Summary
The genetic basis of sleep regulation remains poorly understood. In C. elegans, cellular stress induces sleep through Epidermal Growth Factor (EGF)-dependent activation of the EGF receptor in the ALA neuron. The downstream mechanism by which this neuron promotes sleep is unknown. Single-cell RNA-seq of ALA reveals that the most highly expressed, ALA-enriched genes encode neuropeptides. Here we have systematically investigated the four most highly enriched neuropeptides: flp-7, nlp-8, flp-24, and flp-13. When individually removed by null mutation, these peptides had little or no effect on stress-induced sleep. However, stress-induced sleep was abolished in the nlp-8; flp-24; flp-13 triple mutant animals, indicating that these neuropeptides work collectively in controlling stress-induced sleep. We tested the effect of overexpression of these neuropeptide genes on five behaviors modulated during sleep –pharyngeal pumping, defecation, locomotion, head movement, and avoidance response to an aversive stimulus – and found that if individually overexpressed, each of three neuropeptides (nlp-8, flp-24, or flp-13) induced a different suite of sleep-associated behaviors. These overexpression results raise the possibility that individual components of sleep might be specified by individual or combinations of neuropeptides.
Keywords: Sleep regulation, RFamides, Neuropeptide modulation, single-neuron RNA-seq, genetic redundancy
eTOC Blurb
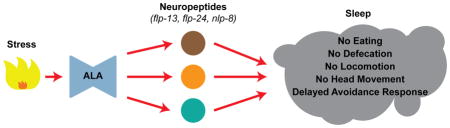
Sleep is a complex behavioral state that requires the coordinated regulation of multiple behaviors. Nath et al. investigate the downstream mechanism by which a neurosecretory cell coordinately promotes sleep in C. elegans. They discovered that sleep emerges from the collective action of three neuropeptide-encoding genes.
Introduction
Sleep is a complex behavioral state that requires the coordinated regulation of multiple behaviors and physiological processes. Sleep is defined as a state of reversible behavioral quiescence, increased arousal threshold, and homeostatic regulation [1,2]. This physiological state has been observed both in invertebrates such as Caenorhabditis elegans and Drosophila melanogaster, as well as in vertebrates such as Danio rerio, Mus musculus, and Homo sapiens [1,2]. Sleep is a genetically encoded state, and key sleep genes are conserved from nematodes to mammals [3–6]. One such sleep regulator is Epidermal Growth Factor Receptor (EGFR), whose activation promotes sleep in both C. elegans and D. melanogaster [7,8], and inhibits locomotion in mammals [9–11].
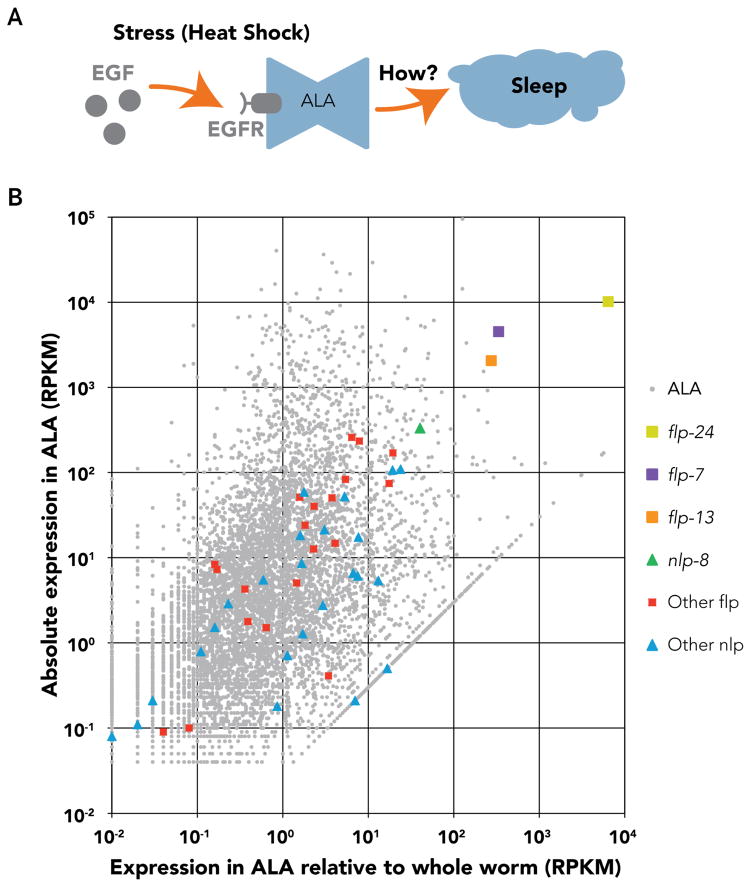
C. elegans sleep has been observed during developmental molting (lethargus), satiety, and Epidermal Growth Factor (EGF)/EGFR signaling [7,12–18]. Here we investigated the C. elegans EGF-induced sleep pathway, thought to represent a distinct molecular pathway from developmentally linked sleep (Figure 1A; [18]). The EGF-induced sleep state occurs in two contexts: by overexpressing the EGF ortholog (LIN-3C; [7]), or by EGF-signaling after stress (such as temperature elevation) in wild-type animals [14]. The EGF receptor ortholog (LET-23) is necessary for EGF-induced sleep and expressed in the ALA neuron [7]. Ablation of ALA demonstrated that it is necessary for EGF-induced sleep [7]. EGF-induced sleep is suppressed by genetic inactivation of the ALA neuron with null mutations of ceh-14 or ceh-17, genes that respectively encode LIM-class and Paired-like homeodomain transcription factors [7,14,19]. These transcription factors control expression of genes in ALA shown to be required for EGF-induced sleep including EGFR [7,14,19]. The mechanism by which the ALA neuron controls animal behavior to induce the sleep phenotype is unknown. Henceforth, we refer to EGF-induced sleep as stress induced-sleep [17].
Figure 1. Single-cell RNA-seq of ALA, the neuron central to C. elegans stress-induced sleep.
(A) Stress-induced sleep is regulated by LIN-3C (EGF) and LET-23 (EGFR) expressed on the surface of ALA. In this work, we study the mechanism of sleep induction downstream of ALA. (B) Single-cell RNA-seq expression data of 8,133 protein-coding genes (grey) collected from two pools of microdissected ALA neurons (four and five cells; see also Figure S1; Table S1) compared with mixed-stage whole larvae. The ratio of expression level of protein-coding genes from the ALA neuron versus whole larvae shows that four neuropeptide-coding genes have ≥10-fold higher expression in ALA than in whole larvae: flp-24, flp-7, flp-13, and nlp-8 (highlighted with colored squares for flps, and a green triangle for nlp-8; see also Figure S2). Expression levels of other flp and nlp coding genes are also highlighted by red squares and blue triangles respectively (see also Table S2). RPKM unit: reads per kilobase of transcript per million mapped reads.
