Abstract
For almost 50 years, the Icahn School of Medicine at Mount Sinai has continually invested in genetics and genomics, facilitating a healthy ecosystem that provides widespread support for the ongoing programs in translational pharmacogenomics. These programs can be broadly cataloged into discovery, education, clinical implementation and testing, which are collaboratively accomplished by multiple departments, institutes, laboratories, companies and colleagues. Focus areas have included drug response association studies and allele discovery, multiethnic pharmacogenomics, personalized genotyping and survey-based education programs, pre-emptive clinical testing implementation and novel assay development. This overview summarizes the current state of translational pharmacogenomics at Mount Sinai, including a future outlook on the forthcoming expansions in overall support, research and clinical programs, genomic technology infrastructure and the participating faculty.
Keywords: : clinical implementation, discovery, education, genetic testing, Icahn School of Medicine at Mount Sinai, pharmacogenetics, pharmacogenomics
The Department of Human Genetics was created at the Icahn School of Medicine at Mount Sinai (ISMMS) in 1993 after almost three decades of academic growth and development as the Division of Medical Genetics in the Department of Pediatrics. In 1967, internationally renowned human geneticist Dr Kurt Hirschhorn [1] was the founding Chief of the Division until he became Chairman of Pediatrics in 1977, which prompted his recruitment of Dr Robert J Desnick as Chief of the Division. Under the leadership of Dr Desnick, the Division expanded in both research and clinical activities, particularly in the areas of molecular and biochemical genetics and the development of treatments for inherited metabolic diseases, which ultimately led to the creation of the Department of Human Genetics. The initial focus of the Department on Mendelian disease research, gene discovery and mutation analyses also catalyzed the formation of the clinical Mount Sinai Genetic Testing Laboratory. The early translational discoveries and departmental infrastructure, including the development of enzyme replacement therapy for lysosomal disorders [2] and establishment of some of the first medical genetics residency, counseling and clinical laboratory fellowship training programs in the country, formed the foundation that subsequently enabled the ISMMS to invest in the emerging field of genomics.
The Department continued to expand its faculty and purview, which was complemented by its rebranding from ‘Human Genetics’ to ‘Genetics and Genomic Sciences’ in 2006. In 2007, the ISMMS established the Charles Bronfman Institute for Personalized Medicine with a central aim of creating and maintaining the BioMe™ Biobank program. Led by Director Dr Erwin Bottinger, the Institute expanded genomic research and implementation efforts that are continued today under current Director Dr Judy H Cho. As Dr Desnick assumed the role of Dean for Genetic and Genomic Medicine in 2012, the ISMMS recruited Dr Eric E Schadt as Chairman of the Department of Genetics and Genomic Sciences and founding Director of the Icahn Institute for Genomics and Multiscale Biology. The following 5 years of continued growth in genomic technologies, computational infrastructure, and research and clinical faculty elevated the Department in 2016 to rank 4th in NIH funding for genetics among medical schools in the USA. These advances in genomics infrastructure also facilitated dramatic growth in the Mount Sinai Genetic Testing Laboratory, led by Director Dr Lisa Edelmann, which prompted the 2017 launch of the Mount Sinai driven venture, Sema4™ (CT, USA), a next-generation health information company dedicated to state-of-the-art clinical genetic testing and data-driven health science discovery. As Dr Schadt assumed the role of Sema4 Chief Executive Officer and Dean for Precision Medicine at the ISMMS, Drs Pamela Sklar and Andrew Kasarskis were appointed Chair of the Department of Genetics and Genomic Sciences and Director of the Icahn Institute for Genomics and Multiscale Biology, respectively.
Throughout this evolution of genetics and genomics at the ISMMS there has been overarching support for scholarship and discovery, clinical and translational research, innovation and entrepreneurship, and the implementation of genetic and genomic medicine. As such, the ongoing research, education and clinical programs dedicated to translational pharmacogenomics at Mount Sinai have been consistently supported broadly across the Institution and collaboratively between Departments and Institutes, Divisions and Laboratories, companies and colleagues.
Discovery & diversity
Collaborative research on pharmacogenomics over the past decade has been driven by a number of investigators at Mount Sinai, and frequently centered on drug response association studies and allele discovery, multiethnic pharmacogenomics and/or cardiovascular pharmacogenomics. In collaboration with Mount Sinai cardiologists, discovery efforts have studied the genetic determinants of anticoagulant and antiplatelet response variability. For example, the CYP2C9*8 allele association with warfarin dosing and its high frequency in the African–American population was originally identified at Mount Sinai [3], as well as multiethnic assessments of warfarin dosing algorithms [4], and the recent exome sequencing identification of B4GALT2 and clopidogrel response variability [5]. Consistent with these studies, Mount Sinai was selected as a clinical site for the multicenter NIH-funded Clarification of Optimal Anticoagulation through Genetics (COAG) warfarin pharmacogenetics clinical trial [6].
Interrogating the diverse patient population of the New York City metropolitan area (Figure 1) has resulted in the discovery of the warfarin-resistant VKORC1 p.D36Y allele in the Ashkenazi Jewish population [7], as well as the novel CYP2C19*4B, which is a unique haplotype that harbors both increased- and loss-of-function variants [8]. Population screening studies performed collaboratively with commercial biotechnology companies have identified multiethnic frequencies of warfarin [9] and clopidogrel [8] response genes, as well as clinically relevant CYP2C haplotypes [10], and novel CYP450 copy number variation alleles [11]. In addition, Mount Sinai investigators have recently made novel pharmacogenomic association discoveries with glucose-insulin-potassium therapy response among acute coronary syndrome patients [12,13] and antipsychotic response among schizophrenia patients [14], as well as ongoing research efforts studying integrative oncology pharmacogenomics [15].
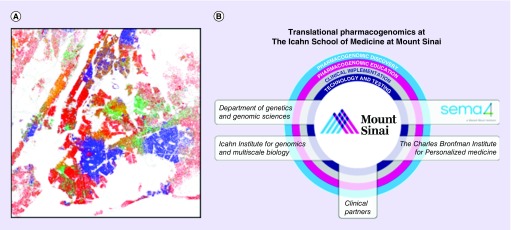
Figure 1. . Translational pharmacogenomics at the Icahn School of Medicine at Mount Sinai in New York City.
(A) Race and ethnicity of the New York City metropolitan area population based on data from census 2010. Each colored dot represents 25 residents: red is Caucasian, blue is African–American, green is Asian, orange is Hispanic and yellow is ‘other’. (B) An illustration of the translational pharmacogenomics programs at the Icahn School of Medicine at Mount Sinai, highlighting the academic and commercial entities in boxes, which in collaboration with clinical partners are jointly invested in pharmacogenomic discovery, education, clinical implementation, and technology and testing.
(A) Adapted with permission from Eric Fischer: Race and Ethnicity 2010 - New York City (5559914315).png (© Eric Fischer, some rights reserved, CC BY-SA 2.0).
Education & implementation
Interwoven with all of the pharmacogenomic efforts at Mount Sinai is an underlying commitment to education with several active venues for students and professionals currently ongoing. Since 2013, all medical students at the ISMMS are offered the option to participate in a personalized genotyping program that allows first year students to receive their own clinical pharmacogenomic test results. This program also incorporates a survey on knowledge and attitudes toward clinical pharmacogenomic testing for all medical students [16]. Similarly, returning personal pharmacogenomic results is a standard module within Mount Sinai's progressive ‘Practical Analysis of Your Personal Genome’ graduate level course [17,18], and additional knowledge and attitude survey studies directed at novel patient cohorts (e.g., BioMe patient subgroups), pharmacists and physician specialists are currently ongoing. A consistent finding from these studies has been the identification of a general lack of knowledge about pharmacogenomics at most levels of medical practice, prompting current projects to also measure best practices for deploying pharmacogenomic education tools among healthcare providers. These studies are already informing the 4–6 week pharmacogenomics rotations offered to New York City area Doctor of Pharmacy (PharmD) students by Dr Aniwaa Owusu Obeng and the Department of Pharmacy at Mount Sinai Hospital.
In addition to education, translating pharmacogenomic discoveries to clinical implementation is a major priority at Mount Sinai, as evidenced by active co-authorship of Clinical Pharmacogenetics Implementation Consortium guidelines and projects [19–23], as well as additional internal programs. In 2013, the Charles Bronfman Institute for Personalized Medicine initiated ‘IPM-PGx’, which enrolled 1000 patients from the BioMe Biobank for pre-emptive pharmacogenomic genotyping through the Mount Sinai Genetic Testing Laboratory [24]. IPM-PGx is enabled by the Clinical Implementation of Personalized Medicine through Electronic Health Records and Genomics (CLIPMERGE) platform [24], which is an independent data management and clinical decision support (CDS) system that communicates with the Mount Sinai electronic health record. A parallel implementation program at Mount Sinai is the NIH-funded eMERGE-PGx, which is a multicenter network project pre-emptively interrogating genes involved in drug response for both research discovery and clinical implementation [25]. Approximately 700 patients were recruited from the Mount Sinai Faculty Practice Associates Primary Care clinic for this program, which employed targeted gene sequencing using the PGRNseq platform, orthogonal clinical genotyping and data management and CDS using CLIPMERGE.
