Abstract
Optogenetic tools offer fast and reversible control of protein activity with subcellular spatial precision. In the past few years, remarkable progress has been made in engineering photoactivatable systems regulating the activity of cellular proteins. In this review, we discuss general strategies in designing and optimizing such optogenetic tools and highlight recent advances in the field, with specific focus on applications regulating protein catalytic activity.
Main text
In the past decade, an emerging class of optogenetic tools have enabled the manipulation of diverse molecular and cellular processes with light, including signal transduction, gene expression, cell migration, and protein and organelle trafficking. In comparison to traditional chemical-genetic methods, light-actuated tools allow for fast and reversible control of transient and local cellular events, enabling study of dynamic cellular processes with high spatiotemporal precision. Design strategies for protein photocontrol utilize photosensory proteins and domains—molecules that undergo conformational changes upon photoexcitation—and fall into two major classes. A first strategy for light-mediated control of protein activity relies on manipulating protein subcellular localization, a regulatory approach commonly used in nature. By inducing changes in subcellular localization with light illumination, target proteins can be brought into proximity with their effectors or substrates, or alternatively tethered or sequestered at subcellular locations away from their effectors, resulting in stimulation or inhibition of activity. A second photocontrol strategy relies on triggering molecular changes within a target protein that are independent of subcellular localization. In this class of tools, light-induced changes within photosensory domains lead, through clustering, complementation, steric, or allosteric mechanisms, to changes in the activity of tethered protein targets. This review focuses on this latter class of optogenetic tools, with specific emphasis on light control of enzymes and enzymatic activity. Included is a discussion of general strategies for direct photocontrol of protein catalytic activity, and a review of recent progress in this area.
1. The building blocks – photosensory domains
Photosensory domains are essential components of all optogenetic tools, and are derived from naturally-occurring photoreceptors or fluorescent proteins [1]. Absorption of light by a photosensory domain is mediated through the chromophore, typically a light-sensitive cofactor, but in some cases an intrinsic residue such as tryptophan. Photon absorption induces a ground-to-activated state transition of the chromophore resulting in conformational changes within the photosensory domain. Such structural rearrangement can directly transmit to a tethered effector domain, resulting in light-dependent changes in activity or binding. Alternatively, conformational changes may alter the tendency for photosensory domains to self-associate or to interact with binding partners. Light-dependent interaction or dissociation of such ‘photodimerizers’ can be used to control the activities of attached target proteins or domains.
Different photosensory proteins have unique properties, showing different activation and reversion kinetics and binding affinities. These differences make them suitable for vastly different applications. For instance, proteins with fast dark reversion kinetics (fast-cycling) are preferable for inducing transient and local activation of signaling pathways, while those with slower reversion kinetics (slow-cycling) are preferred for inducing sustained enzymatic activity requiring minimal light input. When used for photodimerization applications, the binding affinity of photosensory domains for partner proteins is critical. In practice, photodimerizers with larger differences between the dissociation constants in dark and light can be easier to optimize for photocontrol applications; however, appropriate expression level of these components is critical. For example, photodimerizers that interact with high affinity may be ideal for use in situations where proteins are expressed at low intracellular levels, but the same molecules may show unwanted interactions in dark and thus reduced dynamic range when expressed at higher levels [2].
In a number of cases, photosensory domains have been engineered and improved for specific applications. For instance, extensive structural and biochemical analysis of the second light-oxygen-voltage sensing domain of Avena sativa phototropin 1 (AsLOV2) [3] has enabled identification of mutations that modulate the interaction between the LOV domain and a C-terminal Jα helix [4–6]. The dynamic range of AsLOV2-based photoswitches has also been improved through a combination of computational design and phage-display screening methods [7]. As another example of photosensory domain optimization, Neurospora crassa Vivid (VVD), a LOV domain that naturally forms homodimers upon light activation, was engineered into two distinct heterodimerizing variants, pMag and nMag (i.e., ‘Magnets’) [8].
Other practical concerns for the choice of photosensory domains, especially for potential adaptation in vivo, include the size of proteins (relevant for packaging into viral vectors and as fusion tags), expression level in targeted tissues, excitation wavelength, and requirement for cofactors. Near-infrared (NIR) light, for example, has been shown to penetrate mammalian tissues more effectively and cause less phototoxicity compared to shorter wavelength light [9]. The plant photoreceptor phytochrome B (PhyB) is responsive to red and far-red light but requires a cofactor that is not present in animal cells, thus constraining the use of PhyB-based optogenetic systems in vivo. An alternative NIR-sensitive photoreceptor, bacterial phytochrome from Rhodopseudomonas palustris (BphP1), uses biliverdin as cofactor, a molecule abundant in animal cells. BphP1 interacts with transcriptional repressor PpsR2 in a light-dependent manner, and has been successfully applied for optogenetic control of transcription in mice [9]. Further engineering of phytochromes may yield variants with altered photokinetics and dynamic ranges, and would provide a novel toolkit for NIR-activatable optogenetic systems.
2. Protein activation via induced oligomerization
Some proteins, particularly signaling proteins, are naturally activated in the cell through the process of dimerization or oligomerization [10]. For example, receptor tyrosine kinases undergo oligomerization in response to ligand binding, which leads to activation of kinase catalytic activity [11]. For such targets, light can be used to modulate protein oligomeric state, thus inducing signaling or other protein activity. To this end, target proteins can be fused to photosensory domains that undergo light-induced homo-dimerization or oligomerization such as VVD [12,13], Arabidopsis thaliana cryptochrome 2 (CRY2) [14–18], or the LOV domains from Vaucheria frigida aureochrome1 (VfAU1-LOV) [19,20] or Phaeodactylum tricornutum aureochrome1a (PtAu1a) [21,22]. Alternatively, two different targets can be fused to domains that heterodimerize in light, such as Arabidopsis thaliana PhyB/PIF [23,24], FKF1/GIGANTEA [25], CRY2/CIB1 [26], TULIPs [6], iLIDs [7], Magnets [8], or BphP1/RpsR2 [9] (Fig. 1A). Homo- and hetero-dimerization strategies have proven to be effective in light-dependent activation of receptor tyrosine kinases [16,19,27], serine/threonine kinases [28], and caspase [13].
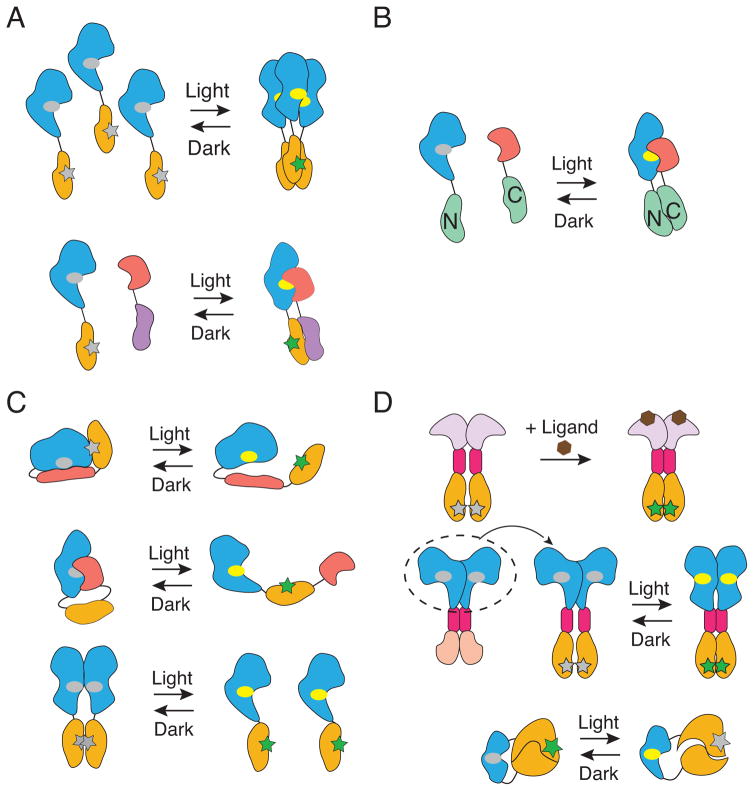
Figure 1. General strategies for photocontrol of protein activity.
