ, was partially JAK3-dependent. At the doses of tofacitinib we chose for our study, there is minimal impact of JAK2 function9, which along with TYK2 transduces both IL-12 and IL-23 signals43. Consistent with this, we found no impact of tofacitinib on human ILC3 production of IL-22 after stimulation with IL-23 or mouse ILC2 production of IL-5 after stimulation with IL-25. Thus, the reduced production of IFN-γ by iILC1 we found is unlikely to be caused by direct blockade of JAK2 signaling, but may be JAK1-dependent.
Interestingly, JAK inhibition with tofacitinb has thus far shown clinical efficacy in RA10 and psoriasis44, but have had surprisingly weak phase-two results in Chron’s disease trials45. Our data that tofacitinib can repress antigen-independent iILC1 IFN-γ production but not ILC3 production of IL-22 suggests that there may also be a differential effect on these cells in a therapeutic context. In the long term, reduced ILC proliferation combined with inhibited iILC1 function may further contribute to the therapeutic efficacy of this drug The selective JAK3 inhibitor PF-06651600 is currently enrolling phase-one clinical trials for multiple autoimmune and autoinflammatory conditions. Based on our data, this compound may more selectively impact ILC numbers than function, leaving responsiveness to inflammatory cytokines intact.
MATERIAL AND METHODS
Mice
B6.Cg-Nr1d1tm1Ven/LazJ, B6;129S4-Jak3tm1Ljb/J, and C57BL6/J mice were purchased from the Jackson Laboratory. All mice were bred and maintained in a pathogen-free facility at Washington University. Age- and sex-matched animals were used in experiments when possible, and were analyzed between 4 and 16 weeks of age. The WUSM Animal Studies Committee approved all experiments.
Antibodies and flow cytometry
Anti-α4β7 (DATK32), anti-B220 (RA3-6B2), anti-CD3ε (145-2C11), anti-CD4 (GK1.5), anti-CD8a (53-6.7), anti-CD11c (N418), anti-CD19 (ebio1D3), anti-CD27 (LG.7F9), anti-CD45 (30-F11), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD62L (MEL-14), anti-CD244 (eBio244F4), anti-GR1 (RB6-8C5), anti-EOMES (Dan11mag), anti-Flt3 (A2F10), anti-KLRG1 (2F1), anti-Ly6C (HK1.4), anti-NK1.1 (PK136), anti-NKp46 (29A1.4), anti-PD-1 (J43), anti-RORgt (AFKJS-9), anti-Ter-119 (TER-119), anti CD3ε), anti-CD18 (HB19), anti-CD127 (eBioRDR5), anti-IFN-γ (eBio45.B3), SAV Pe-Cy7, SAV APC-Fluor 780, and 123count eBeads were obtained from eBioscience. Anti-Ly6G (1A8), anti-CD19 (6D5), anti-CD25 (PC61), anti-CD122 (TM-β1), anti-CD127 (A7R39), anti-Ly6G (1A8), anti-Siglec H (551), anti-CD34 (581) and anti-CD161 (HP-3G10) were obtained from BioLegend. Anti-CCR6 (140706), anti-CD11b (M1/70), anti-CD44 (IM7), anti-CD45 (30-F11), anti-GATA3 (L50-823), anti-IL-5 (TRFK5), SAV BV421, anti-CD56 (B159), and anti-NKp44 (p44.8.1) were obtained from BD Biosciences. Anti-IL-22 (142928) was obtained from R&D. Anti-CD34 (QBEnd10 +Immu133 +Immu409) was obtained from Beckman Coulter. Anti-pSTAT5 (C7185) was obtained from Cell Signaling. Anti-Jak3 (D7B12) was obtained from Cell Signaling. LIVE/DEAD Fixable Aqua was from Life Technologies. Anti-NKp46 (CS96) was produced and biotinylated by the Colonna lab at WUSM25. Fc receptors were blocked before staining with supernatant from hybridoma cells producing monoclonal antibody to CD32 (HB-197; ATCC). For intracellular transcription factor staining, the FOXP3 staining kit (eBioscience) was used. For intracellular cytokine staining, the BD Cytofix/Cytoperm kit was used. For phosphoflow, cells were fixed using 2% paraformaldehyde (Electron Microscopy Sciences) for 10 minutes at 37o, washed three times with PBS, and lysed for 30 minutes on ice with 90% methanol before staining. Data were acquired on a BD FACSCanto II or BD FACSCalibur and analyzed with FlowJo software (Treestar).
Cell isolation, culture, and cytokine measurements
From mice, siLP lymphocytes were isolated by Collagenase 4 digestion after two EDTA washes to remove intraepithelial lymphocytes. Lymphocytes were enriched from digested cells by Percoll gradients. From human, tonsillar ILC were isolated and either expanded in culture or directly stimulated ex-vivo, as previously described38. Human adult peripheral blood CD34+ cells were first enriched with CD34+ microbeads (Miltenyi Biotech), then purified to 99% by FACS. Cells were expanded in SCF (100 ng, Peprotech), IL-7 (100 ng, Peprotech), and IL-15 (50 ng, Peprotech) for 14 days. Tofacitinib (Sigma) or PF-06651600 (Pfizer) was added to cultured tonsillar cells at the indicated concentration 48-hours before stimulation, to cultured CD34+ cells for 72-hours before analysis, or ex-vivo at the time of stimulation. Human cells were stimulated for 7–8 hours and mouse cells for 5 hours before analyzing protein by intracellular stain or CBA of supernatants.
Sequencing and analysis
Whole exome sequencing was performed by BGI Americas using an Agilent platform. Briefly, the genomic DNA sample was randomly fragmented into 200 to 250bp pieces, ligated to adapters, and purified by the Agencourt AMPure SPRI beads. Fragments with insert size ~200bp were excised, amplified by ligation-mediated polymerase chain reaction (LM-PCR), purified, and hybridized to the SureSelect Biotinylated RNA Library (BAITS) for enrichment with streptavidin beads. Captured LM-PCR product was loaded on Illumina Hiseq4000 platform for high-throughput sequencing. After sequencing, data were preprocessed, aligned to the C57BL6/J genome using Burrows-Wheeler Aligner (BWA) software, and SNP and InDel variants were called using the Genome Analysis Toolkit HaplotypeCaller (Broad Institute). Variants were hard-filtered to generate a high-confidence dataset, which were then annotated to identify protein-coding changes. For Sanger sequencing, the region around the putative mutation in Jak3 was amplified by PCR using the following primers: Jak3 F: 5’- CACCCACTATACCCCGTGTC-3’ and Jak3 R: 5’- TGAGCCTCCTGGAGACATTC-3’. PCR products were purified (Qiagen) and sequenced using an internal sequencing primer, 5’- GGACCCTTTGTGCAAAGGTGACC-3’ (GeneWiz). Data were visualized using ApE.
Statistics
Prism 7 (GraphPad Software) was used for all statistical analyses. All graphical data show mean ± SEM.
Acknowledgments
Supported by the US National Institutes of Health (1U01AI095542, R01DE021255 and R21CA16719 to the Colonna laboratory; 1F30DK107053-01 to M.L.R.). This research was also supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Conflict of interests: Robinette no conflicts. Cella no conflicts. Telliez is an employee of Pfizer. Ulland no conflicts. Barrow no conflicts. Capuder no conflicts. Gilfillan no conflicts. Lin is an employee of Pfizer. Notarangelo no conflicts. Colonna no conflicts.
AUTHOR CONTRIBUTIONS
M.L.R. identified phenotype and performed pedigree analysis; M.L.R. and T.K.U. performed mouse experiments; M.Ce performed human experiments; M.L.R, M.Ce, and A.D.B. analyzed data. K.C. and L.D.N. performed WES analysis. M.L.R. and S.G. maintained mice. J.B.T. and L.L.L provided critical reagents. M.L.R., M.Ce, and M.Co designed studies. M.L.R. and M.C. wrote the paper.
References
- 1.Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary Immunodeficiency Diseases: an Update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35(8):696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pai SY, Logan BR, Griffith LM, Buckley RH, Parrott RE, Dvorak CC, et al. Transplantation outcomes for severe combined immunodeficiency, 2000–2009. N Engl J Med. 2014;371(5):434–446. doi: 10.1056/NEJMoa1401177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sponzilli I, Notarangelo LD. Severe combined immunodeficiency (SCID): from molecular basis to clinical management. Acta Biomed. 2011;82(1):5–13. [PubMed] [Google Scholar]
- 4.Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol Rev. 2004;202:67–83. doi: 10.1111/j.0105-2896.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 5.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36(4):515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377(6544):65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 7.Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, et al. Defective lymphoid development in mice lacking Jak3. Science. 1995;270(5237):800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 8.Pesu M, Candotti F, Husa M, Hofmann SR, Notarangelo LD, O'Shea JJ. Jak3, severe combined immunodeficiency, and a new class of immunosuppressive drugs. Immunol Rev. 2005;203:127–142. doi: 10.1111/j.0105-2896.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- 9.Ghoreschi K, Jesson MI, Li X, Lee JL, Ghosh S, Alsup JW, et al. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550) J Immunol. 2011;186(7):4234–4243. doi: 10.4049/jimmunol.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann R, Kremer J, Cush J, Schulze-Koops H, Connell CA, Bradley JD, et al. Placebo-controlled trial of tofacitinib monotherapy in rheumatoid arthritis. N Engl J Med. 2012;367(6):495–507. doi: 10.1056/NEJMoa1109071. [DOI] [PubMed] [Google Scholar]