Little is known about the sleep-promoting molecules downstream of ALA; but they may include neuropeptides, which have been implicated in regulating a wide range of behavioral states, including sleep [20–24]. We hypothesized that ALA serves as a neurosecretory cell that releases neuropeptides to modulate sleep-associated behaviors based on two experimental results. First, mutation of unc-31, which encodes a protein important for dense core vesicle (DCV) fusion [25], inhibits stress-induced sleep [7], suggesting that neuropeptide release is necessary for this state. Second, genetic axotomy of ALA does not inhibit stress-induced sleep [7], indicating that the axon of ALA, and thus neurotransmission, is dispensable for this state, and providing additional support for the hypothesis that neuropeptides mediate stress-induced sleep.
Only a few neuropeptide-encoding genes are known to be expressed in ALA, and little is known about their physiological roles. One such gene, flp-7, encodes a FMRFamide-like peptide not required for stress-induced sleep [19], whereas another, flp-13, was previously shown to be partially required for stress-induced sleep [13]. To identify novel genes that regulate sleep, we performed single-neuron RNA-seq of ALA, and observed that this neuron transcribed several genes encoding neuropeptides. We systematically characterized the four most highly expressed, ALA-enriched neuropeptides: flp-7, nlp-8, flp-24, and flp-13. Null mutation of each individual neuropeptide had little or no effect on stress-induced sleep, while a triple knockout (nlp-8; flp-24; flp-13) was fully defective. Overexpression experiments showed that these three neuropeptide genes had an effect on the five behaviors that are modulated during sleep: pharyngeal pumping, defecation, locomotion, head movement, and avoidance response. Each neuropeptide (nlp-8, flp-24, or flp-13) induced a different suite of sleep-associated behaviors. Taken together, these results demonstrate that the collective action of three neuropeptide genes results in stress-induced sleep.
Results
Identification of neuropeptide-coding genes enriched in the ALA neuron
To identify sleep-promoting neuropeptides expressed in ALA, we used microdissection-based single-cell RNA-seq [26] for transcriptomic analysis. We dissected individual ALA neurons from transgenic fourth-stage larval worms (L4) expressing GFP in the ALA neuron, reverse-transcribed mRNA to cDNA, and amplified the cDNA using PCR (Figure S1). Using this procedure we made one pool from four cells and another pool from five cells, and performed deep sequencing. We mapped 17.8 million reads to 8,133 expressed protein-coding genes (Figure 1B; Table S1). Four genes encoding neuropeptides (flp-24, flp-7, flp-13, and nlp-8) were among the most highly expressed and enriched in ALA compared to whole larvae (Figure 1B; Table S2). flp-24 and flp-13 were previously found in the ALA neuron of Ascaris suum by single neuron mass spectrometry [27]. The C. elegans genome contains 122 neuropeptide genes whose mature products contain over 250 distinct neuropeptides [28]. RNA-seq analysis indicated that ALA expresses 23 of the 31 C. elegans FMRFamide-like neuropeptide encoding genes (flp), five of which were expressed at least 10-fold more abundantly in ALA than in whole larvae (Table S2). ALA also expressed 25 of the 51 C. elegans neuropeptide-like-coding genes (nlp), of which five were expressed over 10-fold more abundantly in ALA than in whole larvae (Table S2). These data support our hypothesis that ALA is a neurosecretory cell. The three most ALA-enriched flp genes were flp-24, flp-7, and flp-13 (in descending order of enrichment), and the most enriched nlp gene was nlp-8. Of these, only flp-7 and flp-13 were previously known to be expressed in ALA [13,19,29]. We verified expression of flp-24 and nlp-8 using GFP reporter constructs (Figure S1E–H). Previous analysis showed that each of these genes encodes a prepropeptide containing one or more mature neuropeptides ([30,31]; Figure S2; Figure S3).
Loss-of-function of three ALA-enriched neuropeptides suppresses stress-induced sleep
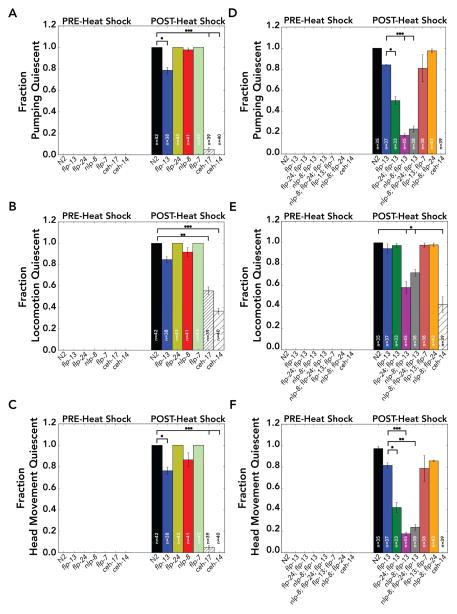
C. elegans sleep has been associated with three behavioral phenotypes: suppression of pharyngeal pumping (a necessary component of eating), suppression of locomotion, and an increased response latency to arousing stimuli [12,14,17,32]. We found that suppression of head movement and defecation are additional sleep-associated behavioral phenotypes. Stress, by heat shock, is sufficient to induce all of these phenotypes (Figure 2; Figure 3; [7,14]). To determine whether ALA-enriched neuropeptides are necessary for stress-induced sleep, we assayed locomotion, head movement, pharyngeal pumping, avoidance response, and defecation before and 30 minutes after heat shock in flp-24, flp-7, flp-13, and nlp-8 single-null mutants (Figure 2; Figure 3; Figure S3; Figure S4; Table S3-S5). Pumping, locomotion, and head movement were repeated in three independent experiments with 10 or more individuals per trial. To score movement we distinguished locomotion, defined as movement of the animal’s centroid in the forward or reverse directions within a 10-second interval, and head movement, defined as dorsal-ventral displacement of the animal’s head from the posterior of the second pharyngeal bulb to the anterior tip.
Figure 2. Double and triple mutants of ALA-enriched neuropeptides suppress pumping, head movement, and locomotion quiescence during stress-induced sleep.