Data from the IPM-PGx and eMERGE-PGx programs are currently being collected and analyzed; however, preliminary results suggest that provider acceptance of pre-emptive pharmacogenomic testing coupled with point-of-care CDS is high. As such, efforts at Mount Sinai are now focused on implementing additional gene/drug pairs through IPM-PGx and designing innovative strategies for implementing medications with pharmacogenomic interactions that are influenced by patient ancestry. For example, a novel strategy for implementing algorithm-guided warfarin dosing across the multiethnic IPM-PGx and eMERGE-PGx cohorts was recently reported by the Mount Sinai implementation investigators [26].
Pharmacogenomic testing & Sema4
All of the translational pharmacogenomic efforts at Mount Sinai have continuously been supported with accessible genomic technologies for all investigators, research and clinical programs. Multiplexed targeted genotyping, commercially available microarray platforms, high-throughput short-read sequencing and third-generation long-read sequencing instrumentation and expertise [27,28] are readily available within the Department and the Mount Sinai Genetic Testing Laboratory, including computational and bioinformatics infrastructure. Translating pharmacogenomic discoveries to clinical genetic tests has been an overarching theme, starting with the 2007 validation and clinical availability of CYP2C9, CYP2C19, CYP2D6 and VKORC1 genotyping [7,29], to more sophisticated CYP2C19 phasing assays [30], and the recent development of single molecule real-time (SMRT) long-read CYP2D6 sequencing using the Pacific Biosciences platform [31]. This novel assay sequences the entire full-length CYP2D6 gene and can differentiate ‘downstream’ and ‘upstream’ copies when a gene duplication event is present. Coupled with this innovative SMRT sequencing pharmacogenomic assay was the development of companion informatics tools, including a third-generation sequencing error correction procedure (Amplicon Long-read Error Correction [ALEC]) [31], and a CYP2D6 star (*) allele translation tool [32]. In addition to assay development, recent collaborative efforts through the Charles Bronfman Institute for Personalized Medicine include thorough comparisons of research sequencing and clinical pharmacogenomic genotyping across the eMERGE-PGx network cohort [33].
When the Mount Sinai Genetic Testing Laboratory transitioned into Sema4 in 2017, pharmacogenomic assay development and technology assessment continued to be supported and prioritized. For example, efforts are now focused on developing expanded clinical pharmacogenomic genotyping and sequencing panels, bioinformatic and reporting software tools, as well as additional novel assays for drug metabolism genes that are difficult to interrogate by targeted genotyping and short-read sequencing platforms. Moreover, expanding the commercial program into pharmacogenomic discovery will be preserved through patient and provider collaboration and data-driven longitudinal research, with the aim of identifying novel genomic and/or clinical signatures that are predictive of medication efficacy, adverse event risk and/or dosing. Consistent with previous work on pharmacogenomics at Mount Sinai, when adequate validity and utility are established, these novel findings will subsequently be used to feed back into the diagnostic laboratory of Sema4.
Future perspective & priorities
An overview of the translational pharmacogenomics programs at the ISMMS is illustrated in Figure 1. As detailed above, these activities can be broadly cataloged into discovery, education, clinical implementation and pharmacogenomic testing. Pharmacogenomic discovery will continue to be fueled by both Mount Sinai and Sema4 investigators, including interrogation of the BioMe Biobank for novel drug response association studies, as well as continued research in cardiovascular drug response variability (and other therapeutic drug classes), and interrogation of the polymorphic human leukocyte antigen region across the diverse Mount Sinai patient population. In addition, collaborative clinical trials are underway with Sema4 and clinical provider partners that are aimed at assessing the clinical utility of pharmacogenomic testing.
