Abstract
The bacterial pathogen Pseudomonas aeruginosa undergoes adaptation and selection over the course of chronic respiratory tract infections which results in repeatedly-observed phenotypic changes that are proposed to enable its persistence. Two of the clinically significant P. aeruginosa phenotypic changes are loss of flagellar motility and modifications to LPS structure, including loss of O-antigen expression. The effect of loss of O-antigen, frequently described as conversion from smooth to rough LPS, and the combined effect of loss of motility and O-antigen on phagocytic susceptibility by immune cells remain unknown. To address this, we generated genetic deletion mutants of waaL, which encodes the O-antigen ligase responsible for linking O-antigen to lipid A-core oligosaccharide, in both motile and non-motile P. aeruginosa strains. With the use of these bacterial strains we provide the first demonstration that, despite a progressive selection for P. aeruginosa with rough LPS during chronic pulmonary infections, loss of the LPS O-antigen does not confer phagocytic resistance in vitro. However, use of the waaLmotABmotCD mutant revealed that loss of motility confers resistance to phagocytosis regardless of the smooth or rough LPS phenotype. These findings reveal how the O-antigen of P. aeruginosa can influence bacterial clearance during infection and expand our current knowledge about the impact of bacterial phenotypic changes during chronic infection.
Keywords: Pseudomonas aeruginosa, phagocytosis, waaL, O-antigen, O-antigen ligase
1. INTRODUCTION
Pseudomonas aeruginosa is a motile Gram-negative bacterial pathogen that causes opportunistic acute and chronic infections in humans [1, 2]. In particular, this pathogen is responsible for high incidences (>80%) of chronic pulmonary infections and morbidity, in adult patients suffering from cystic fibrosis (CF) [1–3]. Chronic P. aeruginosa infections are highly tolerant to standard-of-care clinical antibiotic treatments and are difficult to eradicate [1, 2, 4], highlighting the importance of studying host-pathogen interactions and bacterial adaptations during chronic infections.
Longitudinal studies of P. aeruginosa isolates from CF patients with chronic airway infections have revealed characteristic bacterial phenotypic changes that suggest adaptation and selection of the pathogen within the CF lung [5]. Some of these include reduced expression of virulence factors, a switch from non-mucoid to mucoid colony morphology and an increase in expression of efflux pumps [5]. Other important temporal changes of P. aeruginosa isolated from the CF lungs include the loss of flagellar motility due to downregulation or absent flagellar gene expression [6, 7], growth of bacteria in microcolonies or biofilms, and modifications of the lipopolysaccharide (LPS) structure [4, 6, 8–10]. P. aeruginosa LPS is an integral component of the bacterial outer membrane, has been established as a major virulence factor in P. aeruginosa, and provides the bacteria with resistance to host defense mechanisms [9, 11]. LPS modifications have been reported as a conserved theme in infections and are apparently important in the adaptation to P. aeruginosa in chronic infections [9, 12, 13]. Importantly, P. aeruginosa isolates from chronic infections have rough colony phenotypes with LPS that has short, or no O side chains, rendering the bacteria non-typable and reducing their immunogenicity [14, 15]. Broadly, these changes have been attributed to prolonged inflammation, increased resistance to antibiotic-mediated clearance, and immune system evasion [9, 16]. Innate immunity in the host, specifically through the functions of professional phagocytic cells, is critical for effective control and clearance of P. aeruginosa. This is supported by the observations that humans and animal hosts that are lacking phagocytic cells (neutrophils and/or macrophages) are highly susceptible to P. aeruginosa infection [17–19].
Although the longitudinal transition to bacteria that lack flagellar motility and/or O-antigen is well established, whether these adaptations confer a selective advantage for phagocytic resistance is incompletely understood. One key insight was the previous demonstration that loss of flagellar motility is a crucial factor that dramatically increases P. aeruginosa resistance to phagocytosis (by ~100-fold) by professional phagocytes of the hosts [20, 21]. Another relevant insight is that LPS modifications that reduce its immunostimulatory potential are reported to be prevalent in the adaptation of P. aeruginosa to chronic infection [12]. LPS is composed of three distinct regions: lipid A, core oligosaccharide, and O-antigen polysaccharide, where common polysaccharide (CPA, homopolymer) and O-specific antigen (OSA, heteropolymer) are the two glycoforms of P. aeruginosa O antigens [11]. Importantly, during LPS biogenesis, CPA and OSA O antigens are attached onto lipid A-core oligosaccharides in the periplasm by the O-antigen ligase encoded by waaL [11, 22]. As a result, waaL deletion mutants produce rough LPS lacking O antigen and accumulate polymerized O antigen-linked undecaprenol-phosphate (Und-P) on the periplasmic surface of the cytoplasmic membrane [9, 11, 22]. Genomic analyses of sequential P. aeruginosa isolates recovered from CF patients over time reveals modifications in genes involved in biosynthesis of both CPA and OSA O antigens [13, 23, 24]. Interestingly, O-antigen ligase encoded by waaL was reported to be one of the few hotspots of gene polymorphisms in an analysis of whole-genome sequence data from P. aeruginosa clinical sputum isolates of 32 patients [25]. Many roles have been attributed to O-antigen in the literature; however, the effect of loss of O-antigen with regard to phagocytic susceptibility has never been examined.
