Abstract
Genetic variation in HIV poses a major challenge for prevention and treatment of the AIDS pandemic. Resistance occurs by mutations in the target proteins that lower affinity for the drug or alter the protein dynamics, thereby enabling viral replication in the presence of the drug. Due to the prevalence of drug-resistant strains, monitoring the genotype of the infecting virus is recommended. Computational approaches for predicting resistance from genotype data and guiding therapy are discussed. Many prediction methods rely on rules derived from known resistance-associated mutations, however, statistical or machine learning can improve the classification accuracy and assess unknown mutations. Adding classifiers such as information on the atomic structure of the protein can further enhance the predictions.
Keywords: : antiretroviral therapy, drug resistance mutations, genotype interpretation, HIV/AIDS, integrase strand transfer inhibitor, non-nucleoside reverse transcriptase inhibitor, nucleoside reverse transcriptase inhibitor, protease inhibitor, supervised machine learning
Increased prevalence of drug resistance is a challenge for successful treatment and prevention of HIV/AIDS [1]. The WHO estimates over 36 million people are infected by the virus and about half of them receive antiretroviral treatment [2]. Antiretroviral therapy usually involves a combination of drugs that inhibit the viral enzymes, reverse transcriptase, protease and integrase, and/or block viral entry or fusion with the host cell. Despite the success of antiretroviral drugs [3,4], about 2 million new infections occur per year [5]. Globally, the virus occurs as two major types, HIV-1 and HIV-2, which are subdivided into several major groups and subtypes. The genetic diversity and high mutability of HIV pose a critical challenge for long-term efficacy of antiretrovirals and for development of effective vaccines [6–9]. The virus mutates readily due to its rapid replication, its error-prone reverse transcriptase, viral recombination and effects of host cell factors, especially APOBEC-mediated gene editing [10,11]. Low levels of adherence to therapy and retention of patients are common clinical problems that escalate the development of resistance. Drug resistance is exacerbated both by prophylactic use of drugs [7,12] and transmitted resistance, especially in urban areas [13]. Resistant variants may persist in viral reservoirs and re-emerge during subsequent therapy. Hence, monitoring resistance is recommended to guide the choice of drugs for new patients and those who are failing therapy [1,7,14].
Resistance may be tested by phenotype or genotype assays of the infecting virus. In the phenotype assay, replication of virus with clinically derived RNA is measured in the presence of different drugs. This assay is expensive and time consuming. The genotype assay involves sequencing the viral genome to identify mutations associated with resistance, as described in [15]. Genotype sequencing and interpretation is the most efficient procedure in cost and speed [16]. Next-generation sequence technologies will likely improve the efficiency of resistance testing and increase the detection of rare variants [16,17].
HIV genetic variation
The genetic variability of HIV complicates the analysis and interpretation of genotype assay for resistance. HIV can be considered a quasispecies, comprising genetic variants classified as HIV-1, HIV-2, their major groups and subtypes within the groups. The different mechanisms producing HIV sequence diversity are reviewed in [10]. In addition to the lack of fidelity of the reverse transcriptase, viral recombination between co-infecting genomes contributes to genetic diversity. Moreover, host cell APOBEC enzymes act to edit genes by cytosine deamination and consequent mutation of G to A. The genome-wide variation has been analyzed for the protein-coding regions of HIV-1, HIV-2, major groups and subtypes from >1700 infected individuals [18]. The protein sequences were more conserved for capsid and the three enzymes, protease, reverse transcriptase and integrase, compared with the other virally encoded proteins, matrix, nucleocapsid, Vif, Vpr, Tat and Rev. The integrase protein had the least variation among the different HIV clades. Viral proteins with the highest genetic variability tended to interact with more human proteins. Most critically, the high variability of the envelope proteins GP120 and GP41 poses a problem for the development of an effective vaccine.
The scale of the problem is demonstrated by a recent comprehensive analysis of >100,000 protease and reverse transcriptase sequences and >10,000 integrase sequences [19]. Natural polymorphisms, mutations due to APOBEC-mediated gene editing and mutations arising under drug pressure were identified. The drug targets exhibited extensive variation. Individual amino acid positions had >1% variants in 34–47% of the enzyme sequences with the protease showing the highest variation. This study identified more than 300 nonpolymorphic mutations associated with drug selection.
Molecular mechanisms of resistant mutants
Resistant strains have been documented for all the drugs used in HIV/AIDS therapy and prevention. A list of individual mutations associated with resistance has been compiled and is updated regularly, as shown by the recent version [20]. These resistance mutations alter the protein sequence and 3D structure to enable viral replication in the presence of the drug. Mutations can be divided into two categories: major mutations associated with resistance to one or more drugs; and minor or accessory mutations observed in conjunction with major mutations. The major mutations are often deleterious for viral replication, however, specific accessory mutations can act to restore viral fitness. Moreover, baseline polymorphisms can act to increase replication capacity of viral variants with major mutations [21].
The molecular mechanisms resulting in drug resistance have been summarized in [22,23]. The primary targets of antiretroviral drugs are the three viral enzymes: reverse transcriptase, protease and integrase. Resistance arises through two general mechanisms: mutations of residues in the inhibitor binding site may alter the size, shape and chemical properties of the cavity, thus decreasing the binding affinity of the protein for the inhibitor; and other mutations may alter the stability or conformational dynamics of the protein and enhance the dissociation of inhibitor. In both cases, the mutant enzyme must retain the capacity to bind substrate and perform its catalytic reaction in order to produce new infectious virus.
Resistance mechanisms have been studied primarily for the drug targets of HIV reverse transcriptase and protease. Inhibitors of these two enzymes have been employed in the clinic since 1995 and the physical and enzymatic properties have been analyzed for many of the resistant mutants.
