Abstract
Neurofibromatosis type 1 (NF1) is a common genetic disorder caused by mutations in the NF1 gene. Recalcitrant bone healing following fracture (i.e. pseudarthrosis) is one of the most problematic skeletal complications associated with NF1. The etiology of this condition is still unclear; thus, pharmacological options for clinical management are limited. Multiple studies have shown the reduced osteogenic potential of Nf1-deficient osteoprogenitors. A recent transcriptome profiling investigation revealed that EREG and EGFR, encoding epiregulin and its receptor Epidermal Growth Factor Receptor 1, respectively, were among the top over-expressed genes in cells of the NF1 pseudarthrosis site. Because EGFR stimulation is known to inhibit osteogenic differentiation, we hypothesized that increased EREG and EGFR expression in NF1-deficient skeletal progenitors may contribute to their reduced osteogenic differentiation potential. In this study, we first confirmed via single-cell mRNA sequencing that EREG over-expression was associated with NF1 second hit somatic mutations in human bone cells, whereas Transforming Growth Factor beta 1 (TGFβ1) expression was unchanged. Second, using ex-vivo recombined Nf1-deficient mouse bone marrow stromal cells (mBMSCs), we show that this molecular signature is conserved between mice and humans, and that epiregulin generated by these cells is overexpressed and active, whereas soluble TGFβ1 expression and activity are not affected. However, blocking either epiregulin function or EGFR signaling by EGFR1 or pan EGFR inhibition (using AG-1478 and Poziotinib respectively) did not correct the differentiation defect of Nf1-deficient mBMSCs, as measured by the expression of Alpl, Ibsp and alkaline phosphatase activity. These results suggest that clinically available pharmacological strategies aimed at inhibiting EGFR signaling are unlikely to have a significant benefit for the management of bone non-union in children with NF1 PA.
Keywords: Neurofibromatosis type 1, epiregulin, EGFR, bone marrow stromal cells, osteoblasts, differentiation, RAS-MAPK signaling
Introduction
Neurofibromatosis type 1 (NF1) is a common genetic disorder that occurs in 1 of 3500 live births [1,2]. It is a pleiotropic condition that affect various organs, including the skin, eyes, nervous system and skeleton [3,4]. In bone, complications include osteopenia, short stature, chest wall deformities, sphenoid wing dysplasia, dystrophic scoliosis and tibia bowing that progresses to fracture and pseudarthrosis (PA, i.e. recalcitrant bone healing/non-union) [2,5]. Patients with NF1 are heterozygous for inherited NF1 mutations, and approximately half of all cases of NF1 occur from spontaneous de novo mutations of the NF1 gene [6]. Second-hit somatic NF1 mutations have been observed in cells from 75% of the NF1 PA biopsies analyzed [7,8]. In other words, one inactivating NF1 variant can be inherited from a parent (or de novo) and the other allele subsequently acquires a somatic inactivating variant in cells of the tibia, leading to loss of NF1 function. Nf1 is expressed in bone cells, including osteoprogenitors [9], differentiated osteoblasts [10], chondrocytes [11,12] and osteoclasts [13]. Evidence from mouse models suggest that NF1 loss of heterozygosity occurs in skeletal progenitor cells of the tibia [10,14,15].
In contrast to most cases of fracture in children, which usually progress to bone union within weeks, 2–5% of children with NF1 present with recalcitrant bone healing despite multiple attempts with surgical stabilization. The condition starts in early childhood with an initial and unilateral bowing of the tibia that often progresses to fracture and non-union. It has the highest morbidity among other NF1 skeletal complications, with little clinical management options [16–18], and often leads to amputation of the affected limb [19]. Bone Morphogenic Proteins (BMPs) are currently used Off-label with variable success, and under clinical investigation for efficacy [20–22], although BMP2 did not show a beneficial effect on its own in preclinical models [10,23]. Hence, finding new therapeutic options for the management of this condition is a significant clinical need.
NF1 encodes neurofibromin, a 280 KDa cytosolic multi-domain protein. The central GTPase-related domain (GRD) of neurofibromin facilitates the conversion of RAS-GTP (active form of RAS) to RAS-GDP (inactive form of RAS) and hence acts as a brake on RAS downstream signaling [24], including the MAPK pathways [11,25,26]. In vitro studies using bony biopsies from children with NF1 PA have shown that periosteal cells from the pseudartrotic site have a blunted response to osteogenic differentiation signals compared to cells from unaffected sites [27,28]. Studies based on the use of murine osteoprogenitor cells have provided further evidence that Nf1 is necessary for proper osteogenic differentiation [10,29]. Bone mesenchymal cells deficient for Nf1, including chondrocytes and osteoblasts, are characterized by high RAS and ERK1/2 activation compared to WT controls [10,11,14,30]. Although this molecular signature was expected to contribute to the impaired differentiation of Nf1-deficient osteoprogenitor cells, two independent studies indicated that MEK blockade was unable to rescue the differentiation phenotype of these cells or to improve bone healing in two mouse models of NF1 PA [10,23]. The exact underlying mechanism of this differentiation phenotype thus remains not well understood, although Rhodes et al reported that excess Tgfβ1 expression in Nf1-deficient mouse osteoblasts might be involved [31]. However, a recent transcriptome analysis using RNA-sequencing of cells cultured from rare biopsies collected from the PA site of children with NF1 did not reveal a change in TGFβ1 expression (Dr. Rios, unpublished data) compared to iliac crest-derived bone cell (herein referred to as NF1+/−). Rather, this study identified a significant upregulation of EGFR and EREG [7]. Epiregulin, encoded by EREG, is one of the seven Epithelial Growth Factor (EGF) family members that preferentially binds to and activates EGFR1 and Erb-B4 forms among the four cloned EGFRs [32–34]. These findings sparked great interest because 1) increased EGFR signaling is known to inhibit osteoprogenitor cell differentiation [35–43]; 2) drugs are clinically available to block EGFR signaling, thus raising the possibility of rapidly repurposing EGFR inhibitors to promote the differentiation of NF1-deficient osteoprogenitors and potentially bone healing in cases of NF1 bone non-union, and 3) the beneficial impact of RAS [44], TGFβ [31] or β-catenin [45] inhibition on bone healing in preclinical models of NF1 PA is expected to take additional effort and time to translate to the clinic. Based on these observations, we hypothesized that sustained EGFR signaling in Nf1-deficient osteoprogenitors contributes to their differentiation defect.
Results
EREG expression is increased in human bone cells characterized by NF1 double hit mutations
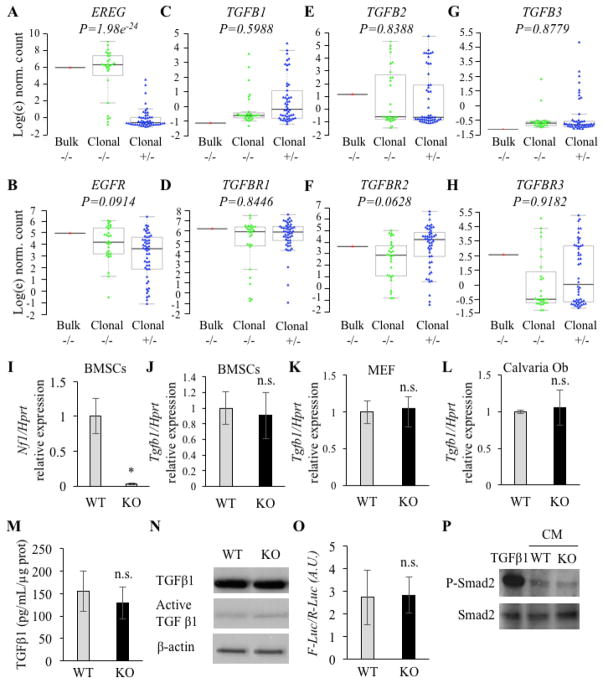
Consistent with previously published data, single-cell sequencing confirmed highly significant upregulation of EREG in NF1−/− clonal cells that harbor a germline p.R461X variant and a somatic p.Asn510_Lys2333del large deletion, compared to patient-matched NF1+/− cells (Figure 1A). EGFR expression was slightly, though not significantly, higher in the NF1-deficient clonal cell line (Figure 1B). No significant differences in gene expression were observed for genes encoding TGFβ ligands nor TGFβ receptors (Figure 1C–H), in contrast to a previous report using Nf1-deficient osteoblasts extracted from the Col12.3kb-cre;Nf1f/f mouse model [31].
Figure 1.
A–H: Gene expression profiling from cells of the NF1 PA site. Bulk: mixed cells of the NF1 PA site; Clonal −/−: Single cells from the NF1 PA site; Clonal +/−: single cells from the iliac crest cultures from the same patient. I: Nf1 expression in WT (Nf1f/f +Ad-GFP) and Nf1-deficient (KO, Nf1f/f +Ad-CRE) mBMSCs (qPCR, n=3). J–L: Tgfb1 expression in murine WT and Nf1-deficient mBMSCs (J), MEF cells (K) and calvaria cells (L)(qPCR, n=3). M–O: TGFβ1 protein expression in WT and Nf1 KO BMSCs using ELISA (M, n=3) and Western blotting (N, n=3). O–P: Measurement of TGFβ-1/SMAD signaling activity in the conditioned medium collected from cultures of WT and -deficient mBMSCs (n=3) using Luciferase assay (O, n=3) and p-SMAd2 level (P, n=3, TGFβ1 positive control: 5ng/ml). n.s: non-significant, *: p < 0.05 between genotypes, qPCR gene expression is normalized by Hprt expression.
These observations led us to assess the expression of Tgfb1 in WT and Nf1-deficient mouse bone marrow stromal cells (mBMSCs). For this purpose, Nf1f/f mBMSCs were cultured and infected with a cre-expressing adenovirus (herein referred to as Nf1-deficient) or a GFP-expressing adenovirus (herein referred to as WT control). Loss of Nf1 gene expression following cre-expressing adenovirus transduction was confirmed by a significant reduction (>90%) in Nf1 gene expression by qRT-PCR compared to Ad-GFP control (Figure 1I). No difference in Tgfb1 expression was found in Nf1-deficient mBMSCs (Figure 1J). Tgfb1 was also expressed at similar levels in WT and Nf1-deficient mouse embryonic fibroblasts (MEFs) isolated from WT and Nf1−/− embryos, considered to be more immature mesenchymal progenitor cells than mBMSCs (Figure 1K) and in WT and Nf1-deficient calvaria-derived cells that are considered more committed to the osteoblast lineage (Figure 1L). No detectable difference in the amount of soluble total TGFβ1 (measured by ELISA, Figure 1M) nor secreted active TGFβ1 (measured by Western Blot, Figure 1N) was observed between the conditioned medium (CM) from WT and Nf1-deficient mBMSCs. Finally, the CM from WT and Nf1-deficient mBMSCs resulted in similar levels of activation of a sensitive SMAD-responsive luciferase reporter MDA231 cell line (limit of detection: 1ng/ml, Supplementary Figure 1A and Figure 1O) [46], and to similar level of p-SMAD2 activation in treated WT BMSCs (Figure 1P). Collectively, these data strongly suggest that increased TGFβ1 production by Nf1-deficient osteoprogenitors is not the main cause of the impaired osteogenic potential of these cells.
Epiregulin is ectopically expressed and active in Nf1-deficient mBMSCs
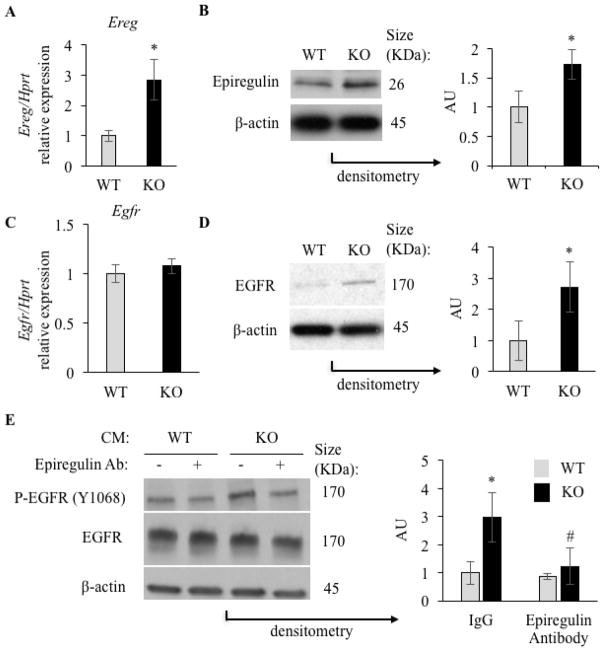
Because single-cell sequencing confirmed that NF1 deficiency in human bone cells was associated with EREG over-expression, we sought to determine whether this phenotype was conserved in mBMSCs. Using the same strategy of ex vivo Nf1 ablation as indicated above, we found Ereg to be expressed in Nf1-deficient mBMSCs at three times the level of WT mBMSCs (Figure 2A). This increase was confirmed at the protein level (Figure 2B). In contrast, expression of Egfr was not altered in Nf1-deficient mBMSCs (Figure 2C), though EGFR protein abundance was higher in these cells (Figure 2D). A similar increase in Ereg expression was also detected in rib primary Nf1f/f chondrocytes infected with Ad-cre compared to Ad-GFP viruses (Supplementary Fig. 1B). The expression of other EGFR ligands, including Betacellulin (Btc), Epidermal Growth Factor (Egf), Transforming Growth Factor a (Tgfa) and Amphiregulin (Areg), was undetectable in both WT and Nf1-deficient mBMSCs (data not shown). These results suggest that neurofibromin signaling represses Ereg expression in both human and mouse BMSCs and that EGFR protein synthesis or stability is regulated by mechanisms that are neurofibromin-dependent and post-transcriptional.
Figure 2.
A: Ereg expression in WT and Nf1-deficient mBMSCs (qPCR, n=3). B: Epiregulin protein expression in WT and Nf1-deficient mBMSCs (Western blot, n=3, Right graph: densitometric analysis). C: Egfr expression in WT and Nf1-deficient mBMSCs (qPCR, n=3). D: EGFR protein expression in WT and Nf1 deficient mBMSCs (Western blot, n=3, Right graph: densitometric analysis). E: Level of phosphorylated EGFR (p-EGFR), EGFR and β-actin in A431 cells treated with the conditioned medium (CM) from WT (grey bar) and Nf1-deficient (KO, black bar) mBMSCs in the presence of IgG control or an epiregulin neutralizing antibody (Western blot, n=3, Right graph: densitometric analysis). * and #: p<0.05 between genotypes and treatments, respectively. qPCR gene expression is normalized by Hprt expression.