In this study, we explored how two of the established temporal trends that occur during chronic infection, loss of motility and O antigen, intersect to alter the host phagocytic response, both individually and in combination. We have previously described the deletion mutant of motABmotCD, which encode partially redundant stator complexes required for flagellar rotation and swimming motility, but not flagellar assembly [20, 21, 26]. Of note, all of the aforementioned phagocytic studies using non-motile P. aeruginosa were performed using bacteria producing wild-type or smooth LPS with O antigen being expressed. Here, we have generated isogenic ΔwaaL deletion mutant and a double phenotypic mutant of waaLmotABmotCD in two independent strains of P. aeruginosa. Quantitative phagocytosis assays were then employed to examine the effect of deletion of waaL on susceptibility to phagocytosis in swimming and non-swimming P. aeruginosa strains.
2. MATERIALS & METHODS
2.1. Bacteria
P. aeruginosa variants on the PA14 and PAK background (wild-type (wt) and motABmotCD) have been previously described and published [20, 21, 26, 27]. Bacteria were cultured overnight at 37°C and subsequently subcultured as indicated for 2 h in lysogeny broth (also commonly called Luria Bertani broth, LB).
2.2. Cells
Bone marrow-derived dendritic cells (BMDCs) were cultured from C57BL/6 WT mice (Charles River Laboratories) as previously described [28]. Six-and seven-day-old BMDCs were used for these studies. Blood was collected from volunteers that include healthy individuals and patients with CF by venipuncture into heparinized tubes. Donor blood samples were procured from three CF patients with the following cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations; R1162X/W1282G (1 patient) and ΔF508/ΔF508 (2 patients). Neutrophils were enriched with a Ficoll-Paque PLUS (GE Healthcare Biosciences) discontinuous gradient followed by dextran sedimentation as previously described, with minor modifications [29]. After Ficoll separation, neutrophils were isolated from the blood pellet using 5% dextran for 75 min and erythrocytes lysed using BD PharmLyse RBC lysing buffer (BD Biosciences).
2.3. Ethics Statement
The Dartmouth Committee for the Protection of Human Subjects (CPHS) approved procurement and use of human cells in this study. All samples were de-identified and obtained with informed consent. This study was conducted in strict compliance with the Guide for the Care and Use of Laboratory Animals of the National Research Council and was approved by the Dartmouth Institutional Animal Care and Use Committee (IACUC).
2.4. Mutagenesis of P. aeruginosa waaL and PCR diagnosis
The waaL gene was deleted from P. aeruginosa PA14 and PAK wt and motABmotCD strains using homologous recombination with flanking sequences on the non-replicative pMQ30 vector [30]. Briefly, upstream flanking region was amplified using waaL-GOI-F-HindIII (5′- TCACTaagcttATTCCGATGTAGGGCTTCAC-3′) and waaL-SOEGOI-R-EcoRI (5′- TGCttgaattcGCGAACATTCCTTGCTCAAT-3′) and the downstream flanking region was amplified using waaL-SOEGOI-F-EcoRI (5′- GTCCgaattcaaCCAAGGAGCACTGGTTTCTG-3′) and waaL-GOI-R-KpnI (5′- TCACTggtaccCCGTGATCATCGCTTCCTAT-3′), where engineered restriction sites are noted with lowercase bases. Both products were cut with EcoRI, KpnI, and HindIII and ligated into HindIII and KpnI cut pMQ30, which was transformed into DH5α and transformants selected for growth on gentamicin. S17λpir was transformed with the resultant plasmid and transformants selected on gentamicin. The resultant S17λpir strain was mixed with the target P. aeruginosa strains for conjugation at 30°C. Single-crossover integrants were selected on PIA (Pseudomonas Isolation Agar) with gentamicin, where the irgosan in PIA selects against the E. coli strain. Strains were grown without gentamicin to allow for survival of cells that underwent double-crossover and these were selected for by growth on LB with no NaCl and 5% sucrose at 30°C. Double crossover yielded two distinct colony sizes and both were screened for deletion of waaL by two PCR reactions. Amplification with waaL-check-f (5′- AGTGCGGCGATACAGTCC-3′) and waaL-check-R-wt (5′- GGATGTAGAAAAGCCGGTGA-3′) yields a 484-bp fragment in wt and no specific product in the mutant, while amplification with waaL-check-f and waaL-check-R-delta (5′- TGGCTACGTCATCGACATTC-3′) yields a 760-bp fragment in the deletion mutant and an 1846 bp fragment in wt. Amplification with the waaL-check-R-wt primer is inefficient in PAK strains due to 3 SNPs within the chosen primer sequence, which was designed with PA14 and PAO1 genomes. Using this PCR diagnostic check, the small colonies from the double crossover step were determined to be waaL deletion mutants (Fig 1a). These strains grow well on LB and appear to only have the small-colony phenotype on the LB with no NaCl and sucrose.
