Abstract
Immunoglobulin G (IgG) consists of four subclasses in humans: IgG1, IgG2, IgG3 and IgG4, which are highly conserved but have unique differences that result in subclass-specific effector functions. Though IgG1 is the most extensively studied IgG subclass, study of other subclasses is important to understand overall immune function and for development of new therapeutics. When compared to IgG1, IgG3 exhibits a similar binding profile to Fcγ receptors and stronger activation of complement. All IgG subclasses are glycosylated at N297, which is required for Fcγ receptor and C1q complement binding as well as maintaining optimal Fc conformation. We have determined the crystal structure of homogenously glycosylated human IgG3 Fc with a GlcNAc2Man5 (Man5) high mannose glycoform at 1.8 Å resolution and compared its structural features with published structures from the other IgG subclasses. Although the overall structure of IgG3 Fc is similar to that of other subclasses, some structural perturbations based on sequence differences were revealed. For instance, the presence of R435 in IgG3 (and H435 in the other IgG subclasses) has been implicated to result in IgG3-specific properties related to binding to protein A, protein G and the neonatal Fc receptor (FcRn). The IgG3 Fc structure helps to explain some of these differences. Additionally, protein-glycan contacts observed in the crystal structure appear to correlate with IgG3 affinity for Fcγ receptors as shown by binding studies with IgG3 Fc glycoforms. Finally, this IgG3 Fc structure provides a template for further studies aimed at engineering the Fc for specific gain of function.
Keywords: immunoglobulin G, glycosylation, high mannose, x-ray crystal structure, Fc receptor
Graphical abstract
1. Introduction
Antibodies serve as a vital link between humoral and cell-mediated immunity against invading pathogens. Immunoglobulin G (IgG) class antibodies are heterotetrameric proteins containing two light chains and two heavy chains. IgGs can be split into two functional fragments, the Fragment antigen binding (Fab), made up of the light chain and the N-terminal half of the heavy chain and the Fragment crystallizable (Fc), made up of a dimer of the C-terminal portion of the heavy chains. The Fab and Fc regions are linked together by a flexible hinge region containing interstrand disulfide bonds (Kuby, 1997). The Fab recognizes specific antigens resulting in formation of an immune complex that can be removed or destroyed by multiple immune effector functions (Kuby, 1997). For antigens bound in an immune complex, outward facing Fc regions direct effector functions such as Antibody Dependent Cellular Cytotoxicity (ADCC), Antibody Dependent Cellular Phagocytosis (ADCP) and Complement Dependent Cytotoxicity (CDC) through interactions with cell-membrane receptors and serum proteins. The Fc contains binding sites for Fcγ receptors present on immune cells and C1q complement protein, whose engagement triggers ADCC/ADCP and CDC respectively (Kuby, 1997). The human IgG class of antibodies consists of four subclasses: IgG1, IgG2, IgG3 and IgG4, which are over 90% homologous in the Fc region, but have unique differences that allow them to elicit subclass-specific effector functions (Vidarsson et al., 2014). Such specificity among the IgG subclasses is driven by their different pattern of Fcγ receptor interactions and their ability to activate complement. For example, the IgG1 and IgG3 subclasses show a relative high affinity towards each human Fcγ receptor (FcγRI, FcγRIIA/B/C, FcγRIIIA/B). In contrast, the IgG2 subclass has only a moderate affinity for FcγRIIAH131, and the IgG4 subclass only has high affinity for FcγRI (Bruhns et al., 2009). In addition, the complement activation capacity of the IgG subclasses differ and is ranked as IgG3 > IgG1 > IgG2 > IgG4 (Vidarsson et al., 2014).
IgG3 is the third most abundant human IgG subclass comprising 5–8% of serum IgG, and is a unique subclass for variety of reasons (Vidarsson et al., 2014). It contains a long hinge region consisting of 11 disulfide bonds compared to the 2 to 4 disulfide bonds found in the other IgG subclasses. The long hinge is thought to provide additional flexibility for antigen binding, which is potentially important at low antigen concentration (Giuntini et al., 2016; Roux et al., 1997). IgG3 also shows extensive polymorphism with the largest number of known allotypes, 13, compared to 4, 1, and 0 allotypes for the IgG1, IgG2, and IgG4 subclasses respectively (Jefferis and Lefranc, 2009). The half-life of IgG3 is remarkably shorter (~7 days) than the other IgG subclasses (~21 days) (Morell et al., 1970). This difference has been attributed to a single amino acid change in the IgG3 Fc region (R435 in IgG3 versus H435 in the other IgG subclasses) (Stapleton et al., 2011). Another difference is that protein A, an Fc binding protein from S. aureus commonly used for IgG purification, doesn't bind to IgG3, while it binds tightly to the other IgG subclasses (Loghem et al., 1982). Recently, IgG3 has also gained attention in the field of HIV vaccines. In the only successful HIV vaccine trial (RV144) IgG3 was found as an immune correlate in vaccine protection. Subsequent analysis suggested a possible role of Fc-mediated effector functions of IgG3 in anti-viral activity (Chung et al., 2014).
Among the IgG subclasses, multiple IgG1 Fc structures alone or in complex with Fc-binding proteins have been published, and IgG2 Fc and IgG4 Fc structures have also recently been reported (see partial list in Table 2). All IgG Fc structures exhibit common structural features having immunoglobulin folds for the CH2 and CH3 domains. The CH2 domains of the Fc homodimer make no direct contact with each other, while the CH3 domains of the dimer strongly interact, thereby holding the dimeric structure together in conjunction with the hinge disulfide bonds. All of the IgG subclasses have a consensus N-glycosylation site at N297 in the CH2 domain, and IgG Fc crystal structures show the N297 glycans occupying the space between the CH2 domains. Glycans are thought to be important for maintaining the optimal ‘open’ conformation of the Fc (Krapp et al., 2003), and removal of glycans results in a ‘closed’ conformation owing to collapse of CH2 domains (Borrok et al., 2012). The ‘open’ conformation of the Fc is believed to be critical for recognition of the Fc by Fcγ receptors and C1q complement, since aglycosylated Fc loses its binding ability (Tao and Morrison, 1989; Walker et al., 1989).
Table 2.