Epiregulin is synthesized as a precursor membrane-bound protein that must be cleaved for biological activity and activation of EGFR [33]. To determine if Nf1-deficient mBMSCs generate higher amount of active epiregulin than WT mBMSCs, a cell line highly sensitive to EGFR ligands (A431 cells) [47] was treated with the CM from WT and Nf1-deficient mBMSCs. Both CMs led to EGFR activation (phosphorylation), but the CM from Nf1-deficient mBMSCs was three times more potent than the one from WT mBMSCs (Figure 2E). In addition, EGFR activity following treatment with the Nf1-deficient CM was blocked following addition of an epiregulin-neutralizing antibody (Figure 2E). These results suggest that the CM of Nf1-deficient mBMSCs contains higher amount of active epiregulin compared to WT mBMSCs.
Inhibition of EGFR signaling fails to rescue the osteogenic differentiation of Nf1-deficient mBMSCs
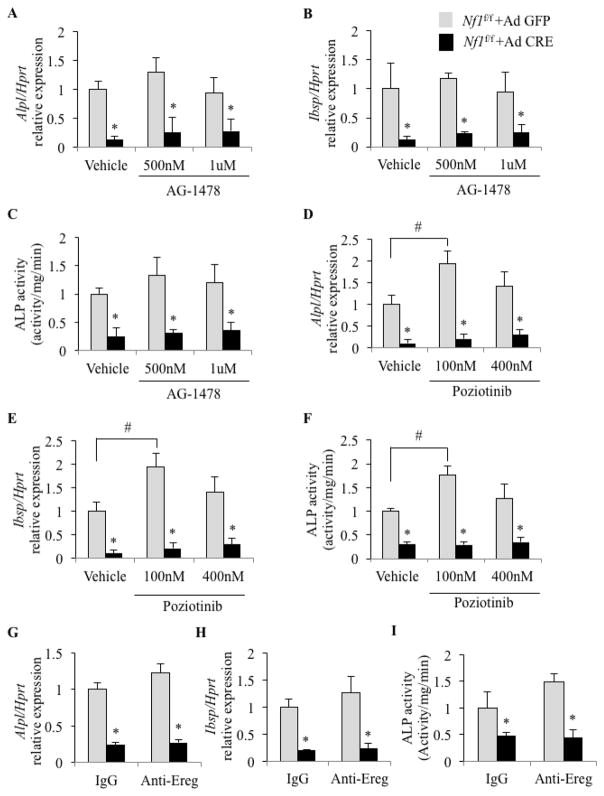
Because chronic activation of EGFR leads to inhibition of osteogenic differentiation (32–33,36–40,42) and Nf1-deficient mBMSCs overexpress both EGFR and its ligand epiregulin, we sought to block EGFR signaling to determine if excessive EGFR signaling contributed to the reduced osteogenic potential of these cells. WT and Nf1-deficient mBMSCs were prepared as described above and treated from the start of differentiation (Day 0) with AG-1478 (0.5 and 1 μM), a potent and selective EGFR kinase inhibitor (IC50=3nM in a cell-free system [48]), for 7 days in osteogenic medium, and early osteogenic differentiation was assessed by measuring Alkaline phosphatase (Alpl) and Integrin binding sialoprotein (Ibsp) expression. As expected, the expression of Alpl and Ibsp in Nf1-deficient mBMSCs was reduced to 10–20% of WT controls (Vehicle in Figure 3A–B). Surprisingly however, inhibition of EGFR signaling with AG-1478 failed to rescue the reduced expression of these genes in Nf1-deficient mBMSCs (Figure 3A, B), which was confirmed by measuring ALP activity (Figure 3C). Poziotinib, an irreversible pan-EGFR inhibitor (IC50=3.2nM for HER1, 5.3nM for HER2 and 23.5nM for HER4 [49]) tested at two concentrations (100nM or 400nM) also failed to increase the expression of Alpl and Ibsp (Figure 3D, E) and ALP activity (Figure 3F) in Nf1-deficient mBMSCs following osteogenic induction. Both AG-1478 and Poziotinib inhibited EGFR activation in A341 reporter cells (Supplementary Figure 1C, D), confirming potent biological activity of these two drugs.
Figure 3.
A–B, D–E and G, H: Expression of early osteoblast marker genes (Alpl, Ibsp) in response to EGFR or Epiregulin inhibition during osteogenic differentiation (A–B: AG-1478, D–E: Poziotinib and G, H: epiregulin-neutralizing antibody) in WT and Nf1-deficient (KO) mBMSCs (qPCR, n=3, * and #: p<0.05 between genotypes and treatments, respectively). C, F and I: ALP activity in response to AG-1478, Poziotinib and Anti-Ereg neutralizing antibodies, respectively (n=3, * and #: p<0.05 between genotypes and treatments, respectively). qPCR gene expression is normalized by Hprt expression.
It remained possible that epiregulin signals via receptors other than EGFR or ERB-B4. To address this hypothesis, WT and Nf1-deficient mBMSCs were grown in osteogenic conditions for 7 days in the presence of an epiregulin-neutralizing antibody (0.4 ug/ml). Although this neutralizing antibody was added to the medium in large excess and successfully blocked EGFR activation in human A341 cells (see Figure 2E), it failed to rescue the osteogenic differentiation of Nf1-deficient mBMSCs (Figure 3G–I).
Together, these results suggest that the increase in epiregulin and EGFR expression in Nf1-deficient osteoprogenitors does not contribute to their defective differentiation potential.
Discussion
Transcriptome profiling of bone cells cultured from a case of NF1 tibial PA indicated that NF1-deficiency was associated with over-expression of EREG, encoding the EGFR ligand Epiregulin [7]. This observation and the known inhibitory effect of EGFR signaling on osteoblast differentiation raised the prospect that clinically available EGFR inhibitors may promote bone union in challenging surgical cases of NF1 PA. Further progress in this direction required pre-clinical studies to determine whether such molecular findings were functionally relevant to the impaired differentiation of NF1-deficient osteoprogenitors and eventually to the recalcitrant bone healing observed in NF1. We show here that Ereg is ectopically overexpressed in Nf1-deficient mouse BMSCs, as observed in human cells with NF1 biallelic mutations. Although evidence for increased epiregulin protein production by Nf1-deficient mBMSCs and EGFR signaling activity were observed, both pharmacological EGFR inhibition and epiregulin ligand blockade failed to correct the differentiation defect of these cells. These results led us to conclude that the upregulation of epiregulin expression and EGFR activation induced by Nf1 deficiency in osteoprogenitor cells is not causal for their impaired osteogenic potential, and indicate that pathways other than EGFR signaling contribute to this phenotype.
The lack of effect of EGFR inhibition in Nf1-deficient BMSCs might appear expected since EGFR signals via the RAS/ERK pathway, whose activity downstream of EGFR is chronically increased in absence of Nf1. However, one should keep in mind that signaling pathways activated by EGFR also include PI3K-Akt, p38, JNK, JAK-Stat, Src, small GTPases such as Rho and Rac, PLC-gamma/Ca2+/PKC and PKD [50]. Hence EGFR blockade could still be pro-osteogenic in Nf1-deficient cells by inhibiting one of these downstream ERK-independent pathways. This is further supported by the lack of effect of MEK inhibitors on the differentiation of these cells [10,23], which suggest the existence of MEK/ERK-independent mechanisms underlying their differentiation defect.
New candidate genes involved in the defective differentiation of Nf1-deficient osteoprogenitors were recently identified, including Tgfb1 [31] and Ctnnb1 (encoding β-catenin) [45]. The finding that the level of TGFβ1 was increased in the culture medium of BMSCs isolated from the Col2.3kb-cre; Nf1flox/− mice is in line with the phenotypic overlap between the cellular abnormalities in NF1 PA and other conditions characterized by excessive TGFβ signaling, including Camurati-Engelmann, Marfan and Loeys-Dietz syndromes [51–53]. It is also in line with the known pro-proliferative and anti-osteogenic differentiation activities of TGFβ [54–59], which mimic the in vitro behavior of Nf1-deficient osteoprogenitors. However, our analyses did not allow us to confirm increased levels of TGFβ1 expression in Nf1-deficient bone cells, including MEFs, BMSCs and calvaria primary cells, and the reason for this may stem in a number of differences between the two studies. One of them is the different cells used between these two studies. Rhodes et al. prepared Nf1−/− MSCs from the bone marrow of periostin-cre; Nf1flox/− mice, and differentiated them after 5–10 passages in osteogenic medium. This is in contrast with our cultures, that were prepared from undifferentiated mBMSCs extracted from Nf1flox/flox mice, infected ex vivo with a GFP- or CRE-adenovirus, and not passaged after infection. Although the adenovirus infection may impact to some extent the behavior of the cultures, this approach has the advantage of comparing clearly-defined genotypes and cultures, whose behavior starts to differ after ex vivo infection, whereas the approach from Rhodes et al. relies on extensively passaged primary cells, whose differentiation and behavior may be impacted in vivo before extraction and plating, and ex vivo because of multiple passages. A consequence from these different experimental conditions is that the two studies may have compared osteoblasts at different differentiation stages, with the Rhodes study based on more differentiated osteoblast cultures than our study, which used undifferentiated, plastic-adherent bone marrow osteoprogenitors and Nf1 ablation induced shortly thereafter before induction of differentiation by confluency and addition of osteogenic medium. This is important to notice because the progressive and long-term nature of tibia bowing and non-union in NF1, and data from genetic mouse models related to this condition, all support the idea that the cell of origin for this condition is a proliferating, undifferentiated mesenchymal progenitor, prior to the expression of Col2 and Osx [10,11]. Hence, the traits and behavior of Nf1-deficient undifferentiated osteoprogenitors are likely to be more clinically relevant than the characteristics of Nf1-deficient mature osteoblasts or osteocytes for instance [60], that are unlikely to be ever generated based on the defective differentiation of Nf1-deficient osteoprogenitors.
It is still important to recognize the beneficial effect of TGFβ blockade on bone mass and fracture healing reported by Rhodes et al., and the clinical relevance of these findings. A similar comment applies to the findings by Ghadakzadeh et al., showing improved bone healing upon use of Nefopam treatment to block the increase in β-catenin expression they detected in Nf1-deficient mBMSCs [45,61]. An important note related to these published studies is that they all use Nf1 conditional floxed cells to achieve gene ablation following cre activity. A caveat with this approach is that gene recombination is rarely complete. Therefore, a detectable increase in osteogenic differentiation in these cultures following treatment can reflect an osteogenic response of non-recombined cells to osteogenic treatments like BMP2, nefopam or blockade of TGFβR. This is supported by the observation that SD-208, a TGFβR inhibitor, increases Alpl expression in both Nf1-deficient and WT mBMSCs (Supplementary Figure 1E) and bone mass in both WT and Col2.3kb-cre; Nf1flox/− mice [31]. Interpretation of results must account for this effect of treatment on non-KO cells, and the extent of this confounding factor should be assessed by the use of appropriate controls, which include treatment of the WT cells. Taking this comment into consideration, we conclude that TGFβ and β-catenin blockade has preclinical value as pharmacological approach to improve bone union in children with NF1 PA, but the stimulatory effect of SD-208 treatment on WT cells and our inability to detect an increase in TGFβ expression in Nf1-deficient bone cells question the contribution of increased TGFβ1 levels to the impaired osteogenic potential of Nf1-deficient BMSCs. Our results also do not support increased Ereg expression and signaling as a major component of the defective differentiation potential of Nf1-deficient osteoprogenitors. The quest for the molecular basis of this phenotype is thus still open in order to identify specific NF1 signaling-related molecular targets/nodes amenable to pharmacological treatment.
Materials and methods
BMSC cultures
The institutional animal care and use committee Baylor College of Medicine approved all the mouse procedures. Mice were housed 2–5 per cage. Mouse BMSCs were extracted from long bones of 2–3 month-old Nf1f/f mice [62] by centrifugation at 3000 g for 3 minutes, as previously described [63]. Extracted marrow was plated in 10 cm dishes in α-MEM medium (without ascorbic acid) supplemented with 10% fetal bovine serum and 100 U/ml Penicillin/Streptomycin (15140-122, ThermoFisher) for three days. At that time, non-adherent cells were discarded by changing the medium. Cells were trypsinized after reaching 80% confluence and were seeded in 6-well plates at 10,000 cells/cm2 for adenovirus transduction. After reaching 60% confluence, cells were incubated with the adenovirus solutions (Ad-GFP or Ad-CRE recombinase, Baylor College of Medicine vector development lab) in the presence of Gene Jammer reagent (Agilent technologies; Cat# 204132), as described previously [64]. Briefly, Gene Jammer was added at a final concentration of 1% to FBS- and antibiotic-free α-MEM medium. The solution was vortexed briefly and incubated for 10 minutes at room temperature before adding the virus at a MOI of 400 and incubating for further 10 minutes. Final mixture was added to each well and cells were incubated with the virus solutions for 24 hours. The media was then changed to fresh complete α-MEM medium containing 10% FBS and Pen/Strep (Thermofisher Cat# 15140122). Mouse BMSCs were differentiated in osteogenic medium containing ascorbic acid (50 μg/ml) and β-glycerophosphate (5mM) in α-MEM medium for 7 days. Medium was changed every other day.
For conditioned medium (CM) collection, mBMSCs infected with either Ad-GFP or Ad-CRE were washed with PBS two times and were grown with FBS-free α-MEM medium for 1 day. The CMs were centrifuged at 1000 g for 5 minutes to remove debris, and the supernatant was collected and were kept at −80°C until use.
A431 cells were grown in DMEM supplemented with 10% FBS and 100 U/ml Penicillin/Streptomycin (15140-122, ThermoFisher). After reaching 80% confluence, cells were starved in serum-free DMEM overnight. Cells were then treated with the conditioned media plus normal goat IgG control (AB-108-C, R&D Systems) or Epiregulin neutralizing antibody (AF1068-SP, R&D Systems) at the final concentration of 0.4 μg/ml. Cell lysates were extracted after ten minutes and the amount of p-EGFR, EGFR (Cat. # 3777S and 4267S from Cell signaling Technology, respectively) and β-actin (A5316 from Sigma) were measured by Western blotting.
To measure Smad2 activation, mBMSC were grown in α-MEM until they reached 80% confluence and were then starved overnight in FBS free medium before treatment with either recombinant activated TGFβ-1 (R&D systems, Cat# 766-MB-005) or the conditioned medium from Nf1 WT and KO BMSCs for 30 minutes. Cells were then scraped in RIPA buffer and after protein extraction, the amount of Smad2,3 (Cat. # 3102, CST) and p-Samd2 (3108, CST) levels were measured.