Figure 1. PCR and western immunoblotting analyses of P. aeruginosa strains.
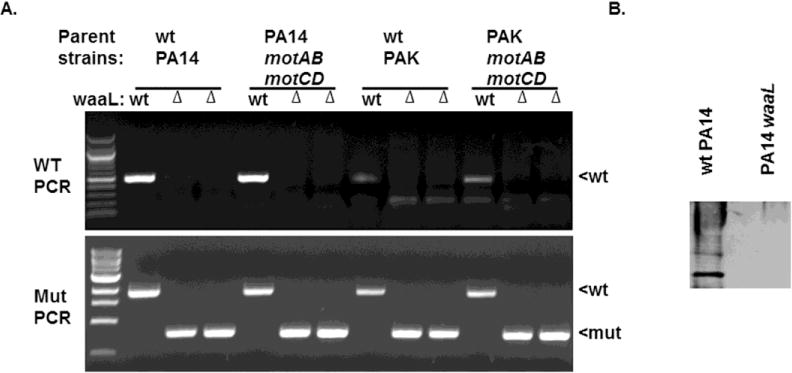
(A) Diagnostic PCR amplification of waaL deletion (waaL) in the wt and motABmotCD backgrounds of the PA14 and PAK strains of P. aeruginosa. Using the primers described in the materials and methods, deletion of the native locus was tested with a PCR reaction containing a reverse primer within the waaL coding sequence (top panel) and one using flanking sequences that shows the decrease in size from the deletion of the waaL coding sequence. (B) LPS from wt PA14 and PA14 waaL was prepared by proteinase-K digestion and probed with rabbit polyclonal antisera raised against the serotype O-19 strain.
2.5. Lipopolysaccharide isolation and immunoblot analysis
Before LPS isolation, 109 bacteria were pelleted from P. aeruginosa wt PA14 and PA14 waaL overnight cultures as measured by the optical density (OD 600nm). The LPS from these strains was prepared by proteinase-K digestion [31]. The bacterial LPS preparations were subjected to SDS-PAGE on 10% polyacrylamide gels. Western immunoblotting of the isolated LPS was performed using rabbit polyclonal antisera raised against the serotype O-19 strain (1:100) (generated by the Lam lab), followed by peroxidase affinipure goat anti-rabbit IgG (H+L) secondary antibody (Jackson ImmunoResearch) diluted to 1:5000 (incubated for 1h). The blot was developed using Pierce ECL substrate (Thermo Scientific) and visualized using autoradiography film (Genesee Scientific).
2.6. Bacterial swimming motility and biofilm formation assays
The phenotypic assay for swimming motility was performed as previously described in 0.3% LB agar plates [21, 32]. Images were collected at 48-h post inoculation to monitor the formation of bacterial halos to assess the relative swimming motilities of the bacterial strains. Biofilm formation assays were performed using 96-well round-bottom microtiter plates (CoStar 2797) as previously described utilizing M63 medium supplemented with 1 mM MgSO4 and arginine [33, 34]. Following incubation inside a humidified chamber at 37°C for 18 h, the supernatants were removed and the wells were stained with 0.1% (w/v) crystal violet solution and rinsed with distilled water [33]. This assay was conducted three times for each strain and the representative images are shown.
2.7. Gentamicin protection assay
Phagocytosis of live bacteria was performed as previously described [21, 28]. Briefly, subcultured bacteria of the indicated genotypes were washed, resuspended in serum-free Hanks balanced salt solution (HBSS) (Corning Cellgro, Manassas, VA), and their concentrations were determined at OD600nm. Where indicated, 10% (vol/vol) of complement-inactivated human serum from a healthy donor was included throughout the assay. A total of 2.5 × 105 cells (BMDCs or neutrophils) were incubated with the indicated bacterial genotype at an MOI of 10 (for BMDCs) or 25 (for human neutrophils) for 45 min at 37°C, followed by incubation with 100 μg/ml gentamicin for 20 min at 37°C. The cells were washed twice in HBSS and lysed with 500 μl 0.1% Triton X-100 solution in 1X Phosphate Buffered Saline (PBS) (HyClone Laboratories, Logan, UT). Lysates were plated on LB plates and incubated overnight at 37°C. CFUs were subsequently enumerated and are represented as percentage of mean wt or waaL bacteria phagocytosis as denoted.
2.8. Akt activation assay
Akt activation assays were performed as previously described [32]. Briefly, a total of 3 × 105 BMDCs were incubated in serum-free HBSS for 90 min at 37°C. The cells were then incubated with the indicated subcultured bacterial strains at MOI = 10 for 45 min at 37°C, followed by blocking for Fc receptor with monoclonal antibody 2.4G2. BMDCs were subsequently fixed in 300 μl of 4% paraformaldehyde in PBