Abstract
CMTM4 is the most conserved member of chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing (CMTM) family on chromosome 16q22.1, a locus that harbors a number of tumor-suppressor genes. In previous studies, CMTM4 was reported to be downregulated and exhibited tumor-suppressor activities by regulating cell growth and cell cycle in clear cell renal cell carcinoma. However, its roles in tumorigenesis of hepatocellular carcinoma (HCC) remain poorly studied. This study first investigated the expression of CMTM4 in HCC, and then examined the association between the expression of CMTM4 with the clinicopathological features and prognosis of HCC patients. It was found that CMTM4 was downregulated in HCC tissues, compared with matched adjacent nontumor tissues, as detected by immunohistochemistry. In addition, Kaplan–Meier survival analysis showed that the negative expression of CMTM4 was associated with decreased overall survival rates in patients with HCC. The results of this study suggest CMTM4 plays a role as a tumor suppressor in HCC and CMTM4 negative expression is a risk factor for poor prognosis of HCC.
Keywords: chemokine-like factor-like MARVEL transmembrane domain-containing 4, hepatocellular carcinoma, immunohistochemistry, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent malignancy in the world and the third most common cause of cancer-related mortality.1 It is estimated that over 748,300 new cases are diagnosed every year, of which about 55% come from People’s Republic of China.2 The majority of patients with HCC are diagnosed in the middle or late stages of the disease and have poor overall survival (OS) rates. Although the curative effect of comprehensive treatment for HCC based on surgery has substantially improved, clinical cure rates and long-term survival rates for HCC remain low.3,4 In addition, 60%–70% of patients with HCC have recurrence or metastasis within 5 years following cancer resection.5 At present, there are extensive studies on the etiology, prognosis, and carcinogenesis of HCC,6–10 but the exact molecular mechanism has not yet been illustrated.
CMTM4 is the most conserved member of the human chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM) on chromosome 16q22.1, a locus that harbors a number of tumor-suppressor genes.11–15 CMTM4 encodes three transcript variants, CMTM4-v1, -v2 and -v3, and codes 234, 208, and 179 amino acids, respectively. However in many types of tissues and cell lines, only CMTM4-v1 and -v2 could be detected. It was reported that overexpression of CMTM4-v1 and -v2 can inhibit HeLa cell growth via G2/M phase accumulation without inducing apoptosis.16 By omic data analysis, Ting Li et al17 found CMTM4 was significantly reduced in 86.9% (53/61) of clear cell renal cell carcinoma (ccRCC) tissues compared with the paired adjacent nontumor tissues, as well as in the 786-O and A498 ccRCC cell lines. Restoration of CMTM4 significantly suppressed 786-O cell growth by inducing G2/M cell cycle arrest and p21 upregulation, and cell migration was also inhibited. Therefore, CMTM4 might be an important gene involved in cell growth and cell cycle regulation.
The aim of this study was to investigate the association between CMTM4 expression and prognosis of HCC. CMTM4 expression levels were detected in 75 paired HCC tissues and adjacent nontumor tissues by immunohistochemistry. And then, the relationship between expression of CMTM4 with clinicopathological features and prognosis of HCC patients was analyzed.
Materials and methods
Patients and samples
All patients were newly diagnosed with HCC by histopathology and recruited from the First Affiliated Hospital of Guilin Medical University from 2007 to 2015. A total of 75 paired cancerous and matched adjacent normal tissues were collected from HCC patients undergoing hepatectomy. Prior to operation, no patient had received any type of treatment. The age of the patients ranged from 18 to 73 years, and the mean age was 49.81±13.29 years. The tissues were prepared into a tissue microarray chip by Guilin Fanpu Biological Technology Co., Ltd. (Guilin, People’s Republic of China). Clinicopathological data, including age, sex, tumor size, tumor number, liver cirrhosis, and serum level of α-fetoprotein were collected from the medical records of each patient. Tumor staging was classified according to the World Health Organization standards, and histological tumor grading was based on Edmondson–Steiner classification. OS was determined from the date of surgery to the time of death or the last follow-up. This study was approved by the ethics committee of Guilin Medical University, and written informed consent was obtained from each patient involved in the study.
Immunohistochemical analysis and scoring
Immunohistochemical analysis was performed on formalin-fixed, paraffin-embedded biopsy samples to detect the expression of CMTM4. A total of 150 paraffin-embedded biopsy samples were first heated at 60°C for 1 hour, dew-axed, rehydrated through decreasing concentrations of EDTA buffer (pH 8.0), and heated at 120°C for 2 minutes to retrieve the antigen. And then the sections were soaked in 3% peroxide solution for 15 minutes to remove endogenous peroxidase. After washing in phosphate-buffered saline (PBS) buffer, the sections were blocked with sheep serum for 20 minutes and incubated with CMTM4 primary antibody (NBP1-84457, 1:100; Novus Biologicals LLC, Littleton, CO, USA) in a humidified chamber at 4°C overnight. The sections were then washed with PBS for 5 minutes and incubated with the anti-rabbit IgG antibody (MaiXin Company, FuZhou, People’s Republic of China) at 37°C for 30 minutes. After washing with PBS, the sections were incubated with 3DAB solution (MaiXin Company) and counterstained with hematoxylin (Beyotime Institute of Biotechnology), which was visualized under a light microscope. All the immunostained sections were evaluated in a blind manner with no knowledge of the clinicopathological information.
For the assessment of CMTM4, five fields in each specimen were randomly selected to determine the mean percentage of immunostained cells relative to the total number of cells. The positive cell staining percentages were scored into five categories: 0 for ≤5%, 1 for 6%–25%, 2 for 26%–50%, 3 for 51%–75%, and 4 for >75% staining. The immunohistochemical staining intensities were also scored into four grades (0, 1, 2, and 3), according to the brown color intensity of the cells: 0 for no color, 1 for light color, 2 for medium color, and 3 for dark brown color. Finally, the percentage and intensity scores were multiplied for the CMTM4 staining score. The total scores were defined: negative expression of CMTM4 for ≤4 and positive expression of CMTM4 for >4.
Statistical analysis
The differences in the distribution of selected variables and expression of CMTM4 were assessed using χ2 test. The Kaplan–Meier estimate was used for survival analysis, and the log-rank test was selected to compare the cumulative survival durations in the HCC patients. All statistical tests were two sided with a 0.05 significance level, and all tests were conducted using SPSS software, version 19.0 (IMB SPSS, Armonk, NY, USA).
Results
Expression of CMTM4 in HCC tissues
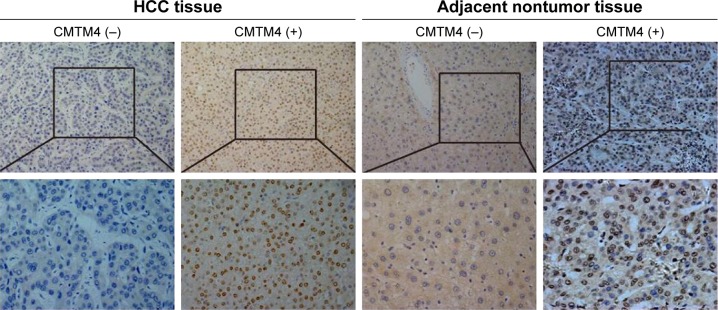
There were 75 pairs of HCC tissues and matched adjacent nontumor tissues included in the study. Immunohistochemical analysis was performed in all 75 paired archival paraffin-embedded samples. Overall, 21 of the 75 (28.00%) cases were positive for the expression of CMTM4 in the cancerous tissues, whereas 49 of the 75 (65.33%) cases were positive for the expression of CMTM4 in the adjacent nontumor tissues. The expression of CMTM4 in HCC tissues was significantly reduced compared with that in the adjacent nontumor tissues (P<0.05, Table 1), and its expression was mainly located in the nucleus (Figure 1).
Table 1.
CMTM4 expression in HCC tissues and adjacent non-tumor tissues
| HCC tissues | Adjacent tissues
|
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 8 | 13 | 21 |
| Negative | 41 | 13 | 54 |
| Total | 49 | 26 | 75 |
Abbreviations: CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HCC, hepatocellular carcinoma.
Figure 1.
The expression of CMTM4 in HCC tissues and adjacent nontumor tissues by immunohistochemistry (magnification, ×200).
Abbreviations: CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HCC, hepatocellular carcinoma.
The association between the expression of CMTM4 and the clinicopathological characteristics of HCC patients
The clinicopathological factors analyzed in relation to the expression of CMTM4 are listed in Table 2. The expression of CMTM4 was significantly associated with tumor size, clinical stage, tumor, node, and metastasis (TNM) staging, and metastasis (P<0.05, Table 2). Nevertheless, no significant association was detected between sex, age, tumor number, pathology classification, liver cirrhosis, α-fetoprotein, and expression of CMTM4 (P>0.05, Table 2). Negative expression of CMTM4 is associated with metastasis in multivariable logistic regression analysis adjusted for sex, age, tumor size, clinical stage, TNM staging, and metastasis (P<0.05, Table 3).
Table 2.
Association between the expression of CMTM4 and clinicopathological features of HCC
| Variables | n | CMTM4
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Sex | 0.508 | 0.476 | |||
| Male | 61 | 16 | 45 | ||
| Female | 14 | 5 | 9 | ||
| Age (years) | 0.585 | 0.445 | |||
| <50 | 34 | 11 | 23 | ||
| ≥50 | 41 | 10 | 31 | ||
| Smoking status | 2.818 | 0.093 | |||
| Never | 42 | 15 | 27 | ||
| Ever | 33 | 6 | 27 | ||
| Alcohol use | 0.861 | 0.353 | |||
| Never | 35 | 8 | 27 | ||
| Ever | 40 | 13 | 27 | ||
| HBV infection | 1.339 | 0.247 | |||
| Negative | 15 | 6 | 9 | ||
| Positive | 60 | 15 | 45 | ||
| Tumor size (cm) | 5.030 | 0.025 | |||
| <5 | 38 | 15 | 23 | ||
| ≥5 | 37 | 6 | 31 | ||
| Tumor number | 2.083 | 0.149 | |||
| 1 | 40 | 14 | 26 | ||
| ≥2 | 35 | 7 | 28 | ||
| Clinical stage | 4.825 | 0.028 | |||
| I + II | 42 | 16 | 26 | ||
| III + IV | 33 | 5 | 28 | ||
| TNM staging | 4.240 | 0.039 | |||
| T1 + T2 | 43 | 16 | 27 | ||
| T3 + T4 | 32 | 5 | 27 | ||
| Pathology classification | 2.045 | 0.153 | |||
| I + II | 33 | 12 | 21 | ||
| III | 42 | 9 | 33 | ||
| Liver cirrhosis | 1.844 | 0.174 | |||
| No | 37 | 13 | 24 | ||
| Yes | 38 | 8 | 30 | ||
| Metastasis | 6.771 | 0.009 | |||
| No | 51 | 19 | 32 | ||
| Yes | 24 | 2 | 22 | ||
| α-fetoprotein (ng/mL) | 2.499 | 0.114 | |||
| <400 | 32 | 12 | 20 | ||
| ≥400 | 43 | 9 | 34 | ||
Notes: Bold values indicate significance. P-values are based on the χ2 test.
Abbreviations: CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; TNM, tumor node metastasis.
Table 3.
Multivariable logistic regression analysis of CMTM4 expression in HCC patients
| Variables | B | Wald | P-value | ORa | 95% CI for OR
|
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Metastasis | 2.531 | 23.769 | 0.005 | 1.654 | 1.136 | 4.463 |
| Tumor size | 1.132 | 8.645 | 0.091 | 1.134 | 0.694 | 1.931 |
Notes:
OR adjusted for age, sex, smoking status, alcohol use, and HBV infection.
Abbreviations: CI, confidence interval; CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; OR, odds ratio.
The association between the expression of CMTM4 and the prognosis of HCC
To validate the prognostic value of CMTM4 in HCC, this study also assessed the association between the expression of CMTM4 and survival rates using Kaplan–Meier analysis with a log-rank test. As shown in Figure 2, the survival rates of the patients with HCC were significantly different between cases positive for CMTM4 expression and cases negative for CMTM4 expression (P=0.037). Negative expression for CMTM4 was an independent risk factor for prognosis of HCC in Cox proportional hazards models (P=0.041, odds ratio =2.433, 95% confidence interval =1.075–5.786, Table 4). In patients with HCC, a high expression level of CMTM4 indicated a better prognosis compared with those with a low expression level of CMTM4, which indicated that CMTM4 played as a tumor suppressor in HCC.
Figure 2.
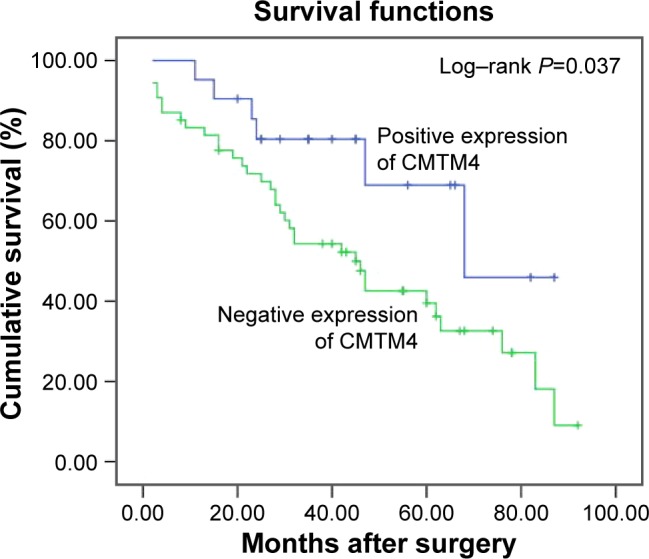
Effect of the expression of CMTM4 on survival rates of HCC patients who underwent resection determined via Kaplan-Meier model analysis.
Abbreviations: CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HCC, hepatocellular carcinoma.
Table 4.
Multivariate analysis of influenced factors for overall survival of HCC patients after operation
| Variables | B | Wald | P-value | OR | 95% CI for OR
|
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| CMTM4 | 0.897 | 4.043 | 0.041 | 2.433 | 1.075 | 5.786 |
Abbreviations: CI, confidence interval; CMTM4, chemokine-like factor-like MARVEL transmembrane domain-containing family 4; HCC, hepatocellular carcinoma; OR, odds ratio.
Discussion
The epigenome is aberrantly reprogrammed leading to a wide range of heritable changes in gene expression such as silencing of tumor-suppressor genes in cancers.18 Methylation of CpG at the promoter region of tumor-suppressor gene results in stable gene silencing through direct inhibition of transcription factor binding or by recruitment of methyl-binding domain proteins, which can cause gene product expression to decline or be absent.19,20 Plate et al17 found that overexpression of CMTM4-v1 and -v2 can inhibit HeLa cell growth via G2/M phase accumulation without inducing apoptosis. Ting Li et al18 found a substantial downregulation of CMTM4 in ccRCC. CMTM4 was significantly reduced in ccRCC tissues compared with the paired adjacent nontumor tissues, as well as in the 786-O and A498 ccRCC cell lines. In this study, immunohistochemistry was used to detect paired HCC and adjacent nontumor tissues, and we found that the expression of CMTM4 in HCC tissues was significantly reduced compared with that in the adjacent nontumor tissues (P<0.05), which is consistent with the previous research findings.17,18 However, the function of CMTM4 in tumorigenesis of HCC remains poorly defined and needs to be further studied.
Despite improved diagnostic and treatment strategies such as surgical resection, transcatheter arterial chemoembolization, and liver transplantation, the OS of patients with HCC remains poor.21–23 Studies found that the clinicopathological features of tumor have very close ties with the prognosis of tumor, including tumor size, tumor number, tumor staging, alpha-fetoprotein levels and metastasis, liver cirrhosis, and vascular invasion often indicates poor prognosis of HCC.24–28 In this study, we found that the expression of CMTM4 was significantly associated with tumor size, clinical stage, TNM staging, and metastasis (P<0.05), and negative expression of CMTM4 was associated with metastasis in multivariable logistic regression analysis. Further analysis found that postoperative survival time of patients with negative expression of CMTM4 was obviously lower than that in positive group, suggesting that CMTM4 played a role as a tumor suppressor in HCC.
Taken together, this study first reported that CMTM4 negative expression was a risk factor for poor prognosis of HCC. It was expected that CMTM4 might be a new molecular marker for early diagnosis and prognosis of HCC combined with other markers. However, the sample size is small and the analysis bias exists; whether it could be used as a single index to prognosis or a marker of tumor invasion and metastasis still needs to be verified in large samples.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (NSFC, 81460515).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Q, Li N, Zeng X, et al. Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget. 2015;6(6):4440–4450. doi: 10.18632/oncotarget.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Miao R, Yang H, et al. Prognostic factors after liver resection for hepatocellular carcinoma: a single-center experience from China. Am J Surg. 2012;203(6):741–750. doi: 10.1016/j.amjsurg.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Lee SC, Tan HT, Chung MC. Prognostic biomarkers for prediction of recurrence of hepatocellular carcinoma: current status and future prospects. World J Gastroenterol. 2014;20(12):3112–3124. doi: 10.3748/wjg.v20.i12.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Q, Wang XY, Zhou J, Fan J. Multiple carcinogenesis contributes to the heterogeneity of HCC. Nat Rev Gastroenterol Hepatol. 2015;12(1):13. doi: 10.1038/nrgastro.2014.6-c1. [DOI] [PubMed] [Google Scholar]