Separation between CH2 domain in IgG3 Fc and reported IgG Fc structures
| PBD id | Fc (glycan) | Space group | Resolution (Å) | P238 (Å) | F241 (Å) | R301 (Å) | P329 (Å) |
|---|---|---|---|---|---|---|---|
| 5W38 | IgG3 Fc (Man5)b | P21 | 1.8 | 23.5 | 26.3 | 35.5 | 29.9 |
| 1FC1 | IgG1 Fc (complex)a | P212121 | 2.9 | 19.6 | 23.5 | 35.2 | 26.8 |
| 1H3T | IgG1 Fc (MN2F)b | P212121 | 2.4 | 18.3 | 21.4 | 32.8 | 22.0 |
| 1H3U | IgG1 Fc (M3N2F)b | P212121 | 2.4 | 19.2 | 22.0 | 32.7 | 24.2 |
| 1H3X | IgG1 Fc (G0F)b | P212121 | 2.4 | 19.7 | 21.9 | 33.9 | 22.6 |
| 1H3V | IgG1 Fc (G2F)b | P212121 | 3.1 | 19.4 | 23.2 | 32.9 | 26.9 |
| 3AVE | IgG1 Fc (complex)a | P212121 | 2.0 | 19.3 | 21.8 | 32.7 | 25.1 |
| 2DTS | IgG1Fc (complex-afucosyl) | P212121 | 2.2 | 18.6 | 21.5 | 32.2 | 24.2 |
| 3DO3 | IgG1 Fc (complex)a | P212121 | 2.5 | 20.1 | 22.4 | 32.8 | 23.5 |
| 2WAH | IgG1 Fc (Man9)b | P212121 | 2.5 | 30.2 | 29.8 | 42.8 | 36.4 |
| 4Q6Y | IgG1 Fc closed (S2G2F)b | P21 | 3.0 | 13.0 | 18.5 | 26.7 | 21.1 |
| 4Q6Y | IgG1 Fc open (S2G2F)b | P21 | 3.0 | 20.2 | 23.9 | 31.9 | NA |
| 4HAF | IgG2 Fc (complex)a | P212121 | 2.0 | NA | 27.8 | 39.5 | NA |
| 4C54 | IgG4 Fc (complex)a | P212121 | 1.9 | 19.7 | 22.1 | 33.7 | 36.1 |
denotes heterogeneously glycosylated Fc with complex-type biantennary N-glycans
denotes homogenously glycosylated Fc with the indicated glycoform
In this work, we report a high-resolution (1.8 Å) crystal structure of human IgG3 Fc containing a homogeneous high mannose, Man5GlcNAc2 (Man5) N-linked glycan at N297. The Man5 glycoform is an early intermediate in N-linked glycoprotein biosynthesis which is found in small amounts in naturally occurring human serum IgG and in significantly larger amounts in many therapeutic mAbs where it can alter effector functions and clearance {Flynn, 2010 #420}{Alessandri, 2012 #421}{Yu, 2012 #417}. To our knowledge, this is the first human IgG3 Fc crystal structure reported and also the first Fc structure from any IgG subclass containing the Man5 glycoform. We have examined structural features of IgG3 Fc and compared it with other IgG subclasses. A comparison of key amino acid difference between IgG3 and the other subclasses provides insight into some of the unusual properties of IgG3, especially those related to differential binding to protein A and protein G, and the FcRn receptor. Additionally, we have studied the binding of IgG3 Fc with Fcγ receptor IIIA (FcγRIIIA) and characterized the effect of high mannose glycan truncation on binding affinity. The Man5-IgG3 Fc structure helps to explain the results of the binding studies since removal of sugar residues involved in protein-glycan interactions observed in the Man5-IgG3 Fc structure result in reduced binding affinity to FcγRIIIA.
2. Material and methods
2.1. Protein expression and purification
The cDNA of the Fc region of hIgG3 (ATCC: MGC-45809, accession: BC033178) was used for PCR amplification. The Fc sequence belongs to IGHG3*11 allele (accession number AJ390247) found in the IMGT database (Bosc, 2003; Vidarsson et al., 2014). The expression construct encodes from T225 to K447 (EU numbering) (Béranger et al., 2001) and consists of only two disulfide bonds in the hinge region from hinge exon1: ELKTPLGDTTHT225CPRCP instead of the naturally occurring eleven hinge disulfide bonds (from additional exons 2, 3 and 4) (Michaelsen et al., 1977).The generally accepted sequence of IgG3 contains N392, however some IGHG3 alleles contain K392 (Vidarsson et al., 2014). The present construct was modified with mutation N392K to delete a potential glycosylation site N392TT. The protein was expressed as previously described in glycosylation-deficient Pichia pastoris (SMD1168) yeast with the OCH1 and PNO1 gene deletions (Okbazghi et al., 2016). The strain was further modified by deleting the BMT1 and BMT2 genes to reduce β-mannosylation (Hopkins et al., 2011). Additionally, the STT3D gene from Leishmania major was added to the strain to improve the glycosylation site-occupancy (Choi et al., 2012). The secreted protein was first purified by Protein G affinity chromatography and then with phenyl sepharose chromatography (supplementary information). The IgG3 Fc was digested with B.t. α-1,2 mannosidase (Bt3990) (Cuskin et al., 2015) and endomannosidase (supplementary information) to convert high mannose glycosylation on the expressed protein to the homogenous GlcNAc2Man5 (Man5) glycoform. Man5-IgG3 Fc was characterized by SDS-PAGE and intact protein mass spectrometry, as per previously described method (Okbazghi et al., 2016).
2.2. Crystallization and Data Collection
Man5-IgG3 Fc was concentrated to 10.8 mg/mL in 150 mM NaCl, 10 mM MES, pH 6.6 for crystallization screening. All crystallization experiments were conducted Compact Jr. (Rigaku Reagents) sitting drop vapor diffusion plates at 20 °C using equal volumes of protein and crystallization solution (0.7 uL) equilibrated against 75 uL of the latter. Crystals that displayed a prismatic morphology were obtained approximately 1 week from the Wizard 3–4 screen (Rigaku Reagents) condition B5 (20% (w/v) PEG 4000, 100 mM sodium citrate/citric acid pH 5.5, 10% (v/v) 2-propanol). The crystals tended to form contact twins but could be readily separated to obtain single crystals for data collection. Samples were transferred to a fresh drop composed of 80% crystallization solution and 20% ethylene glycol and stored in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source IMCA-CAT beamline 17-ID using a Dectris Pilatus 6M pixel array detector.
2.3. Structure Solution and Refinement
Intensities were integrated using XDS (Kabsch, 1988; Kabsch, 2010) via Autoproc (Vonrhein et al., 2011) and the Laue class analysis and data scaling were performed with Aimless (Evans, 2011) which indicated that the highest probability was 2/m (P2 or P21). Structure solution was conducted by molecular replacement with Phaser (McCoy et al., 2007) using a single subunit form previously determined structure of IgG1 Fc (PDB: 4DZ8) (Strop et al., 2012) as the search model (Weerawarna et al., 2016). The top solution was obtained in the space group P21 that contained one homodimer in the asymmetric unit. The model was further improved by automated model building with ARP/wARP (Langer et al., 2008). Structure refinement and manual model building were conducted with Phenix (Adams et al., 2010) and Coot (Emsley et al., 2010) respectively. Disordered side chains were truncated to the point for which electron density could be observed. Structure validation was conducted with Molprobity (Chen et al., 2010) and figures were prepared using the CCP4MG package (Potterton et al., 2004). Full data processing and refinement statistics are provided in Table 1. Coordinates and structure factors have been deposited to the Worldwide Protein Databank with the accession code 5W38.
Table 1.
Crystallographic data and refinement statistics for IgG3 Fc
| Data Collection | |
|---|---|
| Unit-cell parameters (Å, °) | a=64.43, b=60.44, c=71.40, β=109.2 |
| Space group | P21 |
| Resolution (Å)1 | 45.00–1.80 (1.84–1.80) |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Observed reflections | 156,654 |
| Unique reflections | 47,374 |
| <I/σ(I)>1 | 12.7 (2.1) |
| Completeness (%)1 | 98.4 (97.7) |
| Multiplicity1 | 3.3 (3.2) |
| Rmerge (%)1,2 | 5.5 (57.5) |
| Rmeas (%)1,4 | 6.6 (69.0) |
| Rpim (%)1,4 | 3.6 (37.8) |
| CC1/21,5 | 0.998 (0.734) |
| Refinement | |
| Resolution (Å)1 | 34.73–1.80 |
| Reflections (working/test)1 | 44,926/2,427 |
| Rfactor/Rfree (%)1,3 | 18.4/22.3 |
| No. of atoms (Protein/Water) | 3,378/351 |
| Model Quality | |
| R.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.020 |
| Average B-factor (Å2) | |
| All Atoms | 28.1 |
| Protein | 27.6 |
| Water | 33.4 |
| Coordinate error(maximum likelihood) (Å) | 0.23 |
| Ramachandran Plot | |
| Most favored (%) | 98.6 |
| Additionally allowed (%) | 1.4 |
Values in parenthesis are for the highest resolution shell.
Rmerge = ƩhklƩi |Ii(hkl) - <I(hkl)>|/ƩhklƩi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Ʃhkl ||Fobs (hkl) | - |Fcalc (hkl)||/Ʃhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Rmeas = redundancy-independent (multiplicity-weighted) Rmerge(Evans, 2006; Evans, 2011). Rpim = precision-indicating (multiplicity-weighted) Rmerge(Diederichs and Karplus, 1997; Weiss, 2001).