Chondrocyte cultures
The cartilaginous ribs from 6 day-old pups were extracted and digested overnight by collagenase (Sigma, Cat# C6885) dissolved in DMEM at a final concentration of 3 mg/ml. On the next day, the digestion medium was centrifuged and pellets were re-suspended in DMEM and filtered through 50 micrometer filters. Resuspended cells were plated in 6 well plates in DMEM supplemented with FBS and supplemented with 10% FBS and 100 U/ml Penicillin/Streptomycin (Cat# 15140-122, ThermoFisher). Cells were infected when they reached 50% confluence with GFP or CRE adenoviruses as indicated above.
Calvaria cultures
The calvariae from 4 day-old Nf1f/f pups were extracted and digested consecutively three times in digestion medium, prepared by dissolving collagenase P at a final concentration of 0.1 mg/ml (Sigma, Cat# 11213865001) in 0.25% Trypsin (ThemoSisher, Cat# 25200-056). After the last digestion, bone fragments were plated in 10 cm dish in α-MEM medium (without ascorbic acid) supplemented with 10% FBS and 100 U/ml Penicillin/Streptomycin (Cat# 15140-122, ThermoFisher). Medium was changed after 4–5 days. Cells were trypsinized after reaching 80% confluence and were replated in 6 well plates before infection with a GFP- or CRE adenovirus as indicated above.
Human primary cell culture and sorting
Human BMSCs isolated from tibial PA of one NF1 patient with an inherited mutation c.1381C>T (p.R461X) and a somatic deletion (c.1642_6999del; p.Asn510_Lys2333del) in the NF1 gene [7] were cultured in α-MEM (without ascorbic acid) supplemented with 10% FBS and 1% PS. Cells were trypsinized and resuspended in 500ul of PBS containing 10% FBS and 2.5mM EDTA. The 7AAD live cell marker dye was added to the cell suspension and live single cells were sorted using an Aria Cell Sorter (BD Biosciences) into 96-well plates containing 100ul of α-MEM media with 20% FBS. 100ul of conditioned media from the original “bulk” culture was added to help with the growth of single cell clones. After reaching confluence, cells were expanded into 6-well plates and cultured again with fresh medium complemented with 50% of bulk culture conditioned media. DNA from clonal lines was extracted using the QiaAmp DNA mini kit (Qiagen) and sequenced for the presence or absence of the deleted allele.
Luciferase assay for TGFβ1 activity measurements
The conditioned medium from mBMSCs was harvested as described above, and the TGFβ1 reporter cell line MDA-scp28 was used to quantify active TGFβ1 in this CM [46]. Briefly, 50,000 MDA-scp28 cells/well were plated in a 96-mutliwell culture plate in high glucose DMEM supplemented with 10% FBS and 100 U/ml Penicillin/Streptomycin. The MDA-scp28 were then starved for 24hr in FBS-free DMEM high glucose medium and were treated with the CM of mBMSCs or recombinant TGFβ1 for 8hr. Luciferase activity was detected by the Dual Luciferase kit (Promega, E1960), following the manufacturer instructions. Firefly luciferase activity was normalized by the Renilla luciferase activity (ratio F-Luc/R-Luc).
TGFβ1 ELISA
Total TGFβ1 in supernatants of WT and Nf1-deficient mBMSCs was quantified by ELISA (R&D, DY1679). Briefly 100 μl of supernatant were acidified with 20 μl of 1N HCl and incubated 10 min at room temperature. Acidity was then neutralized by the addition of 20 μl of 1.2N NaOH/0.5M HEPES. Total TGFβ1 concentrations of prepared samples were measured according to the manufacturer’s instructions.
Single-cell mRNA sequencing and analysis
Single cells were isolated and cDNAs were generated using the Fuidigm C1 instrument and SMARTer Ultra Low RNA Kit (Clontech Cat#634834). Sequencing libraries were generated using the Nextera XT Library Preparation Kit (Illumina ref#15032354) and sequenced using the Illumina HiSeq 2500 generating paired-end 100bp reads. A single bulk sample of 100–200 clonal NF1−/− cells were isolated and processed in the same manner, except that the BioRad Thermal Cycler was used in place of the C1.
FASTQ sequence reads were trimmed using Flexbar read trimmer (PMID=24832523) and mapped to the human reference genome (GRCh38) using HISAT2 (PMID=25751142). Mapped reads were compared to the GENCODE transcriptome (version 24) and counted using HTSeq [65]. Following filtering, 78 cells (N=50 iliac crest NF1+/− and N=28 clonal NF1−/−) were used for differential gene expression analysis using DESeq2 [66]. One bulk sample of clonal NF1−/− cells were also included for comparison. Log counts per million (CPM) mapped reads was calculated and visualized using the R package beeswarm. Significance values (p-value) are adjusted for multiple comparisons.
Drugs
AG-1478 (Selleckchem Cat# S2728), Poziotinib (Selleckchem, Cat# S7358) SD208 (Sigma, Cat# S7071) and U0126 (Cell Signaling Technology, Cat. # 9903) were reconstituted in DMSO (Vehicle).
ALP activity
Cells were washed with PBS, harvested and lysed in 250 μl of 0.05% Triton plus two cycles of freezing/thawing at −80°C/37°C. Cell lysate were then centrifuged for 20 minutes at 16,000g at 4°C and supernatants were used for protein (BCA method; Life Technologies, Cat# 23225) and ALP activity measurements. ALP activity was measured using a colorimetric assay. Briefly, a PNPP ((4-nitrophenyl phosphate disodium salt hexahydrate, Sigma Cat# P4744) solution was prepared in water and was mixed with AMP (2-amino-2-methyl-1-propanol, Sigma Cat # A65182) buffer. Cells lysate were added to the mix (1:5) and incubated at 37°C for 30 minutes. Absorbance was read at 405 nm and normalized to protein content.
Gene expression assays
Total RNA was extracted using TRIzol (Thermofisher, Cat# 15596026), and contaminating genomic DNA was digested by treatment with DNAse I (Promega, Cat# M6101). cDNAs were synthesized from 1ug RNAs using the high capacity cDNA reverse transcription kit (Thermofisher, Cat# 4368814),). Quantitative qRT-PCR was performed using the following TaqMan primers/probes: Ccnd1 (Mm00432359_m1), Ibsp (Mm00492555_m1), Egfr1 (Mm01187858_m1), Alpl (Mm00475834_m1), Tgfb1 (Mm03024053_m1) and the normalizer Hprt (Mm03024075_m1) from Thermofisher, or SYBR green primers: Nf1 (forward: GTATTGAATTGAAGCACCTTTGTTTGG; reverse: CTGCCCAAGGCTCCCCCAG); Ereg (forward: TTGTGCTGATAACTGCCTGTAGAA; reverse: CACCGAGAAAGAAGGATGGAGAC). SYBR qPCR specificity of amplification was verified by the presence of a single peak on the dissociation curve.