(A) Oligomerization. Light-induced conformational changes within photosensory domains (blue; the chromophore within the photosensory domain is indicated by an ellipse: grey, ground state; yellow, light activated state) increase their tendency for self-association or interaction with binding partners (red), which can be utilized for homo- (upper panel) or hetero- (lower panel) oligomerization of tethered protein targets (yellow and purple). The active site within the protein target is indicated by a star: grey, inactive; green, active).
(B) Reconstituting split proteins. A target protein (green) is split into N- and C- terminal halves, which are reconstituted through light-dependent interaction between the photosensory domain and a binding partner.
(C) Steric effects. The active site (indicated by a star: grey, inactive; green, active) of a target protein (yellow) can be caged via intra- (top and middle panels) or inter- (bottom panel) molecular interaction controlled by light.
(D) Allosteric regulation. If a similar allosteric module (magenta) is shared between a photosensory protein (blue, photosensory domain; pale red, effector domain) and an allosterically regulated enzyme (pale purple, regulatory domain; yellow, catalytic domain), domain swapping could preserve the allosteric switching (upper panel). In other cases, a photosensory domain is inserted into a target protein at coupled allosteric sites to regulate its molecular dynamics (lower panel).
For optogenetic control of cellular processes, one important consideration is how to target endogenous components of a specific pathway. A unique approach, ‘Clustering Indirectly using Cryptochrome 2′ (CLICR), was proposed for light-inducible oligomerization of endogenous proteins [29]. In this design, CRY2 is not directly fused to the protein of interest, but to an adaptor that binds the target protein with low affinity in its monomeric form. Blue light induces oligomerization of the CRY2-fused adaptor, which increases its effective concentration and avidity for the target protein. The oligomeric adaptor protein is recruited to the target (often a receptor) and induces oligomerization, leading to activation. As proof of principle, the authors used the SH2 domain from PLC-γ as an adaptor protein to mediate the oligomerization and activation of endogenous platelet-derived growth factor receptor. The modular design of CLICR makes it applicable for a large variety of photosensory domains, adaptors, and target proteins. In particular, engineered orthogonal adaptors, such as nanobodies that target specific endogenous proteins [30], may further expand the range and versatility of such systems.
3. Reconstitution of split proteins
While only a subset of proteins can be activated by dimerization or oligomerization, a more broadly applicable strategy for protein photocontrol relies on use of photodimerizers to reconstitute split protein fragments (Fig. 1B). In this case, a protein of interest is split into two inactive fragments that are each attached to photodimerizers. Light-induced dimerization facilitates interaction between the fragments and reconstitutes the activity of split protein. Often, the targeted proteins are split at a solvent-exposed loop to minimize perturbation of protein structure, and multiple split sites are screened for optimal light-dependent activity. Many additional factors, including domain orientation, linker lengths, and photodimerizer affinities, also affect the efficiency of reconstitution, and require optimization on a case-by-case basis.
One of the first examples using photodimerizers to reconstitute a split enzyme was a photoactivatable vacuolar ATPase intein [31]. In this system, the red light-induced interaction between Arabidopsis thaliana PhyB and PIF3 was used to bring the N- and C-terminal intein halves together, allowing for induction of protein splicing in yeast. In another example, split Cre DNA recombinase was reconstituted through blue light-inducible dimerization between CRY2 and CIB1 [26]. While this first generation photoactivatable Cre showed only modest levels of recombination and required extended light treatment, two recent studies have applied different strategies to improve this split system. Additional optimization and use of a slow-cycling CRY2 variant in place of wild-type CRY2 resulted in a photoactivatable Cre recombinase showing >20% recombination with a single pulse of light [32]. Alternatively, splitting Cre at another site and reconstituting with a different pair of photodimerizers, Magnets, also greatly improves the dynamic range [33].