(A–C) The fraction of single-null mutants pumping, locomotion, and head movement quiescent before (PRE) and 30 minutes after (POST) heat shock (a 35°C heat shock was used). N2 are wild-type animals, and ceh-14 and ceh-17 mutants serve as negative controls because they have defective ALA neurons which are deficient in EGF-signaling. (D–F) The fraction of double- or triple-null mutants pumping, locomotion, and head movement quiescent before (PRE) and 30 minutes after (POST) heat shock. (A) & (D) C. elegans were scored as quiescent for pumping if there was no pharyngeal grinder movement during 10 seconds of observation. Mutation of flp-13 weakly suppressed pumping quiescence, which was enhanced by mutation of flp-24 or nlp-8. (B) & (E) C. elegans were scored as quiescent for locomotion if there was no centroid movement during 10 seconds of observation. The negative controls ceh-14 and ceh-17 had background locomotion quiescence, and no suppression of locomotion quiescence was observed in single-null mutants of neuropeptides. (C) & (F) C. elegans were scored as quiescent for head movement if there was no head movement in the dorsal-ventral directions during 10 seconds of observation. Mutation of flp-13 weakly suppressed head movement quiescence, which was enhanced by mutation of flp-24 or nlp-8. Data represents the fraction of animals quiescent from three independent assays, where n=total number of C. elegans. Data shown as mean±SEM, *p<0.05; **p<0.01; ***p<0.001; unpaired t-test with Bonferonni correction for multiple comparison. Statistical comparisons are indicated for comparisons to N2 (A–C) & (E) and to flp-13 (D) & (F). See also Figure S3, Figure S4, and Table S3.
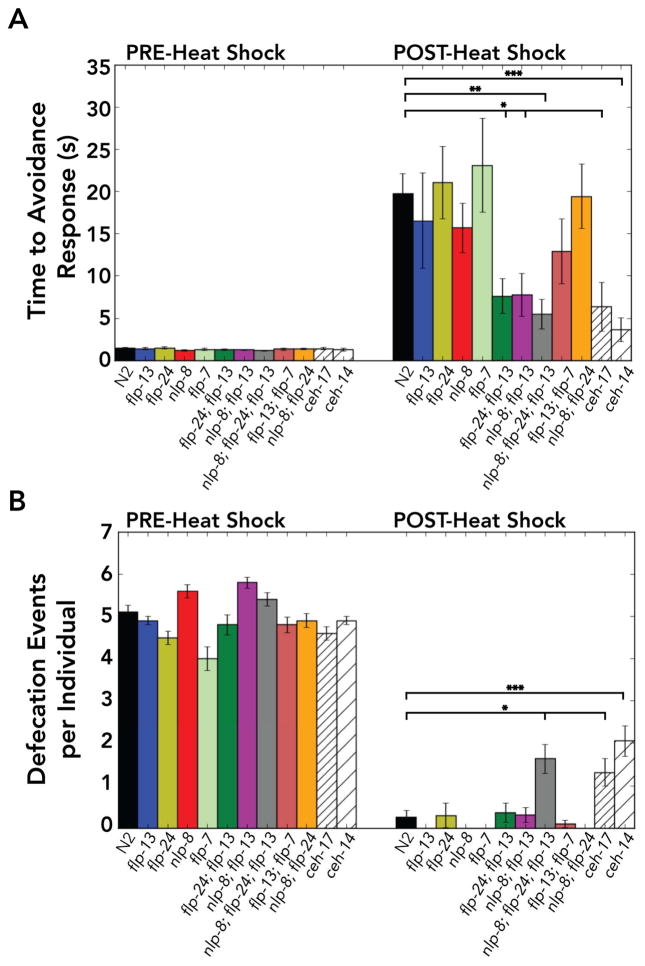
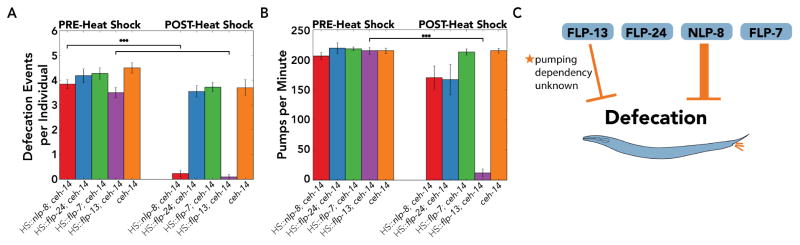
Figure 3. Double and triple mutants of ALA-enriched neuropeptides suppress the increased response latency to aversive stimuli, while only the triple mutant suppresses defecation quiescence during stress-induced sleep.
(A) C. elegans were presented with 30% 1-octanol and avoidance behavior was scored by video recordings before (PRE) and 30 minutes after (POST) heat shock (a 35°C heat shock was used). If there was no response 60 seconds after stimulus delivery, the individuals were classified as non-responsive (Table S4). The nlp-8; flp-13 double mutant, flp-24; flp-13 double mutant, and nlp-8; flp-24; flp-13 triple mutant were resistant to the increased response latency observed during stress-induced sleep. (B) Average number of defecation events for individuals five minutes before (PRE) and 30 minutes after (POST) heat shock. A 33°C heat shock was used for more consistent results (Supplemental Information). We found that our negative controls ceh-14 and ceh-17 had background suppression of defecation at 30 minutes (Table S5). Only the triple mutant (nlp-8; flp-24; flp-13) was resistant to suppression of defecation during stress-induced sleep, and this was statistically indistinguishable from ceh-14 and ceh-17 (p>0.4). Data shown as mean±SEM; n≥10 C. elegans for each strain (see also Table S4–S5); *p<0.05; **p<0.01; ***p<0.001; unpaired t-test with Bonferonni correction for multiple comparison. Statistical comparisons are indicated for comparisons to N2.
flp-24, flp-7, and nlp-8 single-null mutants were indistinguishable from wild type with respect to pumping, locomotion, and head movement after heat shock (p>0.5; Figure 2). flp-13 mutants were slightly resistant to pumping quiescence after heat shock (flp-13: 79±3% pumping quiescent, compared to N2: 100±0%; p<0.05; Figure 2A), confirming the results of Nelson et al. [13]. Resistance to pumping quiescence after heat shock in flp-13 mutants was much weaker than the negative controls, ceh-14 and ceh-17, suggesting that flp-13 is not the only neuropeptide necessary for pumping quiescence during stress-induced sleep (flp-13: 79±3% pumping quiescent compared to ceh-14: 0±0% and compared to ceh-17: 5±3%; p<0.001; Figure 2A).
flp-13 mutants were partially resistant to head movement quiescence after heat shock (flp-13: 76±4% head movement quiescent, compared to N2: 100±0% head movement quiescent; p<0.05; Figure 2C), but we did not observe statistically significant resistance to locomotion quiescence after heat shock in flp-13 mutants, not fully consistent with results reported by Nelson et al. (flp-13: 85±4% locomotion quiescent compared to N2: 100±0% locomotion quiescent; p=0.1; Figure 2B; [13]). ceh-14 and ceh-17 mutants, previously shown to be strongly resistant to heat shock [7, 14], displayed locomotion quiescence after heat shock (ceh-14: 0±0% locomotion quiescent before heat shock compared to ceh-14: 36±4% locomotion quiescent after heat shock, and ceh-17: 0±0% locomotion quiescent before heat shock compared to ceh-17: 56±5% locomotion quiescent after heat shock; p<0.001; Figure 2B). The difference in our results could be due to differences in heat shock protocol or scoring (Supplemental Information). Our data indicate that flp-13 mutants are partly defective for pumping and head movement quiescence after heat shock, but are not defective for locomotion quiescence after heat shock.