Western blot
Proteins were extracted from cell cultures using RIPA buffer. Protein concentration was measured using BCA assay (Thermo-Fisher). Ten μg of total protein was run on SDS gel before transfer to a nitrocellulose membrane. Membranes were blocked using 5% non-fat powder milk in TBST buffer. Epiregulin antibody (AF1068-SP, R&D Systems), β-actin: (A5316, Sigma), and Cell Signaling Technologies antibodies Smad2,3 (3102), p-Samd2 (3108), EGFR (4267S) and p-EGFR (3777S) were diluted in blocking buffer at 1:1000 to 1:2000 dilution and incubated with the membranes overnight at 4°C. Following washing, membranes were then incubated with an HRP-conjugated secondary antibody (goat anti mouse Santa Cruz Cat # sc-2005, goat anti-rabbit Santa Cruz Cat# sc-2030) diluted in blocking buffer at room temperature for one hour. Membranes were washed and incubated with ECL solution for 2 minutes and exposed to photographic film.
Statistical analyses
For comparison between WT and KO cells, a student t-test was performed. For multiple treatments, a two-way analysis of variance (ANOVA) was used to determine whether there was a statistically significant difference in treated vs. non-treated cells between genotypes. P-value less than 0.05 was considered significant. Statistical analysis was performed using Graph Pad PRISM (v6.0a, La Jolla, CA, USA). Data are provided as mean +/− SD.
Supplementary Material
Highlights.
The increase in EREG expression previously observed in human bone cells characterized by double hit mutations in NF1 is conserved between mice and humans.
Mouse Nf1-deficient osteoprogenitors produce higher level of epiregulin and EGFR than WT cell, but normal level of TGFβ1.
EGFR inhibition does not correct the impaired osteogenic potential of Nf1-deficient osteoprogenitors.
Therefore, excessive EGFR signaling is not the major mechanism underlying the reduced differentiation potential of Nf1-deficient osteoprogenitors.
Acknowledgments
Funding: Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R56AR055966 (FE), by the Texas Scottish Rite Hospital for Children and the Pediatric Orthopaedic Society of North America (JR), Children Tumor Foundation Young Investigator Award Number 2015-01-015 (SET). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We wish to thank Dr. Bhuminder Singh for providing the A431 cell line and his expert guidance, Dr. Harry Kim, Dr. Nibuhiro Kamiya and the Texas Scottish Rite Hospital for Children Tissue Bank Repository.
Authors’ role: Study design: SET, GC, FE. Study conduct, data analysis and collection: SET, CG, NP, JG, FL, XW, JJR, FE. Data interpretation: SET, JJR, FE. Drafting and revision manuscript: SET, JJR, FE. Approval final version of the manuscript: SET, CG, NP, JG, FL, XW, JJR, FE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huson SM, Compston DA, Clark P, Harper PS. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. I. Prevalence, fitness, mutation rate, and effect of parental transmission on severity. J Med Genet. 1989;26:704–11. doi: 10.1136/jmg.26.11.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elefteriou F, Kolanczyk M, Schindeler A, Viskochil DH, Hock JM, Schorry EK, Crawford AH, Friedman JM, Little D, Peltonen J, Carey JC, Feldman D, Yu X, Armstrong L, Birch P, Kendler DL, Mundlos S, Yang FC, Agiostratidou G, Hunter-Schaedle K, Stevenson DA. Skeletal abnormalities in neurofibromatosis type 1: approaches to therapeutic options. Am J Med Genet A. 2009;149A:2327–38. doi: 10.1002/ajmg.a.33045. [DOI] [PubMed] [Google Scholar]
- 3.Stumph D. Neurofibromatosis. Conference statement. National Institutes of Health Consensus Development Conference. Arch Neurol. 1988;45:575–8. [PubMed] [Google Scholar]
- 4.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–61. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 5.Vitale MG, Guha A, Skaggs DL. Orthopaedic manifestations of neurofibromatosis in children: an update. Clin Orthop Relat Res. 2002:107–18. doi: 10.1097/00003086-200208000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyaya M, Osborn M, Maynard J, Harper P. Characterization of six mutations in exon 37 of neurofibromatosis type 1 gene. Am J Med Genet. 1996;67:421–3. doi: 10.1002/(SICI)1096-8628(19960726)67:4<421::AID-AJMG20>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Paria N, Cho TJ, Choi IH, Kamiya N, Kayembe K, Mao R, Margraf RL, Obermosser G, Oxendine I, Sant DW, Song MH, Stevenson DA, Viskochil DH, Wise CA, Kim HKW, Rios JJ. Neurofibromin deficiency-associated transcriptional dysregulation suggests a novel therapy for tibial pseudoarthrosis in NF1. J Bone Miner Res. 2014;29:2636–42. doi: 10.1002/jbmr.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevenson DA, Zhou H, Ashrafi S, Messiaen LM, Carey JC, D’Astous JL, Santora SD, Viskochil DH. Double inactivation of NF1 in tibial pseudarthrosis. Am J Hum Genet. 2006;79:143–8. doi: 10.1086/504441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Nyman JS, Ono K, Stevenson DA, Yang X, Elefteriou F. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Hum Mol Genet. 2011;20:3910–3924. doi: 10.1093/hmg/ddr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Ndong JC, Stevens DM, Vignaux G, Uppuganti S, Perrien DS, Yang X, Nyman JS, Harth E, Elefteriou F. Combined MEK inhibition and BMP2 treatment promotes osteoblast differentiation and bone healing in Nf1OSX (−/−) mice. J Bone Miner Res. 2014 doi: 10.1002/jbmr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono K, Karolak MR, de la J, Ndong C, Wang W, Yang X, Elefteriou F. The ras-GTPase activity of neurofibromin restrains ERK-dependent FGFR signaling during endochondral bone formation. Hum Mol Genet. 2013;22:3048–62. doi: 10.1093/hmg/ddt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karolak MR, Yang X, Elefteriou F. FGFR1 signaling in hypertrophic chondrocytes is attenuated by the Ras-GAP neurofibromin during endochondral bone formation. Hum Mol Genet. 2015;24:2552–64. doi: 10.1093/hmg/ddv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang FC, Chen S, Robling AG, Yu X, Nebesio TD, Yan J, Morgan T, Li X, Yuan J, Hock J, Ingram DA, Clapp DW. Hyperactivation of p21ras and PI3K cooperate to alter murine and human neurofibromatosis type 1-haploinsufficient osteoclast functions. J Clin Invest. 2006;116:2880–91. doi: 10.1172/JCI29092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Nyman JS, Ono K, Stevenson DA, Yang X, Elefteriou F. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Hum Mol Genet. 2011;20:3910–24. doi: 10.1093/hmg/ddr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Croix Ndong J, Makowski AJ, Uppuganti S, Vignaux G, Ono K, Perrien DS, Joubert S, Baglio SR, Granchi D, Stevenson DA, Rios JJ, Nyman JS, Elefteriou F. Asfotase-α improves bone growth, mineralization and strength in mouse models of neurofibromatosis type-1. Nat Med. 2014;20:904–10. doi: 10.1038/nm.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevenson DA, Little D, Armstrong L, Crawford AH, Eastwood D, Friedman JM, Greggi T, Gutierrez G, Hunter-Schaedle K, Kendler DL, Kolanczyk M, Monsell F, Oetgen M, Richards BS, Schindeler A, Schorry EK, Wilkes D, Viskochil DH, Yang F-C, Elefteriou F. Approaches to treating NF1 tibial pseudarthrosis: consensus from the Children’s Tumor Foundation NF1 Bone Abnormalities Consortium. J Pediatr Orthop. 2013;33:269–75. doi: 10.1097/BPO.0b013e31828121b8. [DOI] [PubMed] [Google Scholar]