Resistance to inhibitors of reverse transcriptase
HIV reverse transcriptase performs the essential function of converting the single-stranded viral genomic RNA into double-stranded DNA for integration into the host cell genome. The reverse transcriptase acts as a heterodimer of 560 residue (p66) and 440 (p51) residue subunits. The smaller p51 subunit has the same polymerase sequence as the p66 subunit, but the C-terminus is truncated before the RNaseH domain. Inhibitors of HIV reverse transcriptase are critical for both therapy and pre-exposure prophylaxis (PrEP) [12]. The inhibitors fall into two classes: nucleotide or nucleoside RT inhibitors (NRTIs) and non-nucleoside RT inhibitors (NNRTIs). The NRTIs are prodrugs that are metabolized into the diphosphate or triphosphate active forms, which are competitive inhibitors of the deoxyribonucleotide substrates. The NNRTIs act as allosteric inhibitors by binding to a separate site on reverse transcriptase. The heterodimer structure in complex with an NNRTI is illustrated in Figure 1A. Resistance arises to both classes of reverse transcriptase inhibitors. The mutations and their effects on enzyme activity are described in [24–26].
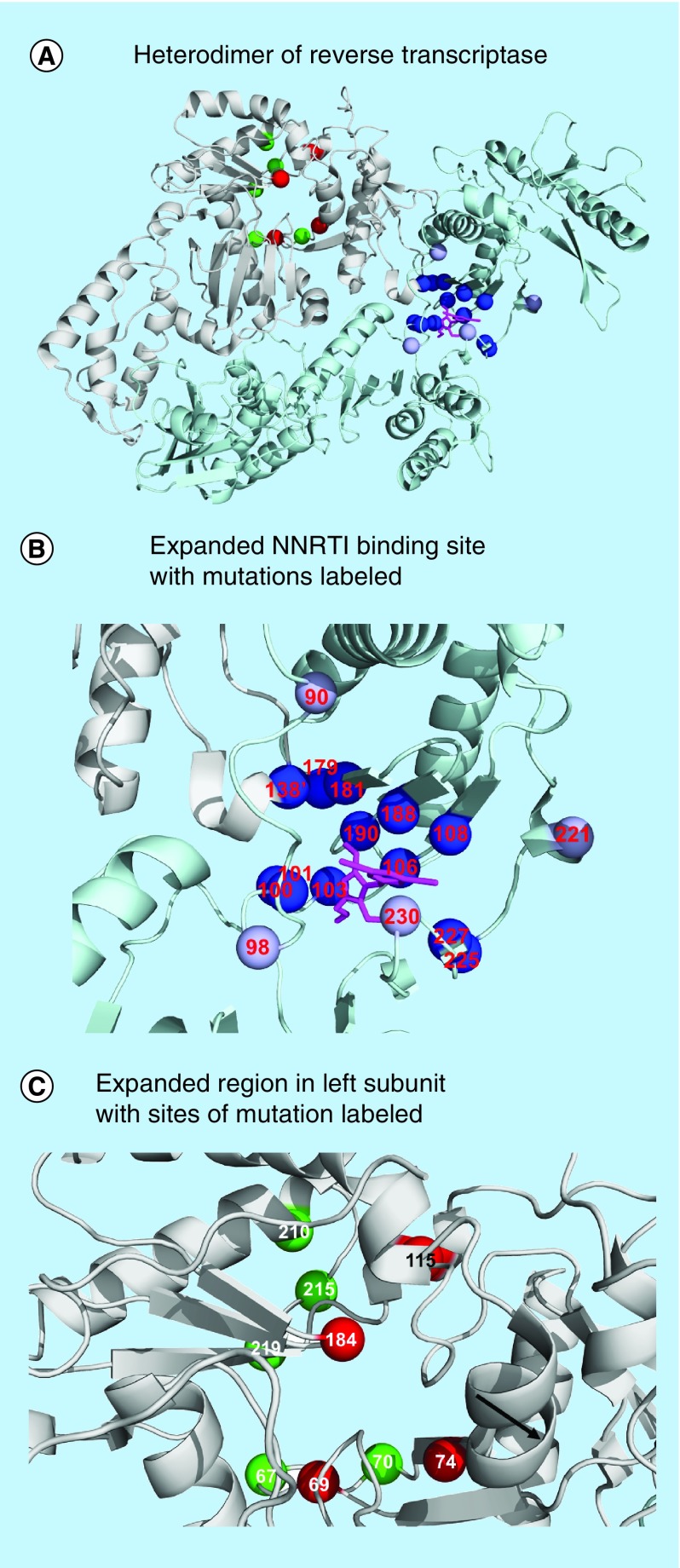
Figure 1. . Reverse transcriptase structure and sites of resistant mutations.
Locations of resistance-associated mutations mapped on the heterodimer of HIV reverse transcriptase in complex with NNRTI lersivirine (PDB: 2WOM). (A) Heterodimer of reverse transcriptase. The two subunits are shown as light gray and dark gray ribbons with NNRTI in black bonds. Sites of mutations associated with resistance to NRTIs are indicated by dark gray spheres on the left subunit. Mutations associated with NNRTI resistance are shown on the right in light gray [20]. (B) Expanded NNRTI binding site with mutations labeled. (C) Expanded region in left subunit with sites of mutations associated with NRTIs labeled.
NNRTI: Non-nucleoside RT inhibitor; NRTI: Nucleoside RT inhibitor; TAM: Thymidine analog-associated mutation.
The NNRTI binding site is adjacent to the polymerase active site in the p66 subunit and comprises residues from both p51 (E138) and p66 (L100, K101, K103, V106, T107, V108, V179, Y181, Y188, V189, G190, P225, F227, W229, L234, P236 and Y318) subunits. Mutations of about 70% of the amino acids in the NNRTI binding site cause resistance (Figure 1B). These mutations act by altering interactions with the drugs, either by removing stabilizing interactions or by blocking the binding of inhibitor and cross-resistance is common [22,27].
Resistance to NRTIs occurs by several mechanisms. There are two main molecular mechanisms for resistance: discriminatory mutations and thymidine analog-associated mutations (TAMs). Discriminatory mutations act to increase the selectivity for binding the natural substrates, while TAMs increase the ability of the enzyme to excise the unnatural NRTI monophosphate incorporated in the blocked DNA primer. The sites of these mutations are indicated in Figure 1C. Specific combinations of mutations confer high level cross resistance to all, or almost all NRTIs. The Q151M complex (mutations A62V, V75I, F77L, F116Y and Q151M) shows improved discrimination against triphosphate derivatives of nucleoside analogs. The 69 insertion complex (M41K, A62V, 69ins, K70R, L210W, T215YF and K219QE) confers resistance to all NRTIS by increasing excision activity, while the combination of TAMs (M41L, D67N, K70R, L210W, T215YF and K219QE) produces resistance to all NRTIs except emtricitabine and lamivudine. In addition, mutations can alter the conformational dynamics of the protein to produce resistance [28]. Combinations of mutations and insertions can produce resistance to multiple NRTIs.