The co-expression of several neuropeptide genes in ALA (Figure 1; Figure S1; Table S2) and the partial requirement for flp-13 in stress-induced sleep suggested that these genes might be functionally redundant. We therefore constructed double- and triple-null mutants (Supplemental Information) and found that nlp-8; flp-13 double mutants, flp-24; flp-13 double mutants, and nlp-8; flp-24; flp-13 triple mutants were more resistant to heat shock-induced pumping quiescence than the flp-13 single mutant (flp-24; flp-13: 51±5% pumping quiescent; nlp-8; flp-13: 18±2% nlp-8; flp-24; flp-13: 24±4% compared to flp-13: 84±0%; p<0.05; Figure 2D), suggesting that flp-24 and nlp-8 enhance the effect of flp-13 and strongly induce pumping quiescence. We found that nlp-8; flp-24 double mutants were not resistant to heat shock-induced pumping quiescence compared to wild type (p=0.4; Figure 2D), suggesting that flp-13 is a key regulator of pumping quiescence for stress-induced sleep. However, not all ALA-enriched neuropeptides enhanced the effect of flp-13 on stress-induced sleep, for instance, flp-13; flp-7 double mutants were phenotypically indistinguishable from flp-13 single mutants (p>0.5; Figure 2D).
Loss of either nlp-8 or flp-24 in the flp-13 knockout background enhanced both head movement and pumping quiescence resistance after heat shock (Figure 2F). The nlp-8; flp-13 double mutants and nlp-8; flp-24; flp-13 triple mutants were resistant to locomotion quiescence after heat shock (nlp-8; flp-13: 58±7% locomotion quiescent; nlp-8; flp-24; flp-13: 72±4% locomotion quiescent; compared to N2: 100±0% locomotion quiescent; p<0.05; Figure 2E). The resistance of nlp-8; flp-13 double mutants and nlp-8; flp-24; flp-13 triple mutants were similar to ceh-14 mutants in locomotion quiescence (ceh-14: 42±9% locomotion quiescent; p=0.1; Figure 2E).
A characteristic feature of sleep is an increased arousal threshold, observed as an increased latency to an aversive stimulus. For example, C. elegans typically respond to 30% 1-octanol by moving backward (a reversal) within 5 seconds, but when the animal is asleep the avoidance response either takes longer or does not occur at all [12,32,33]. We defined avoidance response as backward locomotion for at least one pharynx length within one minute of stimulus delivery. No single mutant was resistant, but the flp-24; flp-13 double mutant, nlp-8; flp-13 double mutant, and nlp-8; flp-24; flp-13 triple mutant were all resistant to the increased time required to avoid aversive stimuli (Figure 3; Table S4; n=10). Therefore, these neuropeptides work collectively to induce the increased latency to avoid aversive stimulus.
Another behavior that is suppressed during sleep in C. elegans is defecation (Figure 3B; Table S5). The defecation motor program comprises posterior body wall contraction, anterior body wall contraction, and expulsion [34]. We scored defecation events using five-minute video recordings before and 30 minutes after heat shock. Some ceh-14 and ceh-17 mutants did not defecate after heat shock (Table S5), suggesting that the ceh-14 and ceh-17 mutants do not completely rescue this aspect of quiescence. The background phenotype could result from either expression of these or other neuropeptides in cells other than ALA, or residual expression of these or other neuropeptides in mutant ceh-14 or ceh-17 ALA neurons. No single or double mutant was resistant to the suppression of defecation (n≥10; p>0.5; Figure 3B). We found that the triple mutant was resistant to the suppression of defecation (nlp-8; flp-24; flp-13: 1.6±0.3 events per individual post-heat shock, n=30, compared to N2: 0.3±0.2 events, n=24; p<0.05; Figure 3B). Resistance to the suppression of defecation in the triple mutant was indistinguishable from ceh-14 and ceh-17 animals (p>0.4; Figure 3B). Taken together, our loss-of-function analyses indicate that nlp-8 and flp-24 enhance the effect of flp-13, and that the collective action of these neuropeptides results in stress-induced sleep.
Experimental design to test the sufficiency of neuropeptides in sleep-associated behaviors
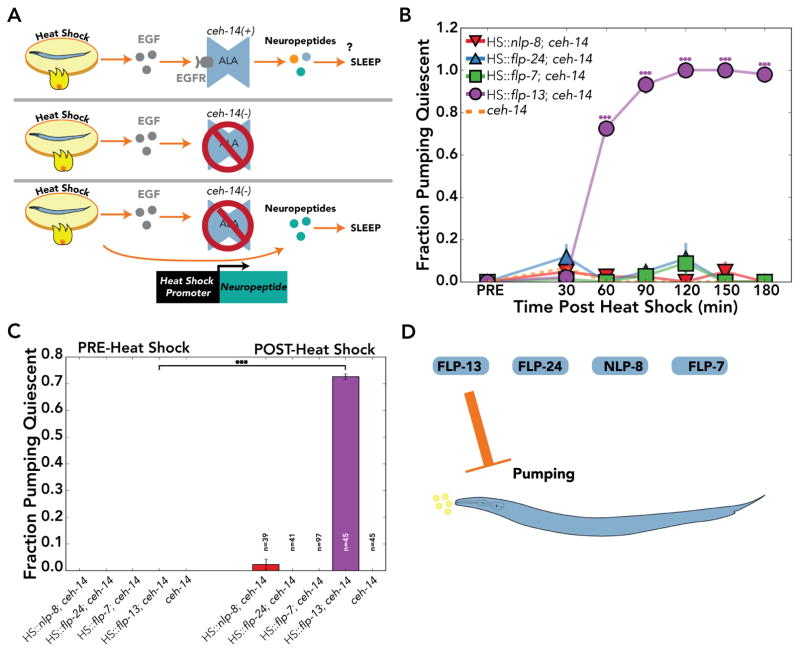
The functions of these candidate sleep-promoting genes were tested using a new overexpression strategy (Figure 4A). To determine if each ALA-enriched neuropeptide was sufficient to induce a sleep-associated behavior, we used a heat shock-inducible promoter to conditionally overexpress each of the four neuropeptide genes (Figure 4A). It is unclear if results from these experiments are hypermorphic or neomorphic, as it is assumed that the neuropeptides are acting at the right targets in physiological concentrations. Such experiments, however, are confounded by the fact that heat shock per se leads to stress-induced sleep [14]. To avoid this possible artifact, all of our overexpression experiments were conducted in ceh-14 mutants, which do not express EGFR (let-23) in the defective ALA neuron, and thus do not exhibit stress-induced sleep (Figure 4A; [7,14,19]).