CC1/2 is the correlation coefficient of the mean intensities between two random half-sets of data (Evans, 2012; Karplus and Diederichs, 2012).
2.4. Binding analysis of IgG3 Fc glycoforms with FcγRIIIA
The high affinity polymorph (V158) of FcγRIIIA was cloned and expressed in yeast P. pastoris and purified using Ni2+-NTA affinity and phenyl sepharose chromatographies, as previously described (Okbazghi et al., 2016; Xiao et al., 2009). Different IgG3 Fc glycoforms were assessed for FcγRIIIA binding with BioLayer Interferometry (BLI) using BLItz. The glycoforms tested were Man8-Man12, Man5, Fc-GlcNAc (prepared with EndoH enzyme treatment) and deglycosylated Fc. The protein G biosensor tip was hydrated with PBS buffer (50 mM sodium phosphate pH 7.4, 150 mM NaCl) with 1 mg/ml casein as a blocking agent for 15 min. The tip was then dipped in PBS buffer for 15 min. The binding experiment was conducted as follows: an initial baseline was established by dipping tip in PBS buffer for 30 sec. Next, the tip was loaded with IgG3 Fc glycovariants at a concentration of 0.88μM for 120 sec to a response level of 2 nm. A new baseline was established for 30 sec in PBS, then an association and dissociation phase for 180 and 360 sec, respectively was measured by dipping the tip in various concentrations of FcγRIIIA and PBS, respectively. The FcγRIIIA concentrations used were from 800 nM to 50 nM in two-fold serial dilution (except 1600 nM to 100 nM for Fc-GlcNAc). Between each run, tip was regenerated using two cycles of 10 mM HCl for 30 sec with one 60 sec cycle of PBS in between. Binding curves were globally fitted to 1:1 binding model and analyzed using BLItz Pro software (ForteBio).
3. Results and Discussion
3.1. Production and characterization of Man5-IgG3 Fc
Human IgG3 Fc consisting of amino acids T225 to K447 (EU numbering) was expressed in a glycoengineered strain of the yeast Pichia pastoris. The expressed protein was secreted into the culture media and was purified first using protein G affinity chromatography and then phenyl sepharose chromatography to remove glycosylation macro-heterogeneity and residual host cell proteins (Fig. S1A, supplementary information). Intact protein mass spectrometry (MS) analysis of the purified protein showed a heterogeneous glycosylation pattern bearing high mannose (HM) glycoforms, mostly consisting of the Man8GlcNAc2 (Man8) glycoform and glycoforms with additional hexose residues added to the initial Man8 structure (Fig. S2A, supplementary information). The HM-IgG3 Fc was converted into the Man5GlcNAc2 (Man5) glycoform by enzymatic digestion with B.t. α-1,2-mannosidase, which specifically cleaves α-1,2-mannose residues (Fig. S1B, supplementary information). Endomannosidase was also added to the digestion reaction to convert any residual glucosylated HM glycans (a biosynthetic precursor to HM glycan and a non-substrate for B.t. α-1,2-mannosidase) into HM glycan (Fig. S3, supplementary information). The resulting Man5-IgG3 Fc was characterized by SDS-PAGE and intact protein MS, which showed a single peak corresponding to Man5 glycoform on IgG3 Fc (Fig.’s S1A, S2B, supplementary information). The abundance of Man5 was estimated to be 96% based on MS peak heights. IgG3 Fc was expressed without loss of the C-terminal lysine (K447). Man5-IgG3 Fc produced and processed by this method was sufficiently pure and homogenously glycosylated, and hence suitable for crystallization trials.
3.2. Overall IgG3 Fc structure
IgG3 Fc crystallized in the space group P21, with one homodimer in the asymmetric unit. Ordered electron density was visible for residues L235-L443 and G237-P445 in chain A and B respectively. No interpretable electron density was observed for hinge disulfide bonds and for residues A231-L234 following the hinge. Both the CH2 and CH3 domains display an immunoglobulin fold, which is stabilized by intra-chain disulfide bonds (C261-C321 in the CH2 and C367-C425 in the CH3). The glycans occupy the space between the CH2 domains, while extensive non-covalent contact exists between the two CH3 domains (Fig. 1). There is a high degree of sequence homology (over 90%) in the Fc region of the IgG subclasses (Fig. 2). Most of the amino acid differences occur on the surface of the Fc, as illustrated for IgG1 and IgG3 (Fig. 1). The following sections describe the influence of key amino acid differences and glycosylation on the structural features of IgG3 Fc as well as on some of the unusual properties of IgG3, especially its interaction with protein A, protein G and the FcRn receptor.
Fig. 1.
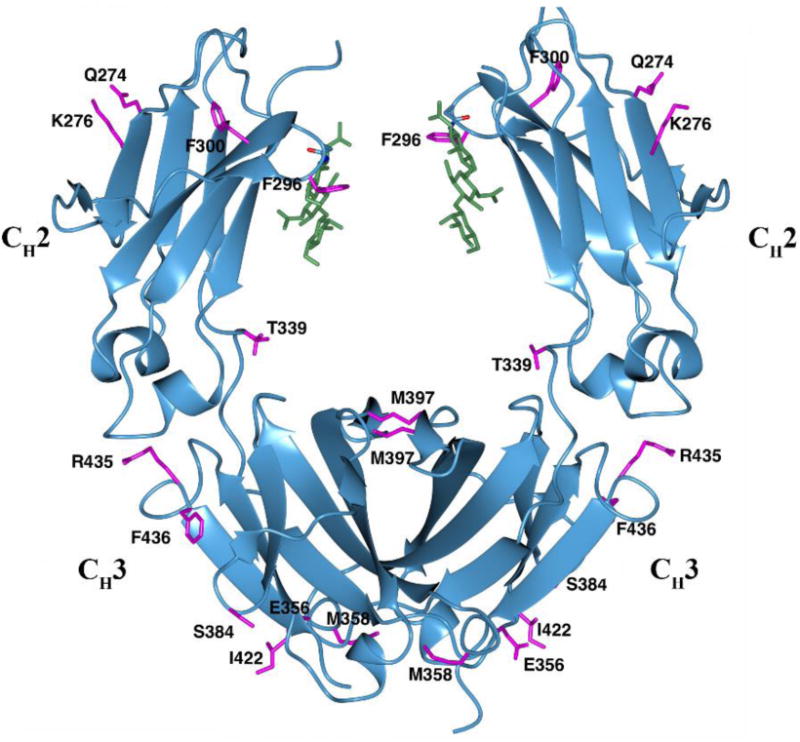
The IgG3 Fc structure. Side chains of IgG3 Fc (magenta) that are different in IgG1 Fc (using PDB 3AVE as reference) are shown in stick representation. The N297 and N-glycans (green) are shown in stick representation.
Fig. 2.
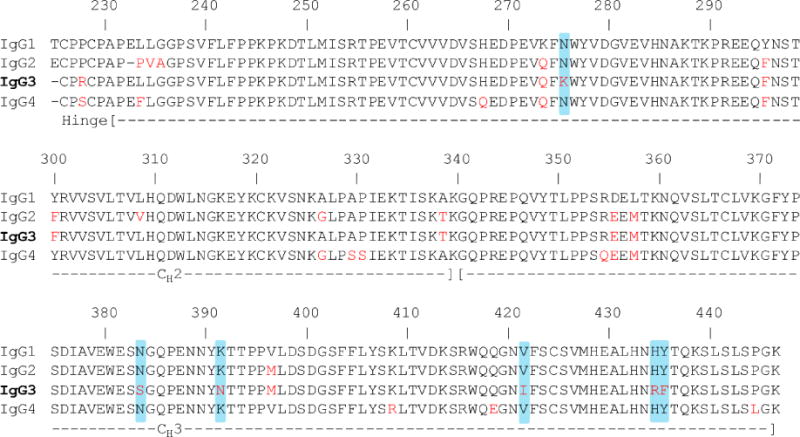
Sequence alignment of IgG1 (3AVE), IgG2 (4HAF), IgG3, and IgG4 (4C54) for the hinge and the Fc region. The IgG3 sequence shown here and used for this study is based on IGHG3*11, *12 allele. The amino acid differences between IgG1 and the other IgG subclasses are shown in red. Amino acid differences unique to IgG3 are highlighted in blue.