- 7.Luo W, Zhu X, Liu W, et al. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget. 2016;7(52):86420–86432. doi: 10.18632/oncotarget.13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X, Wang Z, Qiu X, et al. Associations between single nucleotide polymorphisms in RYBP and the prognosis of hepatocellular carcinoma in a Chinese population. Carcinogenesis. 2017;38(5):532–540. doi: 10.1093/carcin/bgx031. [DOI] [PubMed] [Google Scholar]
- 9.Chua HH, Tsuei DJ, Lee PH, et al. RBMY, a novel inhibitor of glycogen synthase kinase 3beta, increases tumor stemness and predicts poor prognosis of hepatocellular carcinoma. Hepatology. 2015;62(5):1480–1496. doi: 10.1002/hep.27996. [DOI] [PubMed] [Google Scholar]
- 10.Chen HA, Kuo TC, Tseng CF, et al. Angiopoietin-like protein 1 antagonizes MET receptor activity to repress sorafenib resistance and cancer stemness in hepatocellular carcinoma. Hepatology. 2016;64(5):1637–1651. doi: 10.1002/hep.28773. [DOI] [PubMed] [Google Scholar]
- 11.Downing TE, Oktay MH, Fazzari MJ, Montagna C. Prognostic and predictive value of 16p12.1 and 16q22.1 copy number changes in human breast cancer. Cancer Genet Cytogenet. 2010;198(1):52–61. doi: 10.1016/j.cancergencyto.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Keck B, Ellmann C, Stoehr R, et al. Comparative genomic hybridization shows complex genomic changes of plasmacytoid urothelial carcinoma. Urol Oncol. 2014;32(8):1234–1239. doi: 10.1016/j.urolonc.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Fu L, Dong SS, Xie YW, et al. Down-regulation of tyrosine aminotransferase at a frequently deleted region 16q22 contributes to the pathogenesis of hepatocellular carcinoma. Hepatology. 2010;51(5):1624–1634. doi: 10.1002/hep.23540. [DOI] [PubMed] [Google Scholar]
- 14.Czarnecka KH, Migdalska-Sek M, Antczak A, et al. Allelic imbalance in 1p, 7q, 9p, 11p, 12q and 16q regions in non-small cell lung carcinoma and its clinical association: a pilot study. Mol Biol Rep. 2013;40(12):6671–6684. doi: 10.1007/s11033-013-2782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Zhang X, Yuan X, Wang L, Xiao Y. MicroRNA-205 inhibits renal cells apoptosis via targeting CMTM4. Iran J Basic Med Sci. 2015;18(10):1020–1026. [PMC free article] [PubMed] [Google Scholar]
- 16.Plate M, Li T, Wang Y, et al. Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol Cells. 2010;29(4):355–361. doi: 10.1007/s10059-010-0038-7. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Cheng Y, Wang P, et al. CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2015;34:122. doi: 10.1186/s13046-015-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raynal NJ, Lee JT, Wang Y, et al. Targeting Calcium signaling induces epigenetic reactivation of tumor suppressor genes in cancer. Cancer Res. 2016;76(6):1494–1505. doi: 10.1158/0008-5472.CAN-14-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60(6):376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 21.Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140(5):1501–1512.e2. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5(3):215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 23.Lam VW, Ng KK, Chok KS, et al. Risk factors and prognostic factors of local recurrence after radiofrequency ablation of hepatocellular carcinoma. J Am Coll Surg. 2008;207(1):20–29. doi: 10.1016/j.jamcollsurg.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Jin J, Niu X, Zou L, et al. AFP mRNA level in enriched circulating tumor cells from hepatocellular carcinoma patient blood samples is a pivotal predictive marker for metastasis. Cancer Lett. 2016;378(1):33–37. doi: 10.1016/j.canlet.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Xiao S, Chang RM, Yang MY, et al. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63(4):1256–1271. doi: 10.1002/hep.28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu AX, Chen D, He W, et al. Integrative biomarker analyses indicate etiological variations in hepatocellular carcinoma. J Hepatol. 2016;65(2):296–304. doi: 10.1016/j.jhep.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Uchino K, Tateishi R, Shiina S, et al. Hepatocellular carcinoma with extrahepatic metastasis: clinical features and prognostic factors. Cancer. 2011;117(19):4475–4483. doi: 10.1002/cncr.25960. [DOI] [PubMed] [Google Scholar]
- 28.Farinati F, Vitale A, Spolverato G, et al. Development and validation of a new prognostic system for patients with hepatocellular carcinoma. PLoS Med. 2016;13(4):e1002006. doi: 10.1371/journal.pmed.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]