Figure 4. Overexpression of FLP-13 inhibits pumping in C. elegans.
(A) (top) Heat shock induces sleep by EGF signaling. EGF binds to its receptor, EGFR, on ALA, which is thought to release neuropeptides that induce sleep. (middle) ceh-14 mutants have defective ALA neurons that do not express EGFR and are resistant to heat shock induced sleep. (bottom) A conditional heat shock promoter (HS) driving neuropeptide expression in the ceh-14 background can be induced upon heat shock without the confounding effects of EGF-induced sleep. This overexpression strategy assumes that the neuropeptides are acting at the right sites in physiological concentrations. It is unclear if results from these experiments are hypomorphic or neomorphic. (B) Time course monitoring the fraction of C. elegans pumping quiescent before heat shock (PRE) and up to three hours after heat shock induced neuropeptide overexpression (POST). (C) Fraction of C. elegans pumping quiescent before (PRE) and one hour after (POST) heat shock. (D) Overexpression of flp-13, but neither flp-24, flp-7, nor nlp-8 inhibited pumping. Data represents the fraction of animals quiescent from three or more independent assays, where n=total number of C. elegans. Data shown as mean±SEM, *p<0.05; **p<0.01; ***p<0.001; for (B) two-way ANOVA comparing transgenic strains to ceh-14 mutants with four post-hoc contrast using Bonferroni correction for multiple comparisons. (C) Fisher’s exact test.
flp-13 overexpression inhibits pharyngeal pumping
Following the heat shock protocol illustrated in Figure 4A, we tested the effects of flp-24, flp-7, flp-13, or nlp-8 overexpression on pumping. Pumping was scored for 10 seconds per worm before and for three hours after heat shock at 30 minute intervals. Experiments were repeated three or more times with 10 or more individuals per trial. Among the four genes tested, only flp-13 overexpression induced pumping quiescence (HS::flp-13; ceh-14: 73±1% pumping quiescent compared to ceh-14: 0±0% pumping quiescent, at one hour post-heat shock; p<0.001; Figure 4B,C). We conclude that overexpression of flp-13, but neither flp-24, flp-7, nor nlp-8, is sufficient to inhibit pumping (Figure 4D).
flp-13 or nlp-8 overexpression inhibits defecation
We tested if overexpression of any of the ALA-enriched neuropeptides was sufficient to suppress defecation, we scored defecation using five-minute video recordings before and one hour after heat shock (Figure 5). Overexpression of flp-13 or nlp-8 suppressed the number of defecation events (HS::flp-13; ceh-14: 0.1±0.1 events per individual post-heat shock compared to 3.5±0.2 events per individual pre-heat shock, n=10; p<0.001; HS::nlp-8; ceh-14: 0.2±0.1 events per individual post-heat shock compared to 3.9±0.2 events per individual pre-heat shock, n=13; p<0.001; Figure 5A; Figure S5A). One hour after heat shock, ceh-14 mutants were defecating, and no difference was observed in the total number of defecation events pre- and post-heat shock in ceh-14 controls, nor in animals overexpressing flp-24 or flp-7 (p>0.05; Figure 5A; Figure S5A). However, we did observe a difference in the time between defecation events (i.e., the defecation interval). While ceh-14 mutants defecated, they exhibited a significantly longer defecation interval (p<0.05; Figure S5B), suggesting that the ceh-14 mutation does not completely eliminate this aspect of quiescence. We treated the effects of heat shock on the ceh-14 defecation interval as a background phenotype. We observed no significant difference between the post-heat shock lengthening of the defecation interval in ceh-14 mutants and those overexpressing flp-24 or flp-7 (p≥0.4; Figure S5B).
Figure 5. Overexpression of NLP-8 inhibits defecation while pumping continues.
(A) Average number of defecation events for individuals five minutes before (PRE) and one hour after (POST) heat shock. Overexpressing flp-13 or nlp-8 inhibited defecation. (B) Pumping rate was scored from 10-second video recordings before and one hour after heat shock. (C) Overexpression of either nlp-8 or flp-13, but neither flp-24 nor flp-7, inhibited defecation. The thick line indicates strong and independent inhibition of defecation, while the thinner line indicates inhibition of defecation that may be a consequence of pumping quiescence. (A) & (B) Data shown as mean±SEM; n≥10 C. elegans for each strain (see also Figure S5A); ***p<0.001; paired t-test.
Since arrested defecation might be a consequence of halted feeding [34,35], we wanted to test if arrested defecation in animals overexpressing flp-13 or nlp-8 was independent of pumping inhibition. To address this question, we measured pumping rates before and after heat shock from the same individuals, including those that ceased defecation (Figure 5B). Consistent with Figure 4, we found that most flp-13 overexpressing animals (7 of 10) did not pump after heat shock. The 3 animals that continued to pump did so at reduced rates 40.0±12.1 pumps per minute (Figure 5B). This experiment was not able to determine if pumping and defecation are controlled separately by flp-13 overexpression. However, our nlp-8 overexpression results indicate that inhibition of defecation does not itself inhibit pumping. These data suggest that defecation and pumping rates, two aspects of the C. elegans sleep state, can be controlled separately by different neuropeptides.