3.3. CH2 domain conformation
The CH2 domain conformation in IgG3 Fc was compared with representative IgG1, IgG2 and IgG4 structures. The overall CH2 domain conformation can be assessed by measuring the distances between Cα atoms of the P238, F241, R301, and P329 residues on both of the Fc chains (Teplyakov et al., 2013). The distances between pairs of P238, F241, R301 and P329 residues in the Man5-IgG3 Fc structure are 23.5 Å, 26.3 Å, 35.5 Å and 29.9 Å respectively. These distances are about 3–4 Å greater than the range (18.6–20.2 Å for P238, 21.5–23.9 Å for F241, 31.9–35.2 Å for R301, and 22.6–26.9 Å for P329) of distances reported in multiple IgG1 Fc structures that bear complex-type glycans (Table 2). The differences in CH2-CH2 domain distances in published IgG Fc structures can be due to subclass differences, crystal packing or the type of glycosylation (Bowden et al., 2012; Teplyakov et al., 2013). IgG3 Fc was crystallized in the P21 space group, whereas a majority of the published IgG1 Fc structures belong to the P212121 space group (Table 2). Generally, a pronounced correlation between the CH2 separation and type of glycosylation is observed. For example, the CH2 domains are more spread apart in Man9-IgG1 Fc than for IgG1 Fc with a complex-type glycan (Crispin et al., 2009). The Man5 glycoform in IgG3 Fc is a truncated form of a Man9 glycan having similar branch points but lacking the additional α-1,2 mannose residues (Fig. 4A). The CH2 distances in the Man5-IgG3 Fc structure were between those found in the Man9-IgG1 Fc structure and the IgG1 Fc with a complex glycan structure (Fig 3, Table 2). Due to the unavailability of a Man5-IgG1 Fc structure (and hence lack of direct comparision), it is not possible to tell whether the CH2 separation in IgG3 Fc was due to amino acid differences in IgG3 and IgG1 and/or the Man5 glycoform on IgG3 Fc.
Fig. 4.
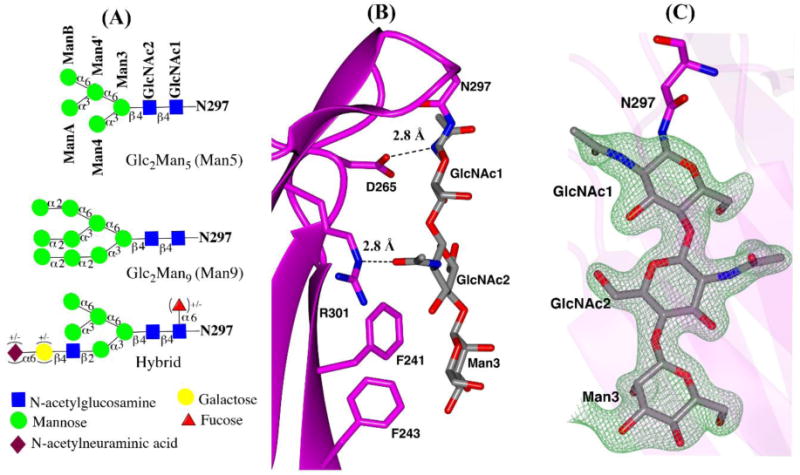
N-glycans in IgG3 Fc structure (A) schematic representation of Man5GlcNAc2 (Man5), Man9GlcNAc2 (Man9) and hybrid glycoform based on the Essentials system of glycan nomenclature (B) close-up view of N297 attached glycans and protein contacts. H-bonds between protein and carbohydrate are shown as black dotted lines. (C) ordered glycans from chain A in stick representation, shown with Fo-Fc electron density difference map contoured at 3σ.
Fig. 3.
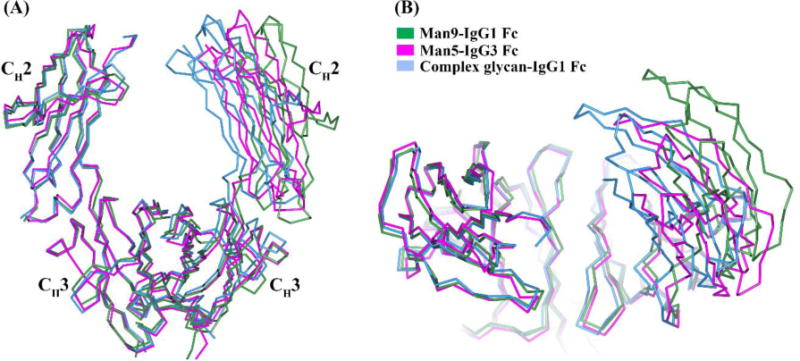
The orientation of the CH2 domain relative to the CH3 domain shown by superposition of Fc structures aligned using only one Fc chain. The compared Fc structures are Man5-IgG3 Fc (magenta), complex glycan-IgG1 Fc (3AVE) (light blue) and Man9-IgG1 Fc (2WAH) (green). The orientation of the Fc is shown in (A) side view and (B) side view rotated by 90° about x-axis.
The IgG3 subclass exhibits the strongest complement dependent cytotoxicity (CDC), a process initiated by binding of C1q complement to IgG Fc. The core of the C1q complement binding site on IgG1 has been mapped to four spatially close residues in the CH2 domain, Asp270, Lys322, Pro329, and Pro331 (Idusogie et al., 2000). Although these four residues are conserved in IgG3, there are other polymorphic differences between IgG1 and IgG3 around the C1q binding site. These differences are believed to result in subclass-dependent variations in complement activation at the level of C1q binding or the antibody-dependent steps of complement cascade. Mutation of one such polymorphic residue, Lys276 in IgG3 to IgG1-like Asn276 was shown to reduce complement activation (Tao et al., 1993). Lys 276 in our Man5-IgG3 Fc structure appears to lie close to Lys322 (Fig. S5, supplementary information), which has been previously determined to be important for IgG3 driven complement activation (Thommesen et al., 2000).
3.4. N-glycosylation
Endogenous IgG3, like the other IgG subclasses bears complex-type glycosylation (Wuhrer et al., 2007). The Man5 glycoform reported in this structure is an intermediate in the biosynthesis of N-linked glycoproteins that is formed just prior to the conversion of high mannose type glycoforms into hybrid and complex glycoforms (Brooks, 2010). Although, the level of the Man5 glycoform found in the normal serum IgG is less than 1% (Flynn et al., 2010), it is present at significantly higher levels (up to 20%) in recombinant mAbs produced in CHO cells (Alessandri et al., 2012). Man5-IgG1 is reported to have higher binding affinity to FcγRIIIA and higher Antibody Dependent Cellular Cytotoxicity (ADCC) when compared to IgG1 bearing complex-type glycoforms (Yu et al., 2012). Additionally, Man5-IgG1 exhibits more rapid clearance than other glycoforms due to uptake by mannose receptors and mannose binding proteins (Goetze et al., 2011). Because of these properties, the level of high mannose glycoforms in mAb therapeutics is considered as a critical quality attribute (CQA) and demonstrating its effect on Fc conformation is important from a Fc glycoengineering perspective (Reusch and Tejada, 2015).