- 17.Stevenson DA, Birch PH, Friedman JM, Viskochil DH, Balestrazzi P, Boni S, Buske A, Korf BR, Niimura M, Pivnick EK, Schorry EK, Short MP, Tenconi R, Tonsgard JH, Carey JC. Descriptive analysis of tibial pseudarthrosis in patients with neurofibromatosis 1. Am J Med Genet. 1999;84:413–9. doi: 10.1002/(sici)1096-8628(19990611)84:5<413::aid-ajmg5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Ippolito E, Corsi A, Grill F, Wientroub S, Bianco P. Pathology of bone lesions associated with congenital pseudarthrosis of the leg. J Pediatr Orthop B. 2000;9:3–10. doi: 10.1097/01202412-200001000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Richards BS, Anderson TD. rhBMP-2 and Intramedullary Fixation in Congenital Pseudarthrosis of the Tibia. J Pediatr Orthop. 2016 doi: 10.1097/BPO.0000000000000789. [DOI] [PubMed] [Google Scholar]
- 20.Anticevic D, Jelic M, Vukicevic S. Treatment of a congenital pseudarthrosis of the tibia by osteogenic protein-1 (bone morphogenetic protein-7): a case report. J Pediatr Orthop B. 2006;15:220–221. doi: 10.1097/01.bpb.0000194439.75378.ac. [DOI] [PubMed] [Google Scholar]
- 21.Lee FY-I, Sinicropi SM, Lee FS, Vitale MG, Roye DP, Choi IH. Treatment of Congenital Pseudarthrosis of the Tibia with Recombinant Human Bone Morphogenetic Protein-7 (rhBMP-7): A Report of Five Cases. JBJS Case Connect. 2006;os-88:627–633. doi: 10.2106/JBJS.D.02201. [DOI] [PubMed] [Google Scholar]
- 22.Fabeck L, Ghafil D, Gerroudj M, Baillon R, Delincé P. Bone morphogenetic protein 7 in the treatment of congenital pseudarthrosis of the tibia. J Bone Jt Surg - Br. 2006;88-B:116–118. doi: 10.1302/0301-620X.88B1.16619. [DOI] [PubMed] [Google Scholar]
- 23.El-Hoss J, Cheng T, Carpenter EC, Sullivan K, Deo N, Mikulec K, Little DG, Schindeler A. A Combination of rhBMP-2 (Recombinant Human Bone Morphogenetic Protein-2) and MEK (MAP Kinase/ERK Kinase) Inhibitor PD0325901 Increases Bone Formation in a Murine Model of Neurofibromatosis Type I Pseudarthrosis. J Bone Joint Surg Am. 2014;96:e117. doi: 10.2106/JBJS.M.00862. [DOI] [PubMed] [Google Scholar]
- 24.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 25.Sharma R, Wu X, Rhodes SD, Chen S, He Y, Yuan J, Li J, Yang X, Li X, Jiang L, Kim ET, Stevenson DA, Viskochil D, Xu M, Yang FC. Hyperactive Ras/MAPK signaling is critical for tibial nonunion fracture in neurofibromin-deficient mice. Hum Mol Genet. 2013;22:4818–28. doi: 10.1093/hmg/ddt333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YH, Gianino SM, Gutmann DH. Neurofibromatosis-1 regulation of neural stem cell proliferation and multilineage differentiation operates through distinct RAS effector pathways. Genes Dev. 2015;29:1677–82. doi: 10.1101/gad.261677.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho TJ, Seo JB, Lee HR, Yoo WJ, Chung CY, Choi IH. Biologic characteristics of fibrous hamartoma from congenital pseudarthrosis of the tibia associated with neurofibromatosis type 1. J Bone Joint Surg Am. 2008;90:2735–44. doi: 10.2106/JBJS.H.00014. [DOI] [PubMed] [Google Scholar]
- 28.Lee DY, Cho TJ, Lee HR, Lee K, Moon HJ, Park MS, Yoo WJ, Chung CY, Choi IH. Disturbed osteoblastic differentiation of fibrous hamartoma cell from congenital pseudarthrosis of the tibia associated with neurofibromatosis type I. Clin Orthop Surg. 2011;3:230–7. doi: 10.4055/cios.2011.3.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Estwick SA, Chen S, Yu M, Ming W, Nebesio TD, Li Y, Yuan J, Kapur R, Ingram D, Yoder MC, Yang FC. Neurofibromin plays a critical role in modulating osteoblast differentiation of mesenchymal stem/progenitor cells. Hum Mol Genet. 2006;15:2837–45. doi: 10.1093/hmg/ddl208. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, He Y, Sharma R, Xing W, Estwick SA, Wu X, Rhodes SD, Xu M, Yang FC. Hyperactive RAS/PI3-K/MAPK Signaling Cascade in Migration and Adhesion of Nf1 Haploinsufficient Mesenchymal Stem/Progenitor Cells. Int J Mol Sci. 2015;16:12345–59. doi: 10.3390/ijms160612345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes SD, Wu X, He Y, Chen S, Yang H, Staser KW, Wang J, Zhang P, Jiang C, Yokota H, Dong R, Peng X, Yang X, Murthy S, Azhar M, Mohammad KS, Xu M, Guise TA, Yang FC. Hyperactive transforming growth factor-β1 signaling potentiates skeletal defects in a neurofibromatosis type 1 mouse model. J Bone Miner Res. 2013;28:2476–89. doi: 10.1002/jbmr.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riese DJ, Cullum RL. Epiregulin: roles in normal physiology and cancer. Semin Cell Dev Biol. 2014;28:49–56. doi: 10.1016/j.semcdb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirakata Y, Komurasaki T, Toyoda H, Hanakawa Y, Yamasaki K, Tokumaru S, Sayama K, Hashimoto K. Epiregulin, a novel member of the epidermal growth factor family, is an autocrine growth factor in normal human keratinocytes. J Biol Chem. 2000;275:5748–53. doi: 10.1074/jbc.275.8.5748. [DOI] [PubMed] [Google Scholar]
- 34.Toyoda H, Komurasaki T, Uchida D, Takayama Y, Isobe T, Okuyama T, Hanada K. Epiregulin. A novel epidermal growth factor with mitogenic activity for rat primary hepatocytes. J Biol Chem. 1995;270:7495–500. doi: 10.1074/jbc.270.13.7495. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Shimizu E, Zhang X, Partridge NC, Qin L. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J Cell Biochem. 2011;112:1749–60. doi: 10.1002/jcb.23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chien HH, Lin WL, Cho MI. Down-regulation of osteoblastic cell differentiation by epidermal growth factor receptor. Calcif Tissue Int. 2000;67:141–50. doi: 10.1007/s00223001128. [DOI] [PubMed] [Google Scholar]
- 37.Qin L, Tamasi J, Raggatt L, Li X, Feyen JHM, Lee DC, Dicicco-Bloom E, Partridge NC. Amphiregulin is a novel growth factor involved in normal bone development and in the cellular response to parathyroid hormone stimulation. J Biol Chem. 2005;280:3974–81. doi: 10.1074/jbc.M409807200. [DOI] [PubMed] [Google Scholar]