flp-24, flp-13, or nlp-8 overexpression inhibits specific aspects of movement
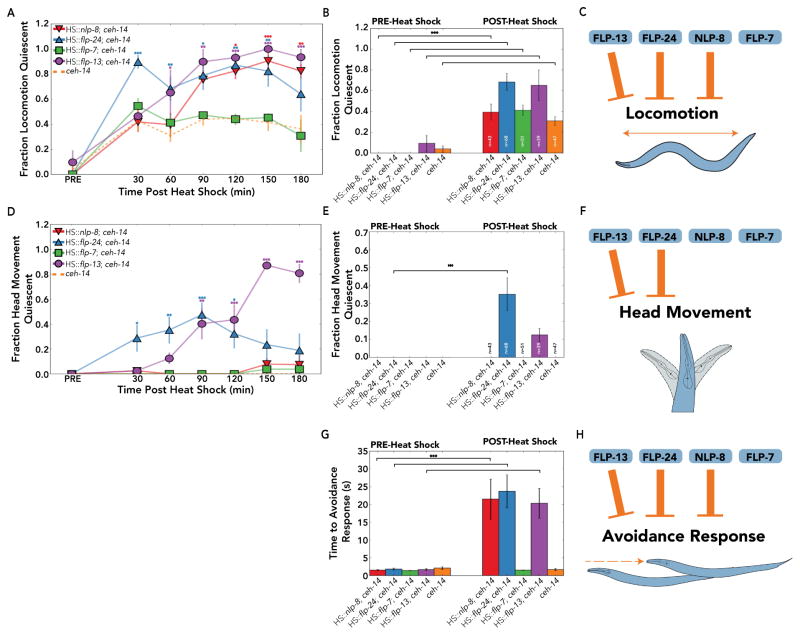
Locomotion quiescence is a canonical sleep-associated behavior [1,2]. Locomotion was scored before heat shock and every 30 minutes for 3 hours after heat shock in a blinded manner. We repeated these experiments three or more times with 10 or more individuals per trial. After heat shock, ceh-14 mutants had less frequent bouts of locomotion (ceh-14: 31±5% locomotion quiescent one hour post-heat shock compared to 4±4% locomotion quiescent pre-heat shock; p<0.001; Figure 6B). For the purposes of this study, we treated the effects of heat shock on ceh-14 locomotion as a background phenotype. flp-7 overexpression did not increase locomotion quiescence compared to ceh-14 (HS::flp-7; ceh-14: 41±6% locomotion quiescent one hour post-heat shock compared to 0±0% locomotion quiescent pre-heat shock; p<0.001; and compared to ceh-14: 31±5% locomotion quiescent one hour post-heat shock; p=0.3; Figure 6A,B). By contrast, worms overexpressing flp-24 showed severe inhibition of locomotion (HS::flp-24; ceh-14: 68±9% locomotion quiescent one hour post-heat shock compared to 0±0% locomotion quiescent pre-heat shock; p<0.001; and compared to ceh-14: 31±5% one hour post-heat shock; p<0.01; Figure 6A,B). flp-24 overexpression suppressed locomotion, but not defecation or pumping, whereas nlp-8 overexpression suppressed locomotion and defecation but not pumping, and flp-13 overexpression suppressed locomotion, defecation, and pumping.
Figure 6. Overexpression of either FLP-13, FLP-24, or NLP-8 inhibit movement and the avoidance response.
(A) & (D) Time-course monitoring the fraction of C. elegans that were locomotion and head-movement quiescent before (PRE) and up to three hours after (POST) heat shock induced neuropeptide overexpression. C. elegans were scored as locomotion quiescent if there was no centroid movement during 10 seconds of observation. C. elegans were scored as head movement quiescent if there was no dorsal-ventral displacement of the worm’s head from the posterior of the second pharyngeal bulb to the anterior tip during 10 seconds of observation. (B) & (E) Fraction of C. elegans locomotion and head movement quiescent pre- and one hour post-heat shock. (C) Overexpression of either flp-24, flp-13, or nlp-8, but not flp-7 inhibited locomotion. (F) Overexpression of either flp-24 or flp-13, but neither flp-7 nor nlp-8 inhibited head movement. (G) C. elegans were presented with 30% 1-octanol and avoidance behavior was scored by video recordings before (PRE) and one hour after heat shock (POST) induced neuropeptide overexpression. If there was no response 60 seconds after stimulus delivery, the individuals were classified as non-responsive (Table S6). (H) Overexpression of either flp-24, flp-13, or nlp-8 strongly increased the response latency to aversive stimuli, while flp-7 overexpression did not. (A–B) & (D–E) Data represents the fraction of animals quiescent from three or more independent assays, where n=total number of C. elegans. Data shown as mean±SEM; *p<0.05; **p<0.01; ***p<0.001; for (A) & (D) two-way ANOVA comparing transgenic strains to ceh-14 mutants with four post-hoc contrast using Bonferroni correction for multiple comparisons. (B) & (E) Fisher’s exact test. (G) Data shown as mean±SEM; n=11 C. elegans for each strain (see also Table S6); ***p<0.001; unpaired t-test with Bonferroni correction for multiple comparisons. See also Video S1–S3.
While scoring locomotion in animals overexpressing flp-24 or flp-13, we noticed a lack of head movement 90 minutes after heat shock, defined as any dorsal-ventral displacement of the worm’s head from the posterior of the second pharyngeal bulb to the anterior tip (Video S1). As with pumping, ceh-14 mutants showed no background quiescence for head movement after heat shock (Figure 6D). Overexpression of either nlp-8 or flp-7 failed to suppress head movement (Figure 6D,E). However, overexpression of either flp-24 or flp-13 inhibited head movement (HS::flp-24; ceh-14: 47±10% head movement quiescent; HS::flp-13; ceh-14: 40±12% head movement quiescent compared to ceh-14: 0±0% head movement quiescent; p<0.01; Figure 5D; Video S2). The movement state of animals overexpressing nlp-8 was unusual; their bodies showed significantly more locomotion quiescence than ceh-14 animals, but their heads continued to move (Figure 6; Video S3). We conclude that overexpression of flp-24, flp-13, or nlp-8 inhibits movement behaviors.
flp-24, flp-13, or nlp-8 overexpression increases latency to avoid aversive stimulus
We tested if overexpression of any of the ALA-enriched neuropeptides was sufficient to increase the latency to avoid 1-octanol. ceh-14 mutants and animals overexpressing flp-7 exhibited normal avoidance behavior one hour after heat shock (Figure 6G). In contrast, overexpression of either flp-24, flp-13, or nlp-8 increased the response time compared to pre-heat shock (HS::flp-24; ceh-14: 23.7±4.6 seconds latency to reversal post-heat shock compared to 1.8±0.3 sec latency to reversal in seconds pre-heat shock, n=9; HS::flp-13; ceh-14: 20.4±4.2 sec latency to reversal post-heat shock compared to 1.7±0.3 sec latency to reversal pre-heat shock, n=2; HS::nlp-8; ceh-14: 21.5±5.6 sec latency to reversal post-heat shock compared to 1.5±0.2 sec mean latency to reversal pre-heat shock, n=6; p<0.001; Figure 6G). Five, two, and nine of eleven young adults overexpressing flp-24, flp-13, and nlp-8, respectively, did not respond to stimulus after 60 seconds, and were classified as non-responsive (Table S6). Thus, overexpression of flp-24, flp-13, or nlp-8, but not of flp-7, inhibited the avoidance response (Figure 6H).
Discussion
Sleep requires the coordinated regulation of multiple aspects of behavior and physiology. However, it is not well understood how disparate processes are coordinately regulated to produce the sleep state. At one extreme, a key factor may affect different aspects of the sleep state, thus ensuring that these processes are coordinately regulated. Alternatively, different processes may be controlled in series, such that one process initiates only if prior steps occur. A third possibility is that different factors may act in parallel to control the sleep state. Our data using a simple model organism supports the latter hypothesis.