Two of the NRTIs, tenofovir disoproxil fumerate and emtricitabine (FTC), are used in PrEP, as well as in postexposure antiretroviral therapy. Studies suggest PrEP use is associated with low levels of acquired resistance, primarily in previously infected individuals [29,30]. Overall, the benefits of preventing viral transmission far outweigh the risks of acquiring resistant viral strains.
Resistance to protease inhibitors
HIV protease plays an essential role in viral replication by processing the Gag and Gag-Pol precursors into the individual viral enzymes and structural proteins. The protease is catalytically active as a dimer of two 99-residue subunits. Due to its small size, the protease is tractable for a variety of biochemical and biophysical studies. Inhibitors of HIV protease bind in the active site cavity as shown in Figure 2. They are potent antiviral agents and a notable success of structure-guided drug design [31,32]. Nine protease inhibitors have been approved for clinical use. Currently, the most widely used are darunavir, lopinavir and atazanavir, which are co-administered with ritonavir to boost their plasma concentration. Clinical resistance is associated with >100 mutations in the protease gene [19] and different drugs elicit distinct patterns of mutations [22,33]. Resistance to protease inhibitors arises by mutations in the protease, and to a lesser extent, in its cleavage sites in the Gag and Gag-Pol polyprotein substrates [20,34]. Major resistance mutations lower the sensitivity to one or more drugs and often decrease viral infectivity, however, accessory mutations in the protease and its cleavage sites in the Gag precursor protein can compensate for loss of viral fitness [34–36]. Mutations in the protease produce resistance by altering the inhibitor binding site, the dimer interface and distal sites (Figure 2A). Recent studies from several groups suggest that distal mutations act synergistically to confer resistance by altering the protease dynamics [37–44].
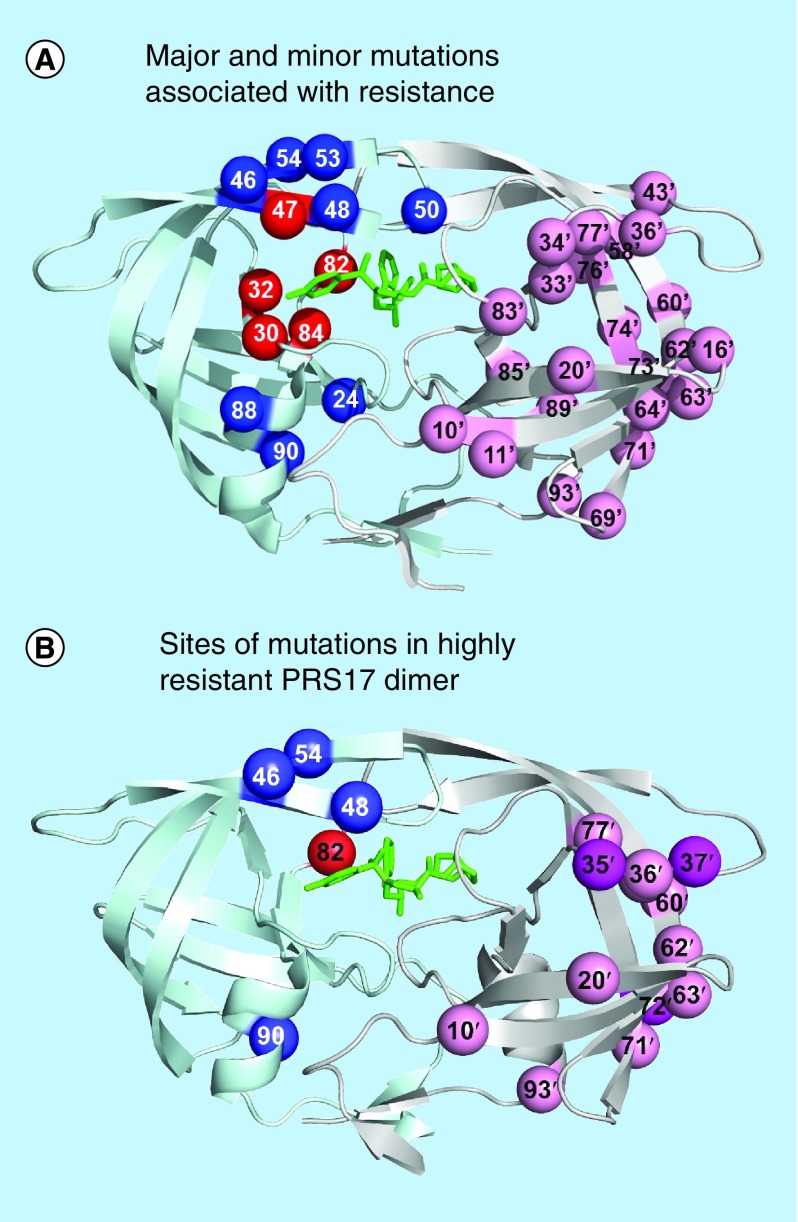
Figure 2. . Protease dimer showing sites of resistance mutations.
(A) Major and minor mutations associated with resistance. Locations of resistance-associated mutations mapped on the HIV protease dimer bound with DRV (PDB: 2IEN). The protease dimer is shown in ribbons with DRV in black sticks. Sites of mutations associated with drug resistance [20] are indicated by spheres. Mutations altering inhibitor binding are in dark gray and mutations affecting dimer stability or flap dynamics are indicated in light gray on the left subunit. Distal mutations with poorly defined effects are shown in gray on the right subunit.
DRV: Darunavir.