- 11.Telliez JB, Dowty ME, Wang L, Jussif J, Lin T, Li L, et al. Discovery of a JAK3-Selective Inhibitor: Functional Differentiation of JAK3-Selective Inhibition over pan-JAK or JAK1-Selective Inhibition. ACS Chem Biol. 2016;11(12):3442–3451. doi: 10.1021/acschembio.6b00677. [DOI] [PubMed] [Google Scholar]
- 12.Thorarensen A, Dowty ME, Banker ME, Juba B, Jussif J, Lin T, et al. Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans. J Med Chem. 2017 doi: 10.1021/acs.jmedchem.6b01694. [DOI] [PubMed] [Google Scholar]
- 13.Robinette ML, Colonna M. Immune modules shared by innate lymphoid cells and T cells. J Allergy Clin Immunol. 2016;138(5):1243–1251. doi: 10.1016/j.jaci.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zook EC, Kee BL. Development of innate lymphoid cells. Nat Immunol. 2016;17(7):775–782. doi: 10.1038/ni.3481. [DOI] [PubMed] [Google Scholar]
- 15.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348(6237):aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15(7):415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 17.Cording S, Medvedovic J, Aychek T, Eberl G. Innate lymphoid cells in defense, immunopathology and immunotherapy. Nat Immunol. 2016;17(7):755–757. doi: 10.1038/ni.3448. [DOI] [PubMed] [Google Scholar]
- 18.Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17(4):451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 19.Robinette ML, Bando JK, Song W, Ulland TK, Gilfillan S, Colonna M. IL-15 sustains IL-7R-independent ILC2 and ILC3 development. Nat Commun. 2017 doi: 10.1038/ncomms14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41(3):366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21(7):698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters CP, Mjosberg JM, Bernink JH, Spits H. Innate lymphoid cells in inflammatory bowel diseases. Immunol Lett. 2016;172:124–131. doi: 10.1016/j.imlet.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016;17(7):765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 24.Juelke K, Romagnani C. Differentiation of human innate lymphoid cells (ILCs) Curr Opin Immunol. 2016;38:75–85. doi: 10.1016/j.coi.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanash AM, Dudakov JA, Hua G, O'Connor MH, Young LF, Singer NV, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37(2):339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270(5237):794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 28.Gozalo-Sanmillan S, McNally JM, Lin MY, Chambers CA, Berg LJ. Cutting edge: two distinct mechanisms lead to impaired T cell homeostasis in Janus kinase 3- and CTLA-4-deficient mice. J Immunol. 2001;166(2):727–730. doi: 10.4049/jimmunol.166.2.727. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol. 2015;16(10):1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3 doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157(2):340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature. 2016;539(7627):102–106. doi: 10.1038/nature20105. [DOI] [PubMed] [Google Scholar]
- 34.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37(4):634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34(12):573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci U S A. 2015;112(16):5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strober B, Buonanno M, Clark JD, Kawabata T, Tan H, Wolk R, et al. Effect of tofacitinib, a Janus kinase inhibitor, on haematological parameters during 12 weeks of psoriasis treatment. Br J Dermatol. 2013;169(5):992–999. doi: 10.1111/bjd.12517. [DOI] [PubMed] [Google Scholar]
- 38.Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U S A. 2010;107(24):10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, et al. Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell. 2016;165(5):1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montaldo E, Teixeira-Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W, et al. Human RORgammat(+)CD34(+) cells are lineage-specified progenitors of group 3 RORgammat(+) innate lymphoid cells. Immunity. 2014;41(6):988–1000. doi: 10.1016/j.immuni.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Chappaz S, Gartner C, Rodewald HR, Finke D. Kit ligand and Il7 differentially regulate Peyer's patch and lymph node development. J Immunol. 2010;185(6):3514–3519. doi: 10.4049/jimmunol.1000665. [DOI] [PubMed] [Google Scholar]
- 42.Vely F, Barlogis V, Vallentin B, Neven B, Piperoglou C, Ebbo M, et al. Evidence of innate lymphoid cell redundancy in humans. Nat Immunol. 2016;17(11):1291–1299. doi: 10.1038/ni.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21(7):719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 44.Bachelez H, van de Kerkhof PC, Strohal R, Kubanov A, Valenzuela F, Lee JH, et al. Tofacitinib versus etanercept or placebo in moderate-to-severe chronic plaque psoriasis: a phase 3 randomised non-inferiority trial. Lancet. 2015;386(9993):552–561. doi: 10.1016/S0140-6736(14)62113-9. [DOI] [PubMed] [Google Scholar]
- 45.Sandborn WJ, Ghosh S, Panes J, Vranic I, Wang W, Niezychowski W, et al. A phase 2 study of tofacitinib, an oral Janus kinase inhibitor, in patients with Crohn's disease. Clin Gastroenterol Hepatol. 2014;12(9):1485–1493. e1482. doi: 10.1016/j.cgh.2014.01.029. [DOI] [PubMed] [Google Scholar]