We investigated how the C. elegans ALA neuron coordinately promotes multiple sleep-associated behaviors. Previous studies suggested that neuropeptides may mediate the sleep-promoting effects of ALA [7]. Using single-cell RNA-seq data of ALA, we observed that 23 flp and 25 nlp neuropeptide genes were highly expressed and enriched in ALA compared to whole larvae. We focused on four neuropeptides (flp-24, flp-7, flp-13, and nlp-8) with the highest level of expression and enrichment in ALA. Given the enrichment of multiple neuropeptide genes in ALA, we considered it unlikely that loss of individual neuropeptides would result in resistance to stress-induced sleep. Indeed, no defects were observed in stress-induced sleep for most neuropeptide single-null mutants. However, we found strong resistance to stress-induced sleep when multiple neuropeptide genes were deleted, indicating that sleep regulation downstream of ALA involves the collective action of multiple neuropeptides.
To determine how each of these neuropeptides induces sleep, we used an experimental paradigm that avoided confounding effects of neuropeptides released by ALA in response to stress. We found that three ALA-enriched neuropeptides, flp-24, flp-13, and nlp-8, were sufficient to induce distinct sleep-associated behaviors, while another, flp-7, showed no behavioral phenotype. For instance, only overexpression of flp-13 inhibited pumping, while worms overexpressing nlp-8 halted defecation even though they continued to eat, and moved their heads but not their bodies. flp-24 overexpression inhibited locomotion and head movement, but eating and defecation continued. In contrast to this specificity, overexpression of flp-24, flp-13, or nlp-8 inhibited locomotion and the avoidance response. The observation that some behaviors were affected by only one of these neuropeptides, while other behaviors were affected by several neuropeptides, has two main implications for sleep regulation: one for multilevel modulation of behavior and the other for the evolution of sleep states.
The behaviors studied here involve multiple cell types (Figure 7). For example, the avoidance response results from sensory neurons, command interneurons, and motor neurons working in series (Figure 7; [32]). Inhibition of any cell type within the neural circuit that regulates the avoidance response should suppress this behavior. Previously we showed that a sleeping worm has dampened sensory neuron activation and asynchronous command interneuron activities [32], both of which contribute to the observed delay in response to an aversive stimulus. Strong neuropeptide modulation (by overexpression of a neuropeptide) of either the sensory neurons or one or more of the command interneurons would lead to the absence of behavioral output, consistent with our observations. On the other hand, this hypothesis suggests that elimination of any one neuropeptide would have a small effect on behavior, consistent with our results. Furthermore, the site of action of these neuropeptides may be redundant at the receptor, cellular, and behavioral level (Figure 7). These results are consistent with the hypothesis that stress-induced sleep is driven by a set of neuropeptides produced by the ALA neuron, each of which independently induced a different suite of sleep-associated behaviors: suppression of eating, defecation, locomotion, head movement, and the avoidance response. We propose that neuropeptides act in parallel to control sleep, and that sleep states could be built during evolution from recruitment of factors controlling pre-existing regulatory pathways.
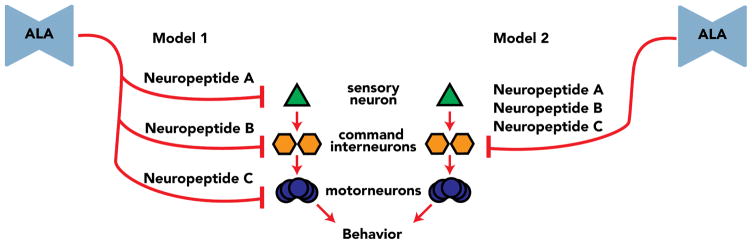
Figure 7. Redundancy models for the collective action of multiple neuropeptides which regulate C. elegans stress-induced sleep.
The neurosecretory ALA is required for stress-induced sleep. ALA transcribes multiple genes encoding neuropeptides, and we have shown that three neuropeptides enriched in ALA collectively regulate C. elegans stress-induced sleep. Given the non-overlapping expression pattern of these neuropeptides in other neurons, it is also possible that these neuropeptides act from neurons which have a minor role in regulating stress-induced sleep. In Model 1 each neuropeptide acts on a distinct neuron within a set of neurons that regulates behavior. In Model 2 each neuropeptide acts on the same neuron within a set of neurons that regulates behavior. The principles of Model 1 and 2 also apply at the receptor and behavioral level. For instance, each neuropeptide may act at a distinct receptor, or all the neuropeptides may act on the same receptor. In addition, strong inhibition of one behavior may inhibit all other behaviors and result in sleep, or there may be shut down of multiple behaviors simultaneously. We predict that these neuropeptides regulate C. elegans stress-induced sleep by some combination of Model 1 and 2 at the cellular, receptor, and behavioral level.
All the neuropeptides studied here have been reported to be expressed in cells other than ALA [13,19,29,31,36–38]. It is possible that these neuropeptides act from neurons other than ALA in stress-induced sleep. Another possibility is that these neuropeptides might have functions outside of sleep. For example, nlp-8 is expressed in specific male sensory neurons [38], and thus might play a role in inhibiting defecation during mating [39]. The multiple and apparently non-overlapping expression patterns of these genes is consistent with this hypothesis. With all the usual caveats of overexpression, the apparently distinct effects of sleep-promoting neuropeptides raises the possibility that the C. elegans sleep state is assembled from pre-existing regulatory pathways. This level of separate molecular control over distinct behaviors associated with sleep would provide evolutionary flexibility to sleep regulation, as unique but overlapping sleep states could be constructed by recruiting modules that regulate specific aspects of sleep. Diverse sleep states are found through out the animal kingdom [1,2], and this diversity may be partially explained by the recruitment of species-specific sleep modules (i.e. a module that shuts down defecation in humans). During the sleep state certain species specific regulatory modules must exist, such as modules that inhibit the avoidance response, defecation, and eating. The mammalian genome contains almost 70 different neuropeptide-encoding genes, many of which have detectable expression in the brain [40], and at least 20 of which may have important functions in sleep-wake regulation [24]. This extensive regulatory capacity is consistent with our view of modular regulatory logic. Testing this hypothesis would require associating each peptide with specific sleep-associated behaviors.
Materials and Methods
Strains were grown on nematode growth medium (NGM) agar Petri plates seeded with E. coli strain OP50 and maintained at 20°C under standard conditions [41]. Our wild-type strain was N2 (Bristol). The following can be found in the Supplemental Experimental Procedures: characterization of behavioral assays, single ALA neuron dissection and transcriptome profiling, data availability, information about strains.
Supplementary Material
Highlights.