The conserved N297 glycan in IgG1 Fc structures maintains extensive contacts with protein (Nagae and Yamaguchi, 2012). In both IgG3 Fc chains, ordered electron density for only the first three reducing terminal monosaccharide residues was detected, corresponding to Manβ1–4 GlcNAcβ1–4GlcNAc linked to N297 (Fig. 4A,C). No interpretable electron density for the terminal mannose residues was detected, presumably due to its higher dynamic nature or disordered state. The protein-carbohydrate contacts for the three visible sugars on both the chains of IgG3 Fc were identical to those previously reported in IgG1 Fc structures (Deisenhofer, 1981; Nagae and Yamaguchi, 2012). The contacts found within hydrogen-bonding distance are between: the carbonyl oxygen of the D265 side chain and amide nitrogen of the N-acetyl group on GlcNAc1, and the Nε atom of the R301 side chain and the amide oxygen of the N-acetyl group on GlcNAc2. Also, π-CH type interactions are possible between phenyl ring of F241 and CH moieties from GlcNAc2 and Man3. (Fig. 4B).
We examined the effect of removal of these interactions on binding of IgG3 Fc to FcγRIIIA, a key receptor involved in ADCC activity using a series of sequentially truncated IgG3 Fc glycoforms. These glycoforms included high mannose (Man8-Man12 glycoforms, potentially having additional protein contacts when compared to Man5), Man5, a single GlcNAc monosaccharide residue (disrupting contacts with Man3 and GlcNAc2) and D297 (deglycosylated by PNGase F treatment, also disrupting contacts with GlcNAc1). Representative binding curves for biolayer interferometry (BLI) based Fc-FcγRIIIA binding assay are shown in Fig. S6, supplementary infomation and the results for these binding experiments are given in Table 3. The binding assay showed similar affinity for the high mannose (Man8-Man12) and Man5 glycoforms. However, a 10-fold reduction in affinity was observed for GlcNAc-IgG3 Fc glycoform and no detectable binding was recorded for the deglycosylated IgG3 Fc using highest receptor concentration of 15 μM. The weaker affinity of the GlcNAc-IgG3 Fc was driven by a faster dissociation rate, with the association rate remaining largely unaffected. A similar trend in FcγRIIIA binding was also seen in IgG1 Fc, as previously reported by our group (Okbazghi et al., 2016) and others (Subedi et al., 2014). These results indicate that protein-glycan interactions of the core pentasaccharide (GlcNAc2Man3) were necessary in maintaining high affinity FcγRIIIA binding, and that the IgG3 Fc interactions with GlcNAc1 are essential for FcγRIIIA binding.
Table 3.
Kinetic parameters obtained for binding of FcγRIIIA with the IgG3 Fc glycovariants
| IgG3 Fc glycoform |
Average kon*105 (1/M.sec) |
Average koff*10−3 (1/sec) |
Average KD (nM) |
|---|---|---|---|
| Man8-Man12 | 1.6 ± 0.1 | 6.5 ± 0.2 | 41.4 ± 2.7 |
| Man5 | 1.5 ± 0.1 | 6.8 ± 0.2 | 46.5 ± 2.4 |
| Fc-GlcNAc | 1.7 ± 0.4 | 85.8 ± 7.7 | 506 ± 113 |
| D297 | nd* | nd* | nd* |
not detected at highest receptor concentration of 15 μ
3.5. CH2-CH3 domain interface
The CH2-CH3 interface in IgG class antibodies maintains the orientation of the CH2 domain relative to the CH3 domain. Moreover, the outer face of the CH2-CH3 domain interface is a binding site for multiple Fc binding proteins like FcRn, protein A, protein G, HSV-1 (herpes simplex virus 1) Fc receptor, rheumatoid factor and TRIM21 (tripartitie motif 21) (Vidarsson et al., 2014). Any structural change at the interface can potentially alter the conformation of the binding site or the CH2 domain orientation. The amino acids at the interface are highly conserved among IgG subclasses with a few exceptions. In IgG1, IgG2 and IgG4 Fc structures, the core of the interface is formed by four stabilizing interactions: two salt bridges between K248-E380 and K338-E430 through side chains and two hydrogen bonds between L251-H435 and K340-Y373 through main chains of L251, K340 and side chains of H435, Y373. All these residues are conserved across IgG subclasses except at position 435, where an arginine is present (R435) in IgG3 rather than histidine (H435) as in the other subclasses (Fig. 2). It must be noted here that only two IgG3 allotypes (G3m (s,t)/G3m15,16) from the thirteen allotypes contain H435 and these are commonly found in Mongoloid population but rare in Caucasoids (Lefranc and Lefranc, 2012).
Both the IgG3 Fc chains showed similar contacts as seen in IgG1 Fc (3AVE), IgG2 Fc (4HAF) and IgG4 Fc (4C54) except for the K248-E380 salt bridge (Fig. 5A). The side chain of E380 on both IgG3 Fc chains adopted a different rotameric conformation that was away from side chain of K248 (Fig. 5B). Interestingly, despite having arginine at position 435, both the IgG3 Fc chains still maintained a H-bond between residues 251 and 435 (through the main chain carbonyl oxygen of L251 and Nε of R435 side chain in place of Nε1 from H435 side chain) (Fig. 5A). The longer arginine 435 side chain in IgG3 Fc is close to the isoleucine 253 side chain (less than 4 Å distance), whereas histidine 435 is located away from isoleucine 253 in the IgG1 Fc structures (Fig. S7A, supplementary information). Isoleucine 253 is a critical residue in the region of IgG Fc where protein A, protein G and FcRn bind (Deisenhofer, 1981; Oganesyan et al., 2014). Arginine (pKa~12.5), unlike histidine (pKa~6.0) remains positively charged at physiological pH and thereby can potentially alter the side chain conformation of I253 or its local conformation. T339 is another residue at the interface that differs across the IgG subclasses. IgG2 and IgG3 contain threonine, while IgG1 and IgG4 have alanine at position 339. In the IgG2 Fc structure, T339 has been shown to maintain contact with D376 either directly or through water mediated H bonds (Teplyakov et al., 2013). In the IgG3 Fc structure, the side chains of T339 and D376 on chain A exists in two conformations, while both residues on chain B adopt a single conformation with no contacts between them in either of the chains (Fig. 5C). Teplyakov et al. have suggested a ball-and-socket joint in the IgG2 Fc structure between a CH2 residue (L251) forming the ball and CH3 residues (M428, H429, E430 and H435) forming the socket that mediate the pivot of the CH2 domain relative to the CH3 domain. (Teplyakov et al., 2013). These residues are conserved in all IgG subclasses except IgG3 where arginine is present at position 435 (Fig. 2). In the IgG3 Fc structure, R453 does not seem to disrupt the socket and L251 in IgG3 Fc was still able to form a similar type of ball-and-socket joint. The longer side chain of R435 can be speculated to extend the socket closer to the CH2 domain owing to the proximity of the R435 and I253 residues and thereby may influence the CH2 domain pivot (Fig. S7B, supplementary information).
Fig. 5.
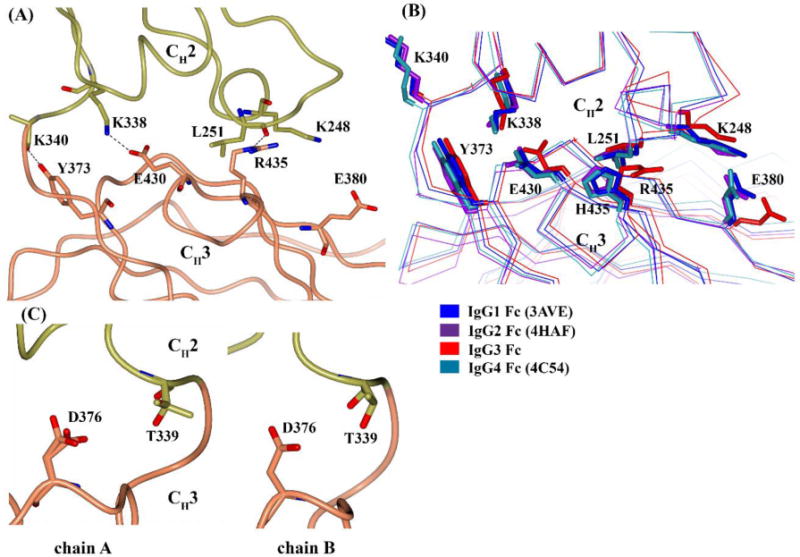
CH2-CH3 interface of IgG3 Fc. (A) H-bonds and salt-bridge present between CH2 (gold) and CH3 (coral) in chain A (B) comparison of CH2-CH3 interface across IgG subclasses. The representative Fc structure include IgG1 Fc (3AVE) (blue), IgG2 Fc (4HAF) (purple), IgG3 Fc (red) and IgG4 Fc (4C54) (cyan). Protein backbone is represented by Cα trace and side chains are shown in stick representation. (C) Conformation of T339 and D376 residues in chains A and B of IgG3 Fc.
A functional significance of the CH2-CH3 interface with respect to Fc receptor binding was demonstrated in an IgG1 mutagenesis study (Shields et al., 2001). In the study, IgG1 with a H435A mutation showed 25% lower binding to FcγRIIA, FcγRIIB and FcγRIIIA, while the mutation increased binding to FcγRI by same amount. An A339T mutation increased FcγRIIIA binding by 34%. Given the distal location of the H435 and A339 from the FcγR binding site, an indirect role of the CH2-CH3 interface in FcγR binding through maintenance of CH2 conformation can be postulated. In the same study, an E380A mutation resulted in an 119% increase in FcRn affinity. E380 doesn't interact directly with FcRn but is a neighboring residue at the binding interface. In summary, the CH2-CH3 interface of IgG3 Fc exhibited minor differences compared to that of IgG1 Fc, which may be responsible for subtle differences in its Fc receptor interaction pattern (Bruhns et al., 2009).
3.6. IgG3-protein A and protein G interactions
IgG3 is the only IgG subclass that does not bind to protein A, whereas all IgG subclasses bind to protein G. All the residues involved in protein A and/or protein G binding are conserved across the IgG subclasses except for IgG3-R435 (Sauer-Eriksson et al., 1995). When the H435R mutation of IgG1 Fc was modeled on the IgG1 Fc structure in the first published protein A-IgG1 Fc complex (1FC2), R435 was unable to be accommodated in the complex (Deisenhofer, 1981). Although an IgG3 Fc structure alone does not provide direct information on the lack of an IgG3-protein A binding interaction, IgG3 can be reasonably compared with IgG1 in complex with protein A, given the highly conserved nature of the binding interface. We used the 1FC2 complex and a more recently reported Rituximab Fc-protein A (1L6X) complex to compare IgG3's structure to IgG1's structure in the complex. In both the complexes, the R435 side chain appears to sterically clash with the phenylalanine (F132/F14 in 1FC2/1L6X) and glutamine (Q129/Q11 in 1FC2/1L6X) residues of protein A and appears to be more conformationally restricted than H435 (Fig. 6A–B). Our analysis is backed by the work of Stapleton et al., who showed that protein A binding ability of IgG3 is restored when Arg is mutated to His (Stapleton et al., 2011). In contrast to protein A, H435 was shown to not make any contact with protein G in the protein G-IgG1 Fc complex (Sauer-Eriksson et al., 1995). When we compare IgG3 Fc to IgG1 Fc in the complex with protein G, the R435 in IgG3 does not appear to sterically clash with protein G and appears to be accommodated at the binding interface (Fig. 6C). Additionally, R435 in IgG3 can be predicted to interact with E27 in protein G through a salt bridge, as the distance between the Nη2 atom of R435 and Oε2 atom of E27 is 3.5 Å. A study investigating binding affinity differences between the IgG subclasses for binding to protein G showed a 3-fold higher affinity for IgG3 compared to IgG1 (Akerström and Björck, 1986). Based on this information, it could be suggested that the longer R435 side chain in IgG3 can potentially form extra contacts with protein G and thereby result in tighter binding. Our IgG3 Fc structure provides some insight on the likely role of the arginine residue at the 435 position in the differential protein A and protein G binding properties of IgG3.
Fig. 6.
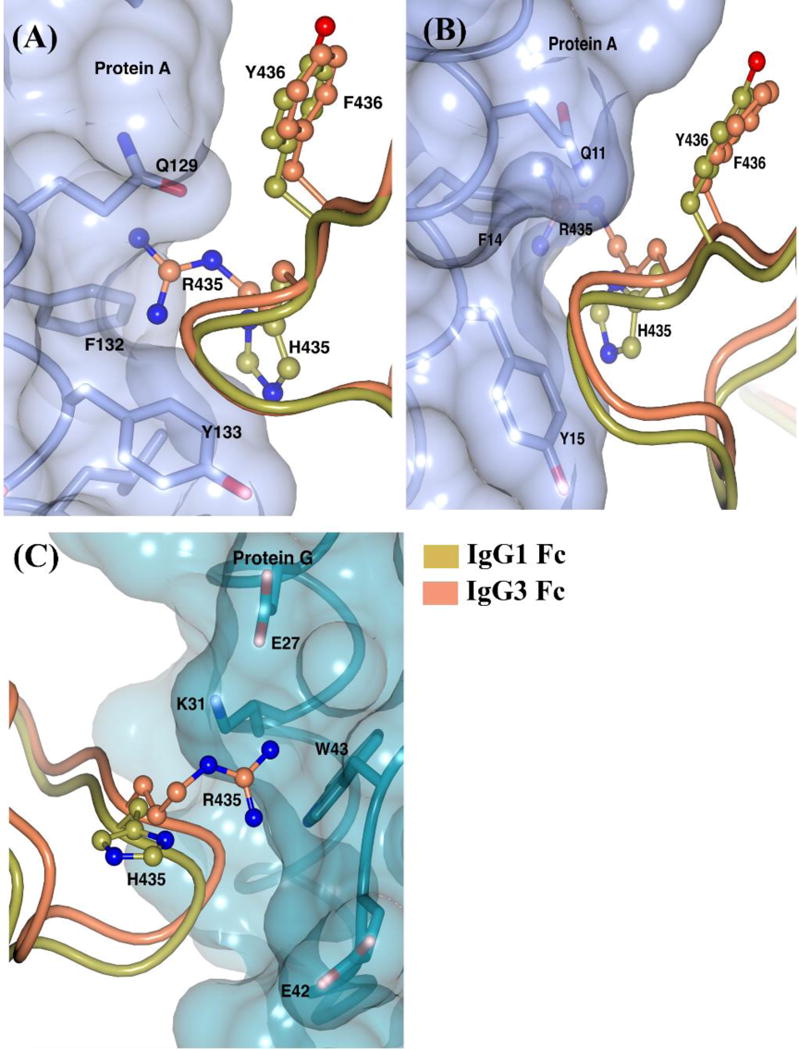
IgG3 Fc at the Fc-protein A/G binding interface. IgG3 Fc is superposed on IgG1 Fc in the IgG1 Fc-protein A complexes from (A) 1FC2 and (B) 1L6X. (C) IgG3 Fc is superposed on IgG1 Fc in the IgG1 Fc-protein G complex (1FCC). Side chains of Fc and protein A/G are represented by ball-and-stick and stick respectively. The transparent surfaces of protein A and G are shown in light purple and dark cyan respectively.
3.7. IgG3-FcRn interactions
The neonatal Fc receptor (FcRn) is responsible for the long half-life of IgG class antibodies by rescuing IgG from endosomal degradation. It binds to IgG at acidic pH in the endosomes of vascular endothelial cells and recycles it back to circulation by dissociating at pH 7.4 (Roopenian and Akilesh, 2007). This pH dependency is mediated by salt bridges involving H310 and H435 of the Fc with the corresponding E115 and D130 residues of human FcRn (Huang et al., 2013; Martin et al., 2001; Oganesyan et al., 2014). The lower half-life of IgG3 (~7 days) has been suggested to be due to an intracellular competition between IgG1 (half-life of ~21 days) and IgG3 during the FcRn recycling process. IgG3 showed a slightly 2-fold weaker affinity at pH 6.0 when compared to IgG1 and an increased binding at pH 7.4 relative to IgG1. These differences were also suggested to play a role in less efficient IgG3 recycling (Stapleton et al., 2011). Also, in the same study, the longer side chain of Arg 435 was shown to be sterically hindered at the binding site by modelling H435R on a rat FcRn-IgG structure. We modeled the probable IgG3-FcRn binding interface by superposing the IgG3 Fc structure on the available IgG1 Fc (YTE)-human FcRn complex (Oganesyan et al., 2014), and analyzed various scenarios in which R435 can influence FcRn binding. The R435 residue appears to be accommodated at the binding interface with some rearrangement (Fig. 7). Moreover, R435 in IgG3 can be predicted to interact with E133 in FcRn through a salt bridge, as the distance between the Nη2 atom of R435 and Oε2 atom of E133 is 3.0 Å. The arginine side chain (free amino acid pKa~12.5) remains positively charged at pH 7.4 and can still maintain ionic interactions with FcRn, unlike the histidine side chain (free amino acid pKa~6.0) which is likely to be less than 10% protonated at pH 7.4. This can contribute to increased binding of IgG3 at physiological pH relative to the other IgG subclasses. Although, the IgG3 Fc structure alone does not provide conclusive evidence pertaining to the IgG3-FcRn interaction, it helps to shed light on the possible role of arginine at position 435 at the binding site.
Fig. 7.
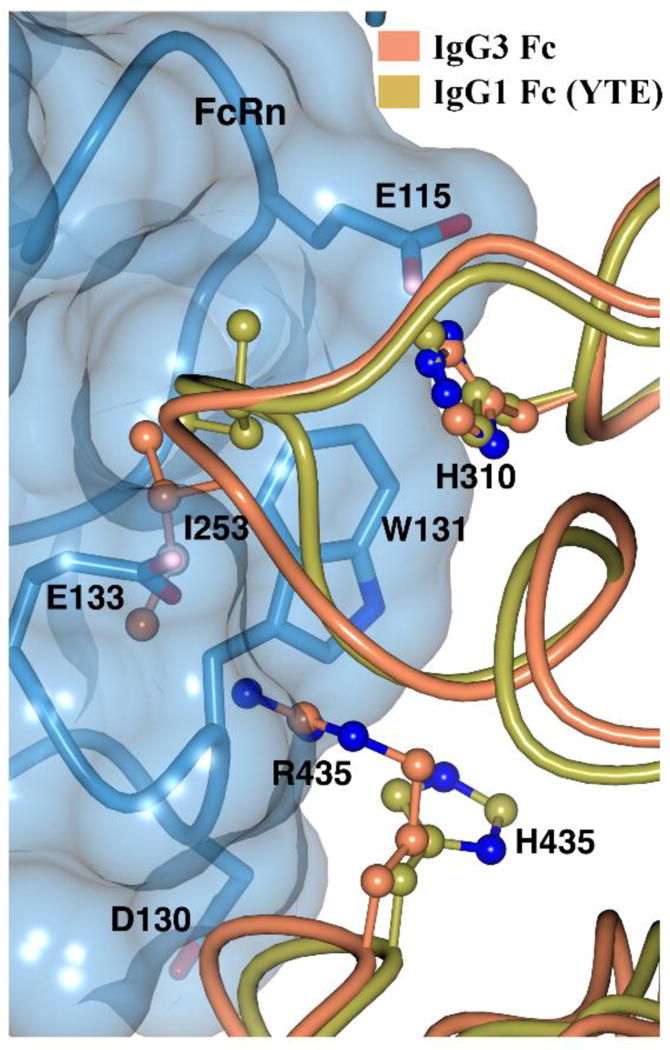
IgG3 Fc at the Fc-FcRn binding interface. IgG3 is superposed on IgG1 Fc (YTE) in the Fc (YTE)-FcRn complex (4N0U). Fc and FcRn residues are shown in ball-and-stick and stick representation respectively. Transparent surface of FcRn is shown in blue.
4. Conclusions
The present study is the first report of an IgG3 Fc crystal structure in the literature and now completes the panel of available Fc structures from all the four human IgG subclasses. The 1.8 Å IgG3 Fc structure is homogenously glycosylated with a high mannose GlcNAc2Man5 (Man5) glycoform and is the first Fc structure from any IgG subclass containing Man5 glycoform. The functional significance of the observed protein-glycan contacts in the structure is demonstrated by reduced binding affinity to FcγRIIIA upon glycan truncation. The subtle differences between IgG3 Fc and the published IgG1 Fc structures with respect to the inter-chain CH2 domain separation and the intra-chain CH2-CH3 domain interface may form the basis for small differences in the Fcγ receptor binding profile of the two IgG subclasses. The conformation of the Arg435 side chain in IgG3 Fc structure provides a probable explanation for the differential protein A and FcRn binding property of IgG3 relative to the other IgG subclasses.
Supplementary Material
Highlights.
The first high-resolution crystal structure of human IgG3 Fc has been solved
Effect of GlcNAc2Man5 (Man5) high mannose type glycosylation on IgG Fc structure was analyzed
Interactions of the N297 glycan with the Fc are demonstrated to be important for binding to FcγRIIIA
IgG3 Fc structure provides basis for IgG3-Arg435 effects on protein A and FcRn binding
IgG3 Fc crystal structure helps to explain some of the unique properties of this IgG subclass
Acknowledgments
This work was supported by NIH grant NIGMS R01 GM090080. The authors also acknowledge financial support of ISS via Howard Rytting Graduate Fellowship from the Department of Pharmaceutical Chemistry, University of Kansas. Use of the University of Kansas Protein Structure Laboratory was supported by a grant the National Institute of General Medical Sciences (P30 GM110761) at the National Institutes of Health. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The authors thank the Gilbert laboratory at Newcastle University for providing the B.t. α-1,2 mannosidase expression plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerström B, Björck L. A physicochemical study of protein G, a molecule with unique immunoglobulin G-binding properties. Journal of Biological Chemistry. 1986;261:10240–10247. [PubMed] [Google Scholar]
- Alessandri L, Ouellette D, Acquah A, Rieser M, LeBlond D, Saltarelli M, Radziejewski C, Fujimori T, Correia I. Increased serum clearance of oligomannose species present on a human IgG1 molecule. MAbs. 2012;4:509–520. doi: 10.4161/mabs.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béranger S, Martinez-Jean C, Bellahcene F, Lefranc MP. Correspondence between the IMGT unique numbering for C-DOMAIN, the IMGT exon numbering, the Eu and Kabat numberings: Human IGHG. IMGT®, the international ImMunoGeneTics information system®. 2001 http://www.imgt.org/IMGTScientificChart/Numbering/Hu_IGHGnber.html.
- Borrok MJ, Jung ST, Kang TH, Monzingo AF, Georgiou G. Revisiting the role of glycosylation in the structure of human IgG Fc. ACS chemical biology. 2012;7:1596–1602. doi: 10.1021/cb300130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosc N. Table of alleles: Human IGHC. IMGT®, the international ImMunoGeneTics information system®. 2003 http://www.imgt.org/IMGTrepertoire/Proteins/taballeles/human/IGH/IGHC/Hu_IGHCall.html.