Combinations of multiple mutations also act to increase the conformational dynamics of the protease dimer as well as by decreasing the binding affinity for the inhibitor. Based on our studies and others, we proposed that high level resistance to protease inhibitors requires up to 20 mutations in the protease [33,45], as exemplified by variants with >1000-fold poorer binding affinity for darunavir and other drugs. These variants exhibit different patterns of 14–20 mutations. Studies of highly resistant mutants suggest that high levels of resistance can evolve by different mechanisms and different combinations of mutations. Several mutants from the clinic show loss of interactions with bound inhibitors, consistent with >1000-fold poorer binding affinity for inhibitors [43,46]. Our detailed structural, biochemical and biophysical characterization of two multiple mutants, PR20 and PRS17, from clinical isolates gave insights into the molecular mechanisms for resistance [43,44,47,48]. PR20 bears 19 mutations relative to wild-type enzyme and exhibits >40 nM binding affinity for tested clinical inhibitors, which is several orders of magnitude worse than wild-type. The mutations act synergistically to enlarge the inhibitor-binding cavity and increase the conformational variability of the flaps, while maintaining efficient catalysis of precursor cleavage. PRS17 with 17 mutations (L10I, K20R, E35D, M36I, S37D, M46L, G48V, I54V, D60E, I62V, L63P, A71V, I72V, V77I, V82S, L90M I93L) was deduced by machine learning algorithms to represent a broad class of highly resistant proteases in a large genotype/phenotype dataset [49]. This mutant showed binding affinity of 11–8000 nM for clinical inhibitors despite having only a single mutation (V82S) in the active site region (Figure 2B) [48]. Structural studies of PRS17 showed no significant change in the interactions with darunavir, however, the flaps had enhanced mobility in the absence of inhibitor [44]. Selection for the open-flap conformation is proposed to alter the folding landscape of highly drug-resistant mutants to avoid a free-energy trap of inhibitor-bound enzyme [42]. Altered dynamic equilibrium between open and closed conformations of the protease dimer may be a common mechanism for high level drug resistance.
Resistance to integrase inhibitors
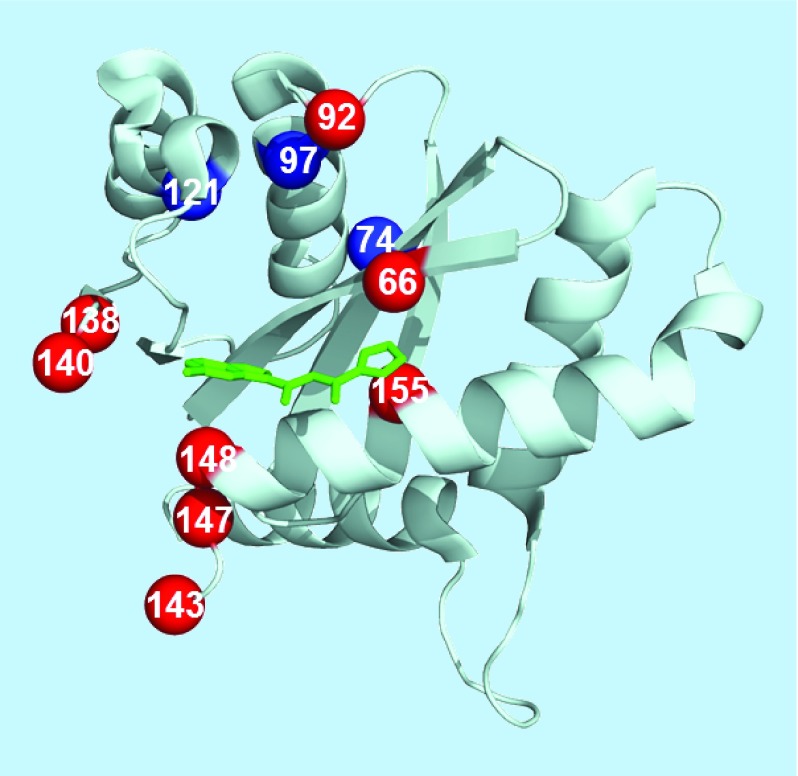
HIV integrase is the enzyme that inserts proviral DNA into the host cell genome. The integrase consists of 288 amino acids that fold into three domains, a central catalytic core domain, flanked by smaller N-terminal and C-terminal domains. One recent class of drugs bind to the catalytic core of the integrase and act as inhibitors of the integrase strand transfer inhibitors (INSTIs) reaction. These drugs are highly effective in therapy, well tolerated with low toxicity and they are recommended for treatment of newly infected patients. Resistance mutations selected by INSTIs are reviewed in [50]. The second-generation INSTI, dolutegravir, is especially promising for successful therapy due to its favorable pharmacokinetic properties and high genetic barrier to resistance [51,52]. Variants with resistance to dolutegravir show loss of replicative capacity, however, this drug was only approved in 2013 and high level resistant variants may not have evolved yet. The structure of the integrase catalytic core domain in complex with inhibitor and the sites of resistance mutations are shown in Figure 3 [53]. Active site interactions of integrase with viral DNA are important for the reaction and resistance to INSTIs [54]. However, our knowledge of the molecular mechanisms for resistance to INSTIs is limited by the lack of a crystal structure of the intact enzyme with its DNA substrates. Instead, the structure of the foamy virus intasome has been used as a model for the HIV integrase complex with viral DNA [55]. The majority of mutations associated with resistance to INSTIs alter residues in the catalytic core domain near catalytic residues D64, D116 and E152 and the bound inhibitor (Figure 3). Therefore, these mutations are likely to influence binding of the inhibitors, the DNA substrates or the catalytic reaction. The recent report of a cryo-electron microscopy structure of HIV-1 integrase with viral DNA in the intasome provides new insight into the integration mechanism and may help in the design of novel inhibitors [56].
Figure 3. . Catalytic core domain of HIV integrase with sites of resistance mutations.
Locations of resistance mutations mapped onto the catalytic core domain of HIV integrase (PDB: 1QS4). The core domain is shown as ribbons with inhibitor in black sticks. Sites of mutations are indicated by spheres with light gray for major mutations and dark gray for minor or accessory mutations.
Predicting resistance from genotype
The vast number and variety of mutations in clinical isolates is a major obstacle in current methods to guide therapy by genotyping the infecting virus [19]. Computational approaches are essential for systematic exploration of the range of sequences leading to highly resistant HIV. The algorithms must be designed to handle the huge number of genome sequences arising from HIV diversity as well as associated metrics for resistance. Hence, analysis of HIV genomes and prediction of resistance falls under the computational challenge of Big Data due to the size of the problem and its inherent variability. Special attention must be placed on data storage, representation and efficiency of analysis. Many statistical analysis methods cannot handle problems of the scale of HIV genotype data with several thousand to over 100,000 sequences and associated metrics for resistance or clinical factors.