- 38.Yu S, Geng Q, Ma J, Sun F, Yu Y, Pan Q, Hong A. Heparin-binding EGF-like growth factor and miR-1192 exert opposite effect on Runx2-induced osteogenic differentiation. Cell Death Dis. 2013;4:e868. doi: 10.1038/cddis.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krampera M, Pasini A, Rigo A, Scupoli MT, Tecchio C, Malpeli G, Scarpa A, Dazzi F, Pizzolo G, Vinante F. HB-EGF/HER-1 signaling in bone marrow mesenchymal stem cells: inducing cell expansion and reversibly preventing multilineage differentiation. Blood. 2005;106:59–66. doi: 10.1182/blood-2004-09-3645. [DOI] [PubMed] [Google Scholar]
- 40.Antosz ME, Bellows CG, Aubin JE. Biphasic effects of epidermal growth factor on bone nodule formation by isolated rat calvaria cells in vitro. J Bone Miner Res. 1987;2:385–93. doi: 10.1002/jbmr.5650020505. [DOI] [PubMed] [Google Scholar]
- 41.Nicolas V, Nefussi JR, Collin P, Forest N. Effects of acidic fibroblast growth factor and epidermal growth factor on subconfluent fetal rat calvaria cell cultures: DNA synthesis and alkaline phosphatase activity. Bone Miner. 1990;8:145–56. doi: 10.1016/0169-6009(90)90117-x. [DOI] [PubMed] [Google Scholar]
- 42.Genetos DC, Rao RR, Vidal MA. Betacellulin inhibits osteogenic differentiation and stimulates proliferation through HIF-1alpha. Cell Tissue Res. 2010;340:81–9. doi: 10.1007/s00441-010-0929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura T, Toita H, Yoshimoto A, Nishimura D, Takagi T, Ogawa T, Takeya T, Ishida-Kitagawa N. Potential Involvement of Twist2 and Erk in the Regulation of Osteoblastogenesis by HB-EGF-EGFR Signaling. Cell Struct Funct. 2010;35:53–61. doi: 10.1247/csf.10001. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Nyman JS, Moss HE, Gutierrez G, Mundy GR, Yang X, Elefteriou F. Local low-dose lovastatin delivery improves the bone-healing defect caused by Nf1 loss of function in osteoblasts. J Bone Miner Res. 2010;25:1658–67. doi: 10.1002/jbmr.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghadakzadeh S, Kannu P, Whetstone H, Howard A, Alman BA. β-Catenin modulation in neurofibromatosis type 1 bone repair: therapeutic implications. FASEB J. 2016 doi: 10.1096/fj.201500190RR. [DOI] [PubMed] [Google Scholar]
- 46.Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15:960–6. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 47.Hunter T, Cooper JA. Epidermal growth factor induces rapid tyrosine phosphorylation of proteins in A431 human tumor cells. Cell. 1981;24:741–52. doi: 10.1016/0092-8674(81)90100-8. [DOI] [PubMed] [Google Scholar]
- 48.Fry DW, Kraker AJ, McMichael A, Ambroso LA, Nelson JM, Leopold WR, Connors RW, Bridges AJ. A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science. 1994;265:1093–5. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 49.Nam HJ, Kim HP, Yoon YK, Hur HS, Song SH, Kim MS, Lee GS, Han SW, Im SA, Kim TY, Oh DY, Bang YJ. Antitumor activity of HM781-36B, an irreversible Pan-HER inhibitor, alone or in combination with cytotoxic chemotherapeutic agents in gastric cancer. Cancer Lett. 2011;302:155–165. doi: 10.1016/j.canlet.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Zeng F, Forrester SJ, Eguchi S, Zhang MZ, Harris RC. Expression and Function of the Epidermal Growth Factor Receptor in Physiology and Disease. Physiol Rev. 2016;96:1025–1069. doi: 10.1152/physrev.00030.2015. [DOI] [PubMed] [Google Scholar]
- 51.Janssens K, Vanhoenacker F, Bonduelle M, Verbruggen L, Van Maldergem L, Ralston S, Guañabens N, Migone N, Wientroub S, Divizia MT, Bergmann C, Bennett C, Simsek S, Melançon S, Cundy T, Van Hul W. Camurati-Engelmann disease: review of the clinical, radiological, and molecular data of 24 families and implications for diagnosis and treatment. J Med Genet. 2005;43:1–11. doi: 10.1136/jmg.2005.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.WILNER HI, FINBY N. SKELETAL MANIFESTATIONS IN THE MARFAN SYNDROME. JAMA. 1964;187:490–5. doi: 10.1001/jama.1964.03060200022003. [DOI] [PubMed] [Google Scholar]
- 53.Kirmani S, Tebben PJ, Lteif AN, Gordon D, Clarke BL, Hefferan TE, Yaszemski MJ, McGrann PS, Lindor NM, Ellison JW. Germline TGF-? receptor mutations and skeletal fragility: A report on two patients with Loeys-Dietz syndrome. Am J Med Genet Part A. 2010;152A:1016–1019. doi: 10.1002/ajmg.a.33356. [DOI] [PubMed] [Google Scholar]
- 54.Ottmann OG, Pelus LM. Differential proliferative effects of transforming growth factor-beta on human hematopoietic progenitor cells. J Immunol. 1988;140 [PubMed] [Google Scholar]
- 55.Fine A, Goldstein RH. The effect of transforming growth factor-beta on cell proliferation and collagen formation by lung fibroblasts. J Biol Chem. 1987;262:3897–902. [PubMed] [Google Scholar]
- 56.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–72. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lian N, Lin T, Liu W, Wang W, Li L, Sun S, Nyman JS, Yang X. Transforming growth factor β suppresses osteoblast differentiation via the vimentin activating transcription factor 4 (ATF4) axis. J Biol Chem. 2012;287:35975–84. doi: 10.1074/jbc.M112.372458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samarakoon R, Dobberfuhl AD, Cooley C, Overstreet JM, Patel S, Goldschmeding R, Meldrum KK, Higgins PJ. Induction of renal fibrotic genes by TGF-β1 requires EGFR activation, p53 and reactive oxygen species. Cell Signal. 2013;25:2198–209. doi: 10.1016/j.cellsig.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Kamiya N, Yamaguchi R, Aruwajoye O, Kim AJ, Kuroyanagi G, Phipps M, Adapala NS, Feng JQ, Kim HK. Targeted Disruption of NF1 in Osteocytes Increases FGF23 and Osteoid With Osteomalacia-like Bone Phenotype. J Bone Miner Res. 2017 doi: 10.1002/jbmr.3155. [DOI] [PubMed] [Google Scholar]
- 61.Baht GS, Nadesan P, Silkstone D, Alman BA. Pharmacologically targeting beta-catenin for NF1 associated deficiencies in fracture repair. Bone. 2017;98:31–36. doi: 10.1016/j.bone.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–76. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulcrone PL, Campbell JP, Clément-Demange L, Anbinder AL, Merkel AR, Brekken RA, Sterling JA, Elefteriou F. Skeletal Colonization by Breast Cancer Cells Is Stimulated by an Osteoblast and β2AR-Dependent Neo-angiogenic Switch. J Bone Miner Res. 2017 doi: 10.1002/jbmr.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fouletier-Dilling CM, Bosch P, Davis AR, Shafer JA, Stice SL, Gugala Z, Gannon FH, Olmsted-Davis EA. Novel compound enables high-level adenovirus transduction in the absence of an adenovirus-specific receptor. Hum Gene Ther. 2005;16:1287–97. doi: 10.1089/hum.2005.16.1287. [DOI] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.