- Bowden TA, Baruah K, Coles CH, Harvey DJ, Yu X, Song BD, Stuart DI, Aricescu AR, Scanlan CN, Jones EY. Chemical and structural analysis of an antibody folding intermediate trapped during glycan biosynthesis. Journal of the American Chemical Society. 2012;134:17554–17563. doi: 10.1021/ja306068g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA. Protein Glycosylation. John Wiley and Sons, Inc; 2010. pp. 1–25. [Google Scholar]
- Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daëron M. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Rihardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BK, Warburton S, Lin H, Patel R, Boldogh I, Meehl M, d’Anjou M, Pon L, Stadheim TA, Sethuraman N. Improvement of N-glycan site occupancy of therapeutic glycoproteins produced in Pichia pastoris. Applied microbiology and biotechnology. 2012;95:671–682. doi: 10.1007/s00253-012-4067-3. [DOI] [PubMed] [Google Scholar]
- Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, Dugast AS, Schoen MK, Rolland M, Suscovich TJ. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Science translational medicine. 2014;6:228ra238–228ra238. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, Jones EY. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. Journal of molecular biology. 2009;387:1061–1066. doi: 10.1016/j.jmb.2009.02.033. [DOI] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9-and 2.8-. ANG. resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans P. Biochemistry. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn GC, Chen X, Liu YD, Shah B, Zhang Z. Naturally occurring glycan forms of human immunoglobulins G1 and G2. Molecular immunology. 2010;47:2074–2082. doi: 10.1016/j.molimm.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Giuntini S, Granoff D, Beernink P, Ihle O, Bratlie D, Michaelsen T. Human IgG1, IgG3, and IgG3 Hinge-Truncated Mutants Show Different Protection Capabilities against Meningococci Depending on the Target Antigen and Epitope Specificity. Clinical and Vaccine Immunology. 2016;23:698–706. doi: 10.1128/CVI.00193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze AM, Liu YD, Zhang Z, Shah B, Lee E, Bondarenko PV, Flynn GC. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology. 2011;21:949–959. doi: 10.1093/glycob/cwr027. [DOI] [PubMed] [Google Scholar]
- Hopkins D, Gomathinayagam S, Rittenhour AM, Du M, Hoyt E, Karaveg K, Mitchell T, Nett JH, Sharkey NJ, Stadheim TA. Elimination of β-mannose glycan structures in Pichia pastoris. Glycobiology. 2011;21:1616–1626. doi: 10.1093/glycob/cwr108. [DOI] [PubMed] [Google Scholar]
- Huang X, Zheng F, Zhan CG. Binding structures and energies of the human neonatal Fc receptor with human Fc and its mutants by molecular modeling and dynamics simulations. Molecular BioSystems. 2013;9:3047–3058. doi: 10.1039/c3mb70231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. The Journal of Immunology. 2000;164:4178–4184. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- Jefferis R, Lefranc MP. Human immunoglobulin allotypes: possible implications for immunogenicity. MAbs. 2009;1:332–338. doi: 10.4161/mabs.1.4.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic indexing of rotation diffraction patterns. Journal of Applied Crystallography. 1988;21:67–72. [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. Journal of molecular biology. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- Kuby J. Immunology. 3rd. W.H. Freeman; New York: 1997. [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Lefranc G. Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods in Molecular Biology. 2012;882:635–680. doi: 10.1007/978-1-61779-842-9_34. [DOI] [PubMed] [Google Scholar]
- Loghem EV, Frangione B, Recht B, Franklin E. Staphylococcal protein A and human IgG subclasses and allotypes. Scandinavian journal of immunology. 1982;15:275–278. doi: 10.1111/j.1365-3083.1982.tb00649.x. [DOI] [PubMed] [Google Scholar]
- Martin WL, West AP, Jr, Gan L, Bjorkman PJ. Crystal structure at 2.8 Å of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Molecular cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelsen TE, Frangione B, Franklin EC. Primary structure of the “hinge” region of human IgG3. Probable quadruplication of a 15-amino acid residue basic unit. Journal of Biological Chemistry. 1977;252:883–889. [PubMed] [Google Scholar]
- Morell A, Terry WD, Waldmann TA. Metabolic properties of IgG subclasses in man. Journal of Clinical Investigation. 1970;49:673. doi: 10.1172/JCI106279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Yamaguchi Y. Function and 3D structure of the N-glycans on glycoproteins. International journal of molecular sciences. 2012;13:8398–8429. doi: 10.3390/ijms13078398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V, Damschroder MM, Cook KE, Li Q, Gao C, Wu H, Dall'Acqua WF. Structural insights into neonatal Fc receptor-based recycling mechanisms. Journal of Biological Chemistry. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbazghi SZ, More AS, White DR, Duan S, Shah IS, Joshi SB, Middaugh CR, Volkin DB, Tolbert TJ. Production, characterization, and biological evaluation of well-defined IgG1 Fc glycoforms as a model system for biosimilarity analysis. Journal of pharmaceutical sciences. 2016;105:559–574. doi: 10.1016/j.xphs.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25:1325–1334. doi: 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nature Reviews Immunology. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. The Journal of Immunology. 1997;159:3372–3382. [PubMed] [Google Scholar]
- Sauer-Eriksson AE, Kleywegt GJ, Uhlén M, Jones TA. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B. High resolution mapping of the binding site on human IgG1 for FcγRI, FcγRII, FcγRIII, and FcRn and design of IgG1 variants with improved binding to the FcγR. Journal of Biological Chemistry. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- Stapleton NM, Andersen JT, Stemerding AM, Bjarnarson SP, Verheul RC, Gerritsen J, Zhao Y, Kleijer M, Sandlie I, de Haas M. Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nature Communications. 2011;2:599. doi: 10.1038/ncomms1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. Journal of molecular biology. 2012;420:204–219. doi: 10.1016/j.jmb.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Subedi GP, Hanson QM, Barb AW. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcγRIIIa binding. Structure. 2014;22:1478–1488. doi: 10.1016/j.str.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao MH, Smith R, Morrison S. Structural features of human immunoglobulin G that determine isotype-specific differences in complement activation. The Journal of experimental medicine. 1993;178:661–667. doi: 10.1084/jem.178.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao MH, Morrison SL. Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. The Journal of Immunology. 1989;143:2595–2601. [PubMed] [Google Scholar]
- Teplyakov A, Zhao Y, Malia TJ, Obmolova G, Gilliland GL. IgG2 Fc structure and the dynamic features of the IgG CH 2–CH 3 interface. Molecular immunology. 2013;56:131–139. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Thommesen JE, Michaelsen TE, Løset GÅ, Sandlie I, Brekke OH. Lysine 322 in the human IgG3 C H 2 domain is crucial for antibody dependent complement activation. Molecular immunology. 2000;37:995–1004. doi: 10.1016/s0161-5890(01)00010-4. [DOI] [PubMed] [Google Scholar]
- Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Frontiers in immunology. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MR, Lund J, Thompson KM, Jefferis R. Aglycosylation of human IgG1 and IgG3 monoclonal antibodies can eliminate recognition by human cells expressing Fc gamma RI and/or Fc gamma RII receptors. Biochemical Journal. 1989;259:347. doi: 10.1042/bj2590347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerawarna PM, Kim Y, Galasiti Kankanamalage AC, Damalanka VC, Lushington GH, Alliston KR, Mehzabeen N, Battaile KP, Lovell S, Chang KO, Groutas WC. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies. Eur J Med Chem. 2016;119:300–318. doi: 10.1016/j.ejmech.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MS. Global indicators of X-ray data quality. Journal of Applied Crystallography. 2001;34:130–135. [Google Scholar]
- Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, Verrips CT, Dolhain RJ, Hokke CH, Deelder AM. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics. 2007;7:4070–4081. doi: 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- Xiao J, Chen R, Pawlicki MA, Tolbert TJ. Targeting a homogeneously glycosylated antibody Fc to bind cancer cells using a synthetic receptor ligand. Journal of the American Chemical Society. 2009;131:13616–13618. doi: 10.1021/ja9045179. [DOI] [PubMed] [Google Scholar]
- Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, Wong A, Stephan JP, Bayer R. Production, characterization and pharmacokinetic properties of antibodies with N-linked mannose-5 glycans. MAbs. 2012;4:475–487. doi: 10.4161/mabs.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