Several groups are developing automated methods to predict drug resistance from HIV genotype sequence data, as reviewed in [57,58]. These methods employ two general techniques: rule-based genotype interpretation systems and statistical machine learning algorithms [57]. Rule-based methods codify their predictions based on a small number of rules that are manually or semi-manually selected to reproduce correlations between sequence data and drug resistance. The widely used genotype interpretation system in HIVdb [59] is an example of a rule-based method. The major drawback of rule-based methods is their inability to generalize in the presence of new data. The rule-based methods used with HIV focus on a limited set of known major mutations. The effects of nonmajor mutations are less clearly understood, and viral subtypes can vary in resistance as demonstrated in several studies [60–62]. Statistical or machine learning is applied to predict resistance in geno2pheno [63,15] and the EuResist prediction engine for therapy [64].
Supervised machine learning extracts a relationship or correlation between the dependent data (labels) and the independent data (features) [65]. The correlation that is extracted can be causal or noncausal, and the art of machine learning lies in the selection of meaningful features so that causal relationships are found. Generally, this means selecting a minimal set of features where there is a good theoretical basis for expecting a relationship. For predicting drug resistance, the minimal set of features would be a genomic sequence and the labels would be the drug resistance. A variety of machine learning algorithms are available and some may perform better than others for a particular problem. One sign of a well-selected set of features is that several distinct machine learning algorithms perform well on those data.
Here, we give several recent examples of applying machine learning to HIV drug resistance. Machine learning with linear support vector machine (SVM) gave somewhat improved predictions for clinical HIV sequences compared with rule-based methods based on known mutations [66]. Beerenwinkel et al. [67] modeled the evolutionary escape dynamics of HIV to predict suppression of viral load. They demonstrated that calculating the individualized genetic barrier with Isotonic Conjunctive Bayesian Networks, a class of probabilistic graphical models, outperformed the expert rule-based genotypic susceptibility score. A recent study of resistance to integrase inhibitors applied n-gram relative frequencies to achieve cross-validated accuracies of 85–89% in classification of mutants as susceptible or resistant [68]. Data for multiple drugs were analyzed in a multitask learning framework with kernel-based Bayesian dimensionality reduction with binary classification to give improved predictive performance for HIV resistance to RT inhibitors [69].
Ensembles of classifiers (e.g., boosting and bagging [70]) can improve the accuracy of prediction over individual classifiers. Cross resistance information was incorporated in multilabel classification models to improve prediction accuracy for resistance to NNRTIs and protease inhibitors [71,72]. Both classifier chains and ensembles of classifier chains were tested with random forests and logistic regression models.
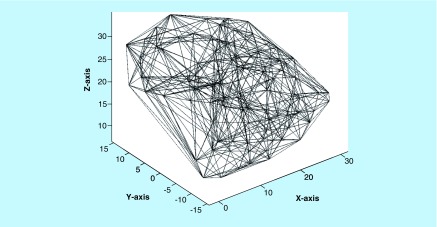
We have explored the concept of incorporating 3D structural information in machine learning with sequence data to improve classification and prediction of resistance. This extends the features beyond the minimal set by including more information. The use of 3D structural information is justified because resistance mutations alter the atomic structure of the protein and the effect of a specific mutation may be influenced by its physical environment. Proteins are folded into large and complex 3D structures, comprising several thousand atoms and generally represented by the x, y and z spatial coordinates of the atomic positions. The specific values of these coordinates depend on the crystallographic reference frame or its equivalent in NMR structures. We tested different ways to encode this spatial information with sequence data in a memory-efficient representation for machine learning. Because the biological molecule has no privileged reference frame, the encoding must be translationally and rotationally invariant. Graph representations are appropriate and can simply state that two atoms or residues are in contact with each other by some criterion. Delauney triangulation [73,74] proved to be the best graph-based encoding of protein sequence and 3D structure, as described in [75,76]. Figure 4 shows the graph for Delauney triangulation of HIV protease. The triangulation graph was summarized into a 210 (the number of unique pairs of amino acids) long feature vector by counting each pair of amino acids for every edge of the graph. The triangulation only had to be calculated one time for each structure as only the labels at the vertexes of the graph (i.e., the amino acids from the sequence) needed to be updated for each mutant sequence. This unified encoding of sequence and structure improved the performance of the machine learning and prediction of resistance [76–79].
Figure 4. . Delauney triangulation of HIV protease.
Graph constructed from the crystal structure of HIV protease as described in [76].
The features produced by this unified encoding algorithm routinely resulted in high levels of accuracy with disparate machine learning algorithms. SVMs [76], artificial neural networks [76,77], sparse dictionary [76,77], Fuzzy Decision Trees [78], k-nearest neighbors and Random forests [79] achieved cross-validated classification accuracies in the 93–99% range for protease instead of 60–87% accuracy with purely sequence-based approaches [57,76]. The prediction accuracy for resistance to the NRTI class of reverse transcriptase inhibitors gave values of 82–99% compared with 73–96% for standard methods. For the NNRTIs, the structural encoding with SVM or artificial neural networks gave accuracies of 98–99% compared with 95–99% for standard methods. This demonstrates that unified encoding accurately summarizes the effects of structure and sequence on drug resistance. This method gave excellent correlation between predicted and observed level of resistance in fivefold cross-validated regression analysis [77,49]. Because the encoding supported regression, the problem was inverted to select a tractable number of mutants for biochemical, biophysical and structural analysis [49]. One mutant was identified with high level resistance to six drugs. This mutant, PRS17 bearing 17 substitutions, was experimentally verified to exhibit inhibition constants of 11–8000 nM for eight tested drugs and the structure has been determined by x-ray crystallography [44,48].
Conclusion
In summary, machine learning can be applied to predict resistance from unknown sequences and to interrogate the phenotype–genotype database for mutants with specified properties. Experimental studies on mutants selected by machine learning have demonstrated the predictive power of the approach. We believe this ‘data-driven’ and rational approach to identifying highly resistant protease variants is an efficient and effective way to maximize the value of experimental studies and to select therapeutic strategies for HIV patients.
Future perspective
The genetic diversity and high propensity of HIV to evolve resistant strains under drug selection pose a severe challenge to effective prevention of viral infection and treatment of AIDS. Due to the large number of potential mutations, computational analysis is critical for interpreting genotype data. Machine learning or statistical learning approaches incorporate more information than rule-based methods, and are generally more accurate. A unified, graph-based encoding of sequence and structure holds promise for fast and accurate predictions of resistance from sequence data and future application in personalized therapy for HIV/AIDS. Furthermore, this approach can be applied to genotype–phenotype data for drug resistance in other diseases when the structure is known for the drug target.
In our current paradigm, high level resistance to several drugs requires the coordinated effects of multiple substitutions to remodel the inhibitor binding site and dynamics of the enzyme [45]. In other words, resistance is not simply an additive effect of several major mutations.
We propose that the 3D atomic structures of drug targets, such as the HIV protease and reverse transcriptase, are key ingredients for improving predictions of resistance from genotype data and hence, optimizing therapy. Our results confirm that machine learning methods derived from structure-based encoding produce superior predictions of resistance compared with using only sequence data. Furthermore, clustering analysis can extract examples for defining the molecular mechanisms of resistance in order to improve the design of new inhibitors. Genotype data can be rapidly analyzed by several computational methods to guide the choice of therapies in the clinic.
Executive summary.
Resistance monitoring to guide therapy
The genetic diversity of HIV leads to rapid selection of drug-resistant virus. Due to this severe problem, resistance testing is recommended.
Computational inference of resistance from sequence data
Drug resistance can be predicted from sequence by rules or expert interpretation based on known mutations. Statistical or machine learning from genotype–phenotype data is effective in predicting drug resistance even for unknown mutations.
Adding protein structural information as a classifier in machine learning to predict resistance
Encoding the atomic structure of target proteins improved the accuracy for predicting resistance and enabled regression and clustering analysis to extract mutants for further study.
Acknowledgements
We thank Y-F Wang, J Agniswamy and S Pawar for assistance with the figures.
Footnotes
Financial & competing interests disclosure
The authors’ research is supported in part by the grant U01 GM062920 awarded by the NIH. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Rhee SY, Jordan MR, Raizes E, et al. HIV-1 drug resistance mutations: potential applications for point-of-care genotypic resistance testing. PLoS ONE. 2015;10(12):e0145772. doi: 10.1371/journal.pone.0145772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization data for HIV. www.who.int/hiv/data/en/
- 3.Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382(9903):1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Passaes CP, Sáez-Cirión A. HIV cure research: advances and prospects. Virology. 2014;454–455:340–352. doi: 10.1016/j.virol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 5.GBD2015: Report from Global Burden of Disease Study 2015: GBD 2015 HIV Collaborators, Estimates of global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2015: the Global Burden of Disease Study 2015. Lancet HIV. 2016;3:e361–e387. doi: 10.1016/S2352-3018(16)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro MM, Perno CF. HIV-1 genetic variability and clinical implications. ISRN Microbiol. 2013:481314. doi: 10.1155/2013/481314. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clutter DS, Jordan MR, Bertagnolio S, Shafer RW. HIV-1 drug resistance and resistance testing. Infect. Genet. Evol. 2016;46:S1567–S1348. doi: 10.1016/j.meegid.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safrit JT, Fast PE, Gieber L, Kuipers H, Dean HJ, Koff WC. Status of vaccine research and development of vaccines for HIV-1. Vaccine. 2016;34:2921–2925. doi: 10.1016/j.vaccine.2016.02.074. [DOI] [PubMed] [Google Scholar]
- 9.Stephenson KE, Barouch DH. A global approach to HIV-1 vaccine development. Immunol. Rev. 2013;254:295–304. doi: 10.1111/imr.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth RP, Davenport MP, Mak J. The origin of genetic diversity in HIV-1. Virus Res. 2012;169:415–429. doi: 10.1016/j.virusres.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd SB, Kent SJ, Winnall WR. The high cost of fidelity. AIDS Res. Hum. Retroviruses. 2014;30(1):8–16. doi: 10.1089/aid.2013.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spinner CD, Boesecke C, Zink A, et al. HIV pre-exposure prophylaxis (PrEP): a review of current knowledge of oral systemic HIV PrEP in humans. Infection. 2016;44:151–158. doi: 10.1007/s15010-015-0850-2. [DOI] [PubMed] [Google Scholar]
- 13.Kassaye SG, Grossman Z, Balamane M, et al. Transmitted HIV drug resistance is high and longstanding in metropolitan Washington, DC. Clin. Infect. Dis. 2016;63(6):836–843. doi: 10.1093/cid/ciw382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, et al. European HIV Drug Resistance Guidelines Panel. European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev. 2011;13:77–108. [PubMed] [Google Scholar]
- 15.Van den Eede P, Van Wesenbeeck L, Verlinden Y, et al. HIV-1 genotyping of the protease-reverse transcriptase and integrase genes to detect mutations that confer antiretroviral resistance. Methods Mol. Biol. 2013;1030:37–55. doi: 10.1007/978-1-62703-484-5_5. [DOI] [PubMed] [Google Scholar]
- 16.Van Laethem K, Theys K, Vandamme AM. HIV-1 genotypic drug resistance testing: digging deep, reaching wide? Curr. Opin. Virol. 2015;14:16–23. doi: 10.1016/j.coviro.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Dudley DM, Bailey AL, Mehta SH, et al. Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq. RetroVirology. 2014;11:122. doi: 10.1186/s12977-014-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, Piampongsant S, Faria NR, et al. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology. 2015;12:18. doi: 10.1186/s12977-015-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee SY, Sankaran K, Varghese V, et al. HIV-1 protease, reverse transcriptase, and integrase variation. J. Virol. 2016;90(13):6058–6070. doi: 10.1128/JVI.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comprehensive analysis of a large number of sequences.
- 20.Wensing AM, Calvez V, Günthard HF, et al. 2017 Update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 2017;24(4):132–141. [PMC free article] [PubMed] [Google Scholar]; • Current list of mutations associated with drug resistance.
- 21.Pingen M, Wensing AM, Fransen K, et al. SPREAD programme. Persistence of frequently transmitted drug-resistant HIV-1 variants can be explained by high viral replication capacity. RetroVirology. 2014;11:105. doi: 10.1186/s12977-014-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menéndez-Arias L. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res. 2013;98:93–120. doi: 10.1016/j.antiviral.2013.01.007. [DOI] [PubMed] [Google Scholar]; • Review of resistance mutations and molecular mechanisms of drug resistance.