Other split photoactivatable enzymes, such as Cas9 [34] and T7 RNA polymerase [35], have been developed using similar approaches. It should be noted that all split systems have a certain level of background activity, due to the intermolecular interaction between the two protein fragments and/or dark interaction between photodimerizers. In many cases, dark background can be reduced by optimizing the binding affinity of photodimerizers or protein expression level [32,33]. Furthermore, other engineering strategies, such as allosteric regulation [35] and subcellular localization [36], can be combined with protein splitting to reduce background in optogenetic systems.
4. Regulation of protein accessibility (steric caging)
Another strategy for light regulation of enzymatic activity, ‘caging’, involves sterically blocking the binding surface or active site of a target protein, which can be reversibly controlled by light-triggered structural changes within photosensory domains (Fig. 1C). In one such design, a GTPase-inactive variant of Rac1 was caged by AsLOV2 in the dark [37]. Blue light triggers the unwinding of the Jα helix between LOV2 and Rac1 [3], exposing the binding surface of Rac1 to downstream effectors. Other proteins, including a calcium channel regulator [38] and transcription factors [39], have also been caged at the C-terminus of Jα helix. Noticeably, the caging efficiency in these systems is largely dependent on the interaction between target protein and the LOV domain [37,39]. Therefore, this strategy seems mostly applicable to small peptides, such as kinase inhibitory peptides [40,41] or protein trafficking peptides [42–46].
As an alternative strategy to cage larger protein domains, photodimerizers can be fused at both N- and C-termini of a target protein. This strategy was used with a variant of Dronpa that tetramerizes in the dark but exhibits light-dependent monomerization, allowing light control of hepatitis C virus protease activity [47]. With its modular design, light-inducible uncaging is a promising strategy for photocontrol. Indeed, the application of this approach has been extended to light-inducible activation of protein kinases (Raf1, MEK1, MEK2, and CDK5), using a dark-dimerized Dronpa variant with improved light response [48]. Other photodimerizers showing light-dependent dissociation, such as Zdk, a synthetic protein engineered for selective binding to the dark state of AsLOV2, could also be used for similar dark caging strategies [49].
In addition to intramolecular caging, the accessibility of target protein active sites and binding interfaces can be controlled through intermolecular interaction, a common natural regulatory mechanism. This strategy has been applied for photo-inhibition of protein activity using a light-dependent oligomerization system, ‘light-activated reversible inhibition by assembled trap’ (LARIAT) [17]. In this approach, CIB1 is fused to a multimeric protein, which enhances the blue light-induced clustering of CRY2 through light-dependent CRY2-CIB1 interaction. A protein of interest can be fused to CRY2 and inactivated by sequestering into clusters with light. Alternatively, protein targets can be brought in to the clusters via a nanobody fused to CRY2, which allows further modularization. This approach was applied to regulate activity of a number of cellular proteins including guanine nucleotide exchange factors (Vav2 and Tiam1), small GTPases (Rac1, Cdc42, and RhoG), and PI3 kinase [17]. Similarly, light-dependent sequestering and inactivation was also achieved by fusing target proteins to CRY2olig, a mutant of CRY2 that forms large clusters on its own and thus does not require co-expression of the CIB1-multimeric protein [18]. In contrast to using photoreceptors that oligomerize with light to disrupt activity, a recent study explored the promise of light-induced monomerization to stimulate activity [50]. In this work, the LOV domain from Rhodobacter sphaeroides (RsLOV), which dissociates into monomers upon blue light stimulation, was inserted in solvent-exposed regions of Cas9 to disrupt activity through dark dimerization. The authors identified a RsLOV-Cas9 variant with activity stimulated by light. While the light-dependent stimulation could only be observed in a cellular context (and not with purified protein) and thus did not represent a cell-autonomous protein switch, this strategy may be applicable for photocontrolling the intrinsic activity of other enzymes.