Knowledge and attitude survey studies continue to be a priority, which are informing the pharmacogenomic educational tools that currently are being developed for Mount Sinai and other healthcare professionals. These efforts are proving to be a foundational component of the expanded pharmacogenomic implementation program that is currently being defined by the Charles Bronfman Institute for Personalized Medicine, as preliminary data indicate that healthcare provider knowledge on pharmacogenomics correlates directly with adoption of pharmacogenomic-guided medication use. Many of these forthcoming research, education and implementation efforts will be supported by the continued technical advances in pharmacogenomic resources at Mount Sinai and Sema4, including expanded clinical sequencing panels, high-throughput and cost-effective targeted genotyping panels, long-read sequencing assays for complex pharmacogenomic regions and haplotypes, and digital product development for electronic patient consenting and longitudinal data acquisition of drug response phenotypes [34]. Additionally, these technology resources will support the future expansion of pharmacogenomic research performed collaboratively with both national and international colleagues with shared interests and aims, including multiethnic pharmacogenomic discovery and implementation.
Conclusion
The ISMMS has been committed to genetics and genomics for almost 50 years, which has resulted in extensive support and enthusiasm for pharmacogenomics at all levels of academic leadership. As detailed above, the interwoven translational pharmacogenomic activities at Mount Sinai involve multiple Departments, Institutes, laboratories and companies, all of which have been elevated and maintained by the strength of the investigators and colleagues among them. At this time of growing enthusiasm for clinical genome sequencing, precision medicine and federally funded genomic medicine network programs, it is these individuals who will continue to define the future landscape of translational pharmacogenomics at Mount Sinai.
Footnotes
Financial & competing interests disclosure
This work was supported in part by the National Institute of General Medical Sciences (NIGMS) of the NIH through grant K23GM104401 (SA Scott), and the National Human Genome Research Institute (NHGRI) of the NIH through grants U01HG008701-02S1 and U01HG006380 (eMERGE Network; EP Bottinger, AO Obeng) and U01HG007278 (IGNITE Network; EP Bottinger, AO Obeng). Funding for The Charles Bronfman Institute for Personalized Medicine was provided by the Andrea and Charles Bronfman Philanthropies and the Icahn School of Medicine at Mount Sinai. SA Scott, MR Botton, Y Yang, ER Scott, R Wallsten, X Zhou, R Chen, P Nicoletti, GA Diaz, L Edelmann, RJ Desnick and EE Schadt are paid employees or consultants of Sema4, which is a commercial company that offers clinical pharmacogenomic testing. EP Bottinger and SB Ellis are named co-inventors on a patent application and have developed software and know-how related to CLIPMERGE, a medical decision support software platform that collects, aggregates and displays clinical data. The platform allows for personalized medical decision making. The CLIPMERGE tool is being utilized in pharmacogenetics research described in this manuscript. The intellectual property related to the software and know-how has been licensed by the Icahn School of Medicine to Ontomics Inc, a privately held company in which EP Bottinger and SB Ellis are equity owners. If the CLIPMERGE technology is commercially successful, EP Bottinger and SB Ellis will benefit financially and, in addition, payments will be made to the Icahn School of Medicine as part of the license agreement. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Hirschhorn K, Hirschhorn R. 2013 Victor A. McKusick Leadership Award addresses. Am. J. Hum. Genet. 2014;94(3):336–337. doi: 10.1016/j.ajhg.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eng CM, Guffon N, Wilcox WR, et al. Safety and efficacy of recombinant human alpha-galactosidase A replacement therapy in Fabry's disease. N. Engl. J. Med. 2001;345(1):9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 3.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African–Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubitz SA, Scott SA, Rothlauf EB, et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J. Thromb. Haemost. 2010;8(5):1018–1026. doi: 10.1111/j.1538-7836.2010.03792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott SA, Collet JP, Baber U, et al. Exome sequencing of extreme clopidogrel response phenotypes identifies B4GALT2 as a determinant of on-treatment platelet reactivity. Clin. Pharmacol. Ther. 2016;100(3):287–294. doi: 10.1002/cpt.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am. J. Hum Genet. 2008;82(2):495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ. Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharmacogenomics J. 2012;12(4):297–305. doi: 10.1038/tpj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11(6):781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martis S, Peter I, Hulot JS, Kornreich R, Desnick RJ, Scott SA. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics J. 2013;13(4):369–377. doi: 10.1038/tpj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martis S, Mei H, Vijzelaar R, Edelmann L, Desnick RJ, Scott SA. Multi-ethnic cytochrome-P450 copy number profiling: novel pharmacogenetic alleles and mechanism of copy number variation formation. Pharmacogenomics J. 2013;13(6):558–566. doi: 10.1038/tpj.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis KL, Zhou Y, Beshansky JR, et al. Genetic modifiers of response to glucose-insulin-potassium (GIK) infusion in acute coronary syndromes and associations with clinical outcomes in the IMMEDIATE trial. Pharmacogenomics J. 2015;15(6):488–495. doi: 10.1038/tpj.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis KL, Zhou Y, Rodriguez-Murillo L, et al. Common variants associated with changes in levels of circulating free fatty acids after administration of glucose-insulin-potassium (GIK) therapy in the IMMEDIATE trial. Pharmacogenomics J. 2017;17(1):76–83. doi: 10.1038/tpj.2015.84. [DOI] [PubMed] [Google Scholar]
- 14.Ruderfer DM, Charney AW, Readhead B, et al. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry. 2016;3(4):350–357. doi: 10.1016/S2215-0366(15)00553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzilov AV, Ding W, Fink MY, et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome Med. 2016;8(1):62. doi: 10.1186/s13073-016-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanderson SC, Scott SA, Schrager N, et al. The 64th Annual Meeting of The American Society of Human Genetics. San Diego, CA, USA: October 19 2014. Personal pharmacogenetic testing in the medical school curriculum: student knowledge and attitudes; abstract number 2311S. Presented at. [Google Scholar]
- 17.Sanderson SC, Linderman MD, Zinberg R, et al. How do students react to analyzing their own genomes in a whole-genome sequencing course?: outcomes of a longitudinal cohort study. Genet. Med. 2015;17(11):866–874. doi: 10.1038/gim.2014.203. [DOI] [PubMed] [Google Scholar]
- 18.Linderman MD, Bashir A, Diaz GA, et al. Preparing the next generation of genomicists: a laboratory-style course in medical genomics. BMC Med. Genomics. 2015;8:47. doi: 10.1186/s12920-015-0124-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriyama B, Obeng AO, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.583. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet. Med. 2017;19(2):215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 2017;102(3):397–404. doi: 10.1002/cpt.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin. Pharmacol. Ther. 2013;94(2):214–217. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 2014;96(4):482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obeng AO, Kaszemacher T, Abul-Husn NS, et al. Implementing algorithm-guided warfarin dosing in an ethnically diverse patient population using electronic health records and preemptive CYP2C9 and VKORC1 genetic testing. Clin. Pharmacol. Ther. 2016;100(5):427–430. doi: 10.1002/cpt.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum. Mol. Genet. 2010;19(R2):R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 28.Kasarskis A, Yang X, Schadt E. Integrative genomics strategies to elucidate the complexity of drug response. Pharmacogenomics. 2011;12(12):1695–1715. doi: 10.2217/pgs.11.115. [DOI] [PubMed] [Google Scholar]
- 29.Scott SA, Edelmann L, Kornreich R, Erazo M, Desnick RJ. CYP2C9, CYP2C19 and CYP2D6 allele frequencies in the Ashkenazi Jewish population. Pharmacogenomics. 2007;8(7):721–730. doi: 10.2217/14622416.8.7.721. [DOI] [PubMed] [Google Scholar]
- 30.Scott SA, Tan Q, Baber U, et al. An allele-specific PCR system for rapid detection and discrimination of the CYP2C19 *4A, *4B, and *17 alleles: implications for clopidogrel response testing. J. Mol. Diagn. 2013;15(6):783–789. doi: 10.1016/j.jmoldx.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Qiao W, Yang Y, Sebra R, et al. Long-read single molecule real-time full gene sequencing of cytochrome P450–2D6. Hum. Mutat. 2016;37(3):315–323. doi: 10.1002/humu.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao W, Wang J, Pullman BS, Chen R, Yang Y, Scott SA. The CYP2D6 VCF Translator. Pharmacogenomics J. 2016;17(4):301–303. doi: 10.1038/tpj.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen-Torvik LJ, Almoguera B, Doheny KF, et al. Concordance between research sequencing and clinical pharmacogenetic genotyping in the eMERGE-PGx study. J. Mol Diagn. 2017;19(4):561–566. doi: 10.1016/j.jmoldx.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan YY, Wang P, Rogers L, et al. The Asthma Mobile Health Study, a large-scale clinical observational study using ResearchKit. Nat. Biotechnol. 2017;35(4):354–362. doi: 10.1038/nbt.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]