- 23.Iyidogan P, Anderson KS. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 2014;6(10):4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das K, Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 1. Curr. Opin. Virol. 2013;3(2):111–118. doi: 10.1016/j.coviro.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das K, Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr. Opin. Virol. 2013;3(2):119–128. doi: 10.1016/j.coviro.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sluis-Cremer N, Wainberg MA, Schinazi RF. Resistance to reverse transcriptase inhibitors used in the treatment and prevention of HIV-1 infection. Future Microbiol. 2015;10(11):1773–1782. doi: 10.2217/fmb.15.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sluis-Cremer N. The emerging profile of cross-resistance among the non-nucleoside HIV-1 reverse transcriptase inhibitors. Viruses. 2014;6(8):2960–2973. doi: 10.3390/v6082960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seckler JM, Leioatts N, Miao H, Grossfield A. The interplay of structure and dynamics: insights from a survey of HIV-1 reverse transcriptase crystal structures. Proteins. 2013;81(10):1792–1801. doi: 10.1002/prot.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baeten JM, Donnell D, Ndase P, et al. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N. Engl. J. Med. 2012;367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parikh UM, Mellors JW. Should we fear resistance from tenofovir/emtricitabine preexposure prophylaxis? Curr. Opin. HIV AIDS. 2016;11(1):49–55. doi: 10.1097/COH.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh AK, Anderson DD, Weber IT, Mitsuya H. Enhancing protein backbone binding – a fruitful concept for combating drug-resistant HIV. Angew. Chem. 2012;51:1778–1802. doi: 10.1002/anie.201102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber IT, Kneller DW, Wong-Sam A. Highly resistant HIV-1 proteases and strategies for their inhibition. Future Med. Chem. 2015;7:1023–1038. doi: 10.4155/fmc.15.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozísek M, Henke S, Sasková KG, et al. Mutations in HIV-1 gag and pol compensate for the loss of viral fitness caused by a highly mutated protease. Antimicrob. Agents Chemother. 2012;56:4320–4330. doi: 10.1128/AAC.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson GJ, Lee SK, Irlbeck DM, et al. Interplay between single resistance-associated mutations in the HIV-1 protease and viral infectivity, protease activity, and inhibitor sensitivity. Antimicrob. Agents Chemother. 2012;56:623–633. doi: 10.1128/AAC.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theys K, Deforche K, Vercauteren J, et al. on behalf of the SPREAD-programme. Treatment-associated polymorphisms in protease are significantly associated with higher viral load and lower CD4 count in newly diagnosed drug-naive HIV-1 infected patients. Retrovirology. 2012;9:81. doi: 10.1186/1742-4690-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai Y, Myint W, Paulsen JL, Schiffer CA, Ishima R, Kurt Yilmaz N. Drug resistance mutations alter dynamics of inhibitor-bound HIV-1 protease. J. Chem. Theory Comput. 2014;10(8):3438–3448. doi: 10.1021/ct4010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter JD, Gonzales EG, Huang X, et al. Effects of PRE and POST therapy drug-pressure selected mutations on HIV-1 protease conformational sampling. FEBS Lett. 2014;588(17):3123–3128. doi: 10.1016/j.febslet.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chetty S, Bhakat S, Martin AJ, Soliman ME. Multi-drug resistance profile of PR20 HIV-1 protease is attributed to distorted conformational and drug binding landscape: molecular dynamics insights. J. Biomol. Struct. Dyn. 2016;34(1):135–151. doi: 10.1080/07391102.2015.1018326. [DOI] [PubMed] [Google Scholar]
- 40.Ragland DA, Nalivaika EA, Nalam MN, et al. Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease. J. Am. Chem. Soc. 2014;136(34):11956–11963. doi: 10.1021/ja504096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen C-H, Chang Y-C, Agniswamy J, Harrison RW, Weber IT. Conformational variation of an extreme drug resistant mutant of HIV protease. J. Mol. Graphics Model. 2015;62:87–96. doi: 10.1016/j.jmgm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis JM, Roche J. Evolution under drug pressure remodels the folding free-energy landscape of mature HIV-1 protease. J. Mol. Biol. 2016;428:2780–2792. doi: 10.1016/j.jmb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agniswamy J, Shen CH, Aniana A, Sayer JM, Louis JM, Weber IT. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51:2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agniswamy J, Louis JM, Roche J, Harrison RW, Weber IT. Structural studies of a rationally selected multi-drug resistant HIV-1 protease reveal synergistic effect of distal mutations on flap dynamics. PLoS ONE. 2016;11(12):e0168616. doi: 10.1371/journal.pone.0168616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber IT, Harrison RW. Tackling the problem of HIV drug resistance. Adv. Biochem. 2016;62:273–279. [PubMed] [Google Scholar]
- 46.Kožíšek M, Lepšík M, Grantz Šašková K, Brynda J, Konvalinka J, Rezacova P. Thermodynamic and structural analysis of HIV protease resistance to DRV - analysis of heavily mutated patient-derived HIV-1 proteases. FEBS J. 2014;281:1834–1847. doi: 10.1111/febs.12743. [DOI] [PubMed] [Google Scholar]
- 47.Louis JM, Aniana A, Weber IT, Sayer JM. Insights into the inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc. Natl Acad. Sci. USA. 2011;108:9072–9077. doi: 10.1073/pnas.1102278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JH, Sayer JM, Aniana A, et al. Binding of clinical inhibitors to a model precursor of a rationally selected multidrug resistant HIV-1 protease is significantly weaker than to the released mature enzyme. Biochemistry. 2016;55:2390–2400. doi: 10.1021/acs.biochem.6b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Weber IT, Harrison RW. Identifying representative drug resistant mutants of HIV. BMC Bioinformatics. 2015;16(Suppl. 17):S1. doi: 10.1186/1471-2105-16-S17-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mesplède T, Wainberg MA. Resistance against integrase strand transfer inhibitors and relevance to HIV persistence. Viruses. 2015;7(7):3703–3718. doi: 10.3390/v7072790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llibre JM, Pulido F, García F, García Deltoro M, Blanco JL, Delgado R. Genetic barrier to resistance for dolutegravir. AIDS Rev. 2015;17(1):56–64. [PubMed] [Google Scholar]
- 52.Laskey SB, Siliciano RF. Quantitative evaluation of the antiretroviral efficacy of dolutegravir. JCI Insight. 2016;1(19):e90033. doi: 10.1172/jci.insight.90033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldgur Y, Craigie R, Cohen GH, et al. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl Acad. Sci. USA. 1999;96(23):13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Métifiot M, Johnson BC, Kiselev E, et al. Selectivity for strand-transfer over 3′-processing and susceptibility to clinical resistance of HIV-1 integrase inhibitors are driven by key enzyme-DNA interactions in the active site. Nucleic Acids Res. 2016;44(14):6896–6906. doi: 10.1093/nar/gkw592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464(7286):232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Passos DO, Li M, Yang R, et al. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 2017;355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]; • New structure of HIV integrase in complex with viral DNA gives insight into reaction mechanism.