5. Other forms of allosteric regulation
In response to light, photosensory proteins often act as allosteric switches, propagating light-induced conformational changes to alter activity of effector domains. This natural strategy has been utilized for design of several artificial engineered switches. To facilitate engineering, groups have exploited structural homology that allows modular domain swapping between naturally occurring photosensory proteins and allosterically regulated enzymes (Fig. 1D). For example, in the design of a light-regulated histidine kinase, the LOV domain of Bacillus subtilis YtvA was fused to the histidine kinase domain from Bradyrhizobium japonicum FixL, joined by a Jα helix that is shared between the two native proteins [51]. The resulting chimeric protein shows blue light inactivation of kinase activity. A similar strategy was applied for the light control of cAMP/cGMP phosphodiesterase [52] and adenylate cyclase [53], in which a related coiled-coil domain was identified between the parental enzymes and bacterial phytochrome. Since the transmission module joining the photosensory domain and the effector is essential for the allosteric effects, this region usually requires extensive optimization to achieve a robust light response [51–53].
Despite the difficulty in predicting long-range structural rearrangement caused by photo-activation of photosensory domains, de novo design of allosteric photoactivatable systems have been greatly facilitated by computational analysis of the structure, molecular dynamics, and evolutionary conservation of targeted proteins. In an early pioneering study, a surface-exposed region of Escherichia coli dihydrofolate reductase (DHFR) was identified to be important for allosteric regulation by statistical coupling analysis, which probes correlated evolution between residues at the active site and distal parts of the protein [54]. Insertion of AsLOV2 domain at this region leads to to blue light-dependent increase of DHFR activity, which was proposed to be due to increased flexibility of the targeted region upon the unwinding of Jα helix. The well-characterized conformational change within AsLOV2 was recently applied in another groundbreaking study to build a generalizable framework for allosteric control of protein activity. In this work, a set of blue light-inactivated proteins (Src kinase, small GTPases, and guanine nucleotide exchange factors) were designed by inserting AsLOV2 into regions where rigidity is required for activity [55]. The authors described detailed computational approaches for predicting insertion sites using molecular dynamic coupling and static contact map analyses to identify surface-exposed regions allosterically linked to enzyme active sites, which provides broadly applicable guidelines for the rational design of single-chain allosteric optogenetic tools [55].
6. Concluding remarks
In the past few years, tremendous progress has been made in photocontrol of protein catalytic activity. To this end, some of the major challenges in the field include developing generalizable strategies to target activity of specific classes of protein, identifying novel photosensory domains and optimizing existing ones, minimizing the perturbation of endogenous cellular processes, and developing improved strategies for tight light control. Tackling these problems will be facilitated by emerging new approaches for protein engineering, including new computational methods for the rational design of photoactivatable proteins and more efficient high-throughput screening strategies for directed evolution. Structural and functional insights into photoactivation mechanisms of newly discovered photosensory domains will also have high impact, enabling creative new design strategies for optogenetic engineering. With rapid methodology advances and an increasing number of optimized, modular approaches available, optogenetic strategies are poised to become essential components of the experimental biologist’s molecular toolkit.
Highlights.
Optogenetic tools allow control of protein activity with spatiotemporal precision.
New photosensory domains provide diverse building blocks for optogenetic systems.
Generalizable design strategies have been proposed for protein photocontrol.
Computational analysis has facilitated the design of optogenetic tools.
High-throughput screening has been instrumental for optimizing engineered systems.
Acknowledgments
This work was supported by grants from the National Institutes of Health [GM100225, EY026363] to C.L.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman SP, Hallett RA, Bourke AM, Bear JE, Kennedy MJ, Kuhlman B. Tuning the Binding Affinities and Reversion Kinetics of a Light Inducible Dimer Allows Control of Transmembrane Protein Localization. Biochemistry. 2016;55:5264–5271. doi: 10.1021/acs.biochem.6b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 4.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •7.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A. 2015;112:112–117. doi: 10.1073/pnas.1417910112. Using computational library design and phage display screening, the authors greatly improved the dynamic range of blue light-induced interaction between AsLOV2-SsrA and SspB. This photodimerizer system, being both small and tunable, has advantageous properties for protein photocontrol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •8.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. Using structure-guided protein design, the authors engineered the Vivid domain, which forms homodimers upon blue light stimulation, into a heterodimerizer pair, pMag and nMag (Magnets). This photodimerizer system has since been used for reconstituting split proteins such as Cas9 and Cre DNA recombinase. [DOI] [PubMed] [Google Scholar]
- ••9.Kaberniuk AA, Shemetov AA, Verkhusha VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods. 2016;13:591–597. doi: 10.1038/nmeth.3864. This red/near-infrared photodimerizer system is based on a bacterial phytochrome that uses biliverdin as chromophore. As biliverdin is synthesized in animal cells, this system can be used in animals and animal cells without addition of exogenous chromophore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 11.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 13.Nihongaki Y, Suzuki H, Kawano F, Sato M. Genetically engineered photoinducible homodimerization system with improved dimer-forming efficiency. ACS Chem Biol. 2014;9:617–621. doi: 10.1021/cb400836k. [DOI] [PubMed] [Google Scholar]
- 14.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 15.Ozkan-Dagliyan I, Chiou YY, Ye R, Hassan BH, Ozturk N, Sancar A. Formation of Arabidopsis Cryptochrome 2 photobodies in mammalian nuclei: application as an optogenetic DNA damage checkpoint switch. J Biol Chem. 2013;288:23244–23251. doi: 10.1074/jbc.M113.493361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KY, Woo D, Jung H, Lee S, Kim S, Won J, Kyung T, Park H, Kim N, Yang HW, et al. Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat Commun. 2014;5:4057. doi: 10.1038/ncomms5057. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Heo WD. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 18.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Ingles-Prieto A, Janovjak H. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 2014;33:1713–1726. doi: 10.15252/embj.201387695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra D, Yang X, Moffat K. Crystal structures of Aureochrome1 LOV suggest new design strategies for optogenetics. Structure. 2012;20:698–706. doi: 10.1016/j.str.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A, Herman E, Kottke T, Essen LO. Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum. Structure. 2016;24:171–178. doi: 10.1016/j.str.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Heintz U, Schlichting I. Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence. Elife. 2016;5:e11860. doi: 10.7554/eLife.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 24.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem Biol. 2014;21:903–912. doi: 10.1016/j.chembiol.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Wend S, Wagner HJ, Muller K, Zurbriggen MD, Weber W, Radziwill G. Optogenetic control of protein kinase activity in mammalian cells. ACS Synth Biol. 2014;3:280–285. doi: 10.1021/sb400090s. [DOI] [PubMed] [Google Scholar]
- •29.Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, Schaffer DV. Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat Commun. 2015;6:6898. doi: 10.1038/ncomms7898. The authors presented a modular and versatile approach for light-inducible oligomerization of endogenous proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 31.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- •32.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. The authors identified variants of CRY2 and CIB1 with smaller sizes and reduced dark interaction, as well as mutants showing different dark reversion kinetics. A slow-cycling variant of CRY2 allowed for optimization of a photoactivatable Cre DNA recombinase with increased dynamic range. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •33.Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol. 2016;12:1059–1064. doi: 10.1038/nchembio.2205. This study also developed a next-generation Cre DNA recombinase, using a different splitting strategy and Magnets as photodimerizers. The resulting system showed improved recombination towards a variety of loxP sites, and was successfully adapted in mice. [DOI] [PubMed] [Google Scholar]
- 34.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 35.Han T, Chen Q, Liu H. Engineered Photoactivatable Genetic Switches Based on the Bacterium Phage T7 RNA Polymerase. ACS Synth Biol. 2017;6:357–366. doi: 10.1021/acssynbio.6b00248. [DOI] [PubMed] [Google Scholar]
- 36.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham E, Mills E, Truong K. A synthetic photoactivated protein to generate local or global Ca(2+) signals. Chem Biol. 2011;18:880–890. doi: 10.1016/j.chembiol.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Paonessa F, Criscuolo S, Sacchetti S, Amoroso D, Scarongella H, Pecoraro Bisogni F, Carminati E, Pruzzo G, Maragliano L, Cesca F, et al. Regulation of neural gene transcription by optogenetic inhibition of the RE1-silencing transcription factor. Proc Natl Acad Sci U S A. 2016;113:E91–100. doi: 10.1073/pnas.1507355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi JJ, Wang H, Vilela M, Danuser G, Hahn KM. Manipulation of endogenous kinase activity in living cells using photoswitchable inhibitory peptides. ACS Synth Biol. 2014;3:788–795. doi: 10.1021/sb5001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunniff B, McKenzie AJ, Heintz NH, Howe AK. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol Biol Cell. 2016;27:2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, Di Ventura B. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun. 2014;5:4404. doi: 10.1038/ncomms5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yumerefendi H, Dickinson DJ, Wang H, Zimmerman SP, Bear JE, Goldstein B, Hahn K, Kuhlman B. Control of Protein Activity and Cell Fate Specification via Light-Mediated Nuclear Translocation. PLoS One. 2015;10:e0128443. doi: 10.1371/journal.pone.0128443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B. Optogenetic control of nuclear protein export. Nat Commun. 2016;7:10624. doi: 10.1038/ncomms10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiltoir JI, Strickland D, Glotzer M, Tucker CL. Optical Control of Peroxisomal Trafficking. ACS Synth Biol. 2016;5:554–560. doi: 10.1021/acssynbio.5b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yumerefendi H, Lerner AM, Zimmerman SP, Hahn K, Bear JE, Strahl BD, Kuhlman B. Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Nat Chem Biol. 2016;12:399–401. doi: 10.1038/nchembio.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •48.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ. Optical control of cell signaling by single-chain photoswitchable kinases. Science. 2017;355:836–842. doi: 10.1126/science.aah3605. The authors engineered the Dronpa protein for improved light response, and used a photodissociable dimeric Dronpa variant to cage kinase activity in mammalian cells and C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •49.Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods. 2016;13:755–758. doi: 10.1038/nmeth.3926. The authors carried out extensive mRNA-display screening and identified Zdark, a small protein that selectively interacts with dark-state AsLOV2. This novel engineered blue light-dissociable dimerizer system was used to sequester cellular proteins at mitochondria, and can be applied for other optogenetic uses such as protein caging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter F, Fonfara I, Bouazza B, Schumacher CH, Bratovic M, Charpentier E, Moglich A. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res. 2016;44:10003–10014. doi: 10.1093/nar/gkw930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasser C, Taiber S, Yeh CM, Wittig CH, Hegemann P, Ryu S, Wunder F, Moglich A. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A. 2014;111:8803–8808. doi: 10.1073/pnas.1321600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen DM, Gomelsky M. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci U S A. 2014;111:10167–10172. doi: 10.1073/pnas.1324301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••55.Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM. Engineering extrinsic disorder to control protein activity in living cells. Science. 2016;354:1441–1444. doi: 10.1126/science.aah3404. The authors presented a generalizable approach for identifying insertion sites within the target protein for AsLOV2-mediated allosteric regulation with blue light. [DOI] [PMC free article] [PubMed] [Google Scholar]