RNA-seq of key sleep neuron ALA reveals enrichment of neuropeptide-encoding genes
A triple-null mutant of three ALA-enriched neuropeptide genes abolishes sleep
Three neuropeptides were each sufficient to induce specific sleep behaviors
The collective action of three neuropeptide genes (nlp-8, flp-24, flp-13) induces sleep
Acknowledgments
We thank the Caenorhabditis Genetics Center (NIHP40OD010440) and S. Mitani for strains, WormBase for bioinformatics support, Gladys Medina for technical support, the Millard and Muriel Jacobs Genetics and Genomics Laboratory for sequencing, and members of our laboratories for discussions. We thank Cheryl Van Buskirk for sharing unpublished observations, and Dr. Daniel Lee, Dr. David Prober, Dr. John Bedbrook, Claire Bedbrook, Dr. Hillel Schwartz and Dr. Mihoko Kato for comments and discussions. We thank Monica Savasta for generating graphics. This work was supported by the Howard Hughes Medical Institute, with which P.W.S. is an investigator, and the National Institutes of Health (GM084389 to P.W.S). R. D. N. was supported by T32GM007616. H.W. is supported by a Della Martin Postdoctoral Fellowship. Strains carrying the tm2427 and tm1583 alleles are available through the National BioResource Project (NBRP), subject to a materials transfer agreement.
Footnotes
The authors declare no competing interests.
Author Contributions
E.S.C., E.M.S., R.D.N., and P.W.S. conceived the project. E.S.C. dissected ALA neurons and sequenced their amplified cDNA. E.M.S. performed the ALA transcriptome analysis. R.D.N., E.S.C. and H.W. performed C. elegans behavior experiments. E.S.C., H.W., and R.D.N. generated reagents. R.D.N., E.S.C., H.W., E.M.S., and P.W.S. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 2.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh K, Chao MY, Somers GA, Komatsu H, Corkins ME, Larkins-Ford J, Tucey T, Dionne HM, Walsh MB, Beaumont EK, et al. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr Biol. 2011;21:825–834. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh K, Ju JY, Walsh MB, Dilorio MA, Hart AC. Deep conservation of genes required for both Drosophila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep. 2014;37:1439–1451. doi: 10.5665/sleep.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman JE, Naidoo N, Raizen DM, Pack AI. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 2008;31:371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 8.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 9.Kushikata T, Fang J, Chen Z, Wang Y, Krueger JM. Epidermal growth factor enhances spontaneous sleep in rabbits. Am J Physiol. 1998;275:R509–R514. doi: 10.1152/ajpregu.1998.275.2.R509. [DOI] [PubMed] [Google Scholar]
- 10.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 11.Snodgrass-Belt P, Gilbert JL, Davis FC. Central administration of transforming growth factor-alpha and neuregulin-1 suppress active behaviors and cause weight loss in hamsters. Brain Res. 2005;1038:171–182. doi: 10.1016/j.brainres.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 13.Nelson MD, Lee KH, Churgin MA, Hill AJ, Van Buskirk C, Fang-Yen C, Raizen DM. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr Biol. 2014;24:2406–2410. doi: 10.1016/j.cub.2014.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill AJ, Mansfield R, Lopez JMNG, Raizen DM, Van Buskirk C. Cellular stress induces a protective sleep-like state in C. elegans. Curr Biol. 2014;24:2399–2405. doi: 10.1016/j.cub.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher T, Kim J, Oldenbroek M, Kerr R, You YJ. ASI regulates satiety quiescence in C. elegans. J Neurosci. 2013;33:9716–9724. doi: 10.1523/JNEUROSCI.4493-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trojanowski NF, Raizen DM. Call it worm sleep. Trends Neurosci. 2016;39:54–62. doi: 10.1016/j.tins.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trojanowski NF, Nelson MD, Flavell SW, Fang-Yen C, Raizen DM. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J Neurosci. 2015;35:14571–14584. doi: 10.1523/JNEUROSCI.1369-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Buskirk C, Sternberg PW. Paired and LIM class homeodomain proteins coordinate differentiation of the C. elegans ALA neuron. Development. 2010;137:2065–2074. doi: 10.1242/dev.040881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. Bioessays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 21.Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marder E. Neuromodulation of neuronal circuits: back to the future. Neuron. 2012;76:1–11. doi: 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DA, Blackshaw S. Feed your head: neurodevelopmental control of feeding and metabolism. Annu Rev Physiol. 2014;76:197–223. doi: 10.1146/annurev-physiol-021113-170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter C, Woods IG, Schier AF. Neuropeptidergic control of sleep and wakefulness. Annu Rev Neurosci. 2014;37:503–531. doi: 10.1146/annurev-neuro-062111-150447. [DOI] [PubMed] [Google Scholar]
- 25.Lin XG, Ming M, Chen MR, Niu WP, Zhang YD, Liu B, Jiu YM, Yu JW, Xu T, Wu ZX. UNC-31/CAPS docks and primes dense core vesicles in C. elegans neurons. Biochem Biophys Res Comm. 2010;397:526–531. doi: 10.1016/j.bbrc.2010.05.148. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz EM, Kato M, Sternberg PW. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:16246–16251. doi: 10.1073/pnas.1203045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode Ascaris suum. ACS Chem Neurosci. 2010;1:505–519. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol. 2004;475:540–550. doi: 10.1002/cne.20189. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 1999;897:239–252. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- 31.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci USA. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho JY, Sternberg PW. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell. 2014;156:249–260. doi: 10.1016/j.cell.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz J, Lewandrowski I, Bringmann H. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr Biol. 2011;21:R983–R984. doi: 10.1016/j.cub.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 34.Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics. 1990;124:855–872. doi: 10.1093/genetics/124.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu DWC, Thomas JH. Regulation of a periodic motor program in C. elegans. J Neurosci. 1994;14:1953–1962. doi: 10.1523/JNEUROSCI.14-04-01953.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan G, Kim K, Li C, Walthall WW. Convergent genetic programs regulate similarities and differences between related motor neuron classes in Caenorhabditis elegans. Developmental Biology. 2005;280:494–503. doi: 10.1016/j.ydbio.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Pretot RF, Burglin TR, Sternberg PW. Distinct roles of transcription factors EGL-46 and DAF-19 in specifying the functionality of a polycystin-expressing sensory neuron necessary for C. elegans male vulva location behavior. Development. 2003;130:5217–5227. doi: 10.1242/dev.00678. [DOI] [PubMed] [Google Scholar]
- 39.Liu KS, Sternberg PW. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron. 1995;14:79–89. doi: 10.1016/0896-6273(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 40.Burbach JPH. What are neuropeptides? Meth Mol Biol. 2011;789:1–36. doi: 10.1007/978-1-61779-310-3_1. [DOI] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.