- 57.Prosperi MC, De Luca A. Computational models for prediction of response to antiretroviral therapies. AIDS Rev. 2012;14(2):145–153. [PubMed] [Google Scholar]
- 58.Lengauer T, Pfeifer N, Kaiser R. Personalized HIV therapy to control drug resistance. Drug Discov. Today Technol. 2014;11:57–64. doi: 10.1016/j.ddtec.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology. 2012;55(2):98–101. doi: 10.1159/000331998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandaranayake RM, Kolli M, King NM, et al. The effect of clade-specific sequence polymorphisms on HIV-1 protease activity and inhibitor resistance pathways. J. Virol. 2010;84:9995–10003. doi: 10.1128/JVI.00505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koning FA, Castro H, Dunn D, Tilston P, Cane PA, Mbisa JL UK Collaborative Group on HIV Drug Resistance. Subtype-specific differences in the development of accessory mutations associated with high-level resistance to HIV-1 nucleoside reverse transcriptase inhibitors. J. Antimicrob. Chemother. 2013;68(6):1220–1236. doi: 10.1093/jac/dkt012. [DOI] [PubMed] [Google Scholar]
- 62.Quashie PK, Oliviera M, Veres T, et al. Differential effects of the G118R, H51Y, and E138K resistance substitutions in different subtypes of HIV integrase. J. Virol. 2015;89(6):3163–3175. doi: 10.1128/JVI.03353-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beerenwinkel N, Däumer M, Oette M, et al. Geno2pheno: estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res. 2003;31:3850–3855. doi: 10.1093/nar/gkg575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zazzi M, Incardona F, Rosen-Zvi M, et al. Predicting response to antiretroviral treatment by machine learning: the EuResist project. Intervirology. 2012;55(2):123–127. doi: 10.1159/000332008. [DOI] [PubMed] [Google Scholar]
- 65.Tarca AL, Carey VJ, Chen XW, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput. Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pironti A, Pfeifer N, Kaiser R, Walter H, Lengauer T. Improved therapy-success prediction with GSS estimated from clinical HIV-1 sequences. J. Int. AIDS Soc. 2014;17(4 Suppl. 3):19743. doi: 10.7448/IAS.17.4.19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beerenwinkel N, Montazeri H, Schuhmacher H, et al. Swiss HIV Cohort Study. The individualized genetic barrier predicts treatment response in a large cohort of HIV-1 infected patients. PLoS Comput. Biol. 2013;9(8):e1003203. doi: 10.1371/journal.pcbi.1003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masso M. Sequence-based predictive models of resistance to HIV-1 integrase inhibitors: an n-grams approach to phenotype assessment. Curr. HIV Res. 2015;13(6):497–502. doi: 10.2174/1570162x13666150624100535. [DOI] [PubMed] [Google Scholar]
- 69.Goenen M, Margolin AA. Drug susceptibility prediction against a panel of drugs using kernelized bayesian multitask learning. Bioinformatics. 2014;30(17):556–563. doi: 10.1093/bioinformatics/btu464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rokach L. Ensemble-based classifiers. Artif. Intell. Rev. 2010;33:1–30. [Google Scholar]
- 71.Heider D, Senge R, Cheng W, Hüllermeier E. Multilabel classification for exploiting cross-resistance information in HIV-1 drug resistance prediction. Bioinformatics. 2013;29(16):1946–1952. doi: 10.1093/bioinformatics/btt331. [DOI] [PubMed] [Google Scholar]
- 72.Riemenschneider M, Senge R, Neumann U, Hüllermeier E, Heider D. Exploiting HIV-1 protease and reverse transcriptase cross-resistance information for improved drug resistance prediction by means of multi-label classification. BioData Min. 2016;9:10. doi: 10.1186/s13040-016-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards FM. The interpretation of protein structures: total volume, group volume distributions and packing density. J. Mol. Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 74.Poupon A. Voronoi and Voronoi-related tessellations in studies of protein structure and interaction. Curr. Opin. Struct. Biol. 2004;14:233–241. doi: 10.1016/j.sbi.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 75.Bose P, Yu X, Harrison RW. Bioinformatics and Biomedicine Workshops (BIBMW) IEEE International Conference; Atlanta, GA, USA: 12–15 November 2011. Encoding protein structure with functions on graphs; pp. 338–344. Proceedings of. [Google Scholar]
- 76.Yu X, Weber IT, Harrison RW. Proceedings of: the SIAM International Conference on Data Mining SDM 2013. SIAM; Austin, TX, USA: 1–4 May 2013. Sparse representation for HIV-1 protease drug resistance prediction; pp. 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu X, Weber IT, Harrison RW. Prediction of HIV drug resistance from genotype with encoded three-dimensional protein structure. BMC Genomics. 2014;15(Suppl. 5):S1. doi: 10.1186/1471-2164-15-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durham EA, Yu X, Harrison RW. IEEE Symposium on Computational Intelligence in Healthcare and e-health (CICARE) Orlando, FL: 9–12 December 2014. FDT 2.0: Improving scalability of the fuzzy decision tree induction tool – integrating database storage 2014. Proceedings of. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen CH, Yu X, Harrison RW, Weber IT. Automated prediction of HIV drug resistance from genotype data. BMC Bioinform. 2016;17(Suppl. 8):278. doi: 10.1186/s12859-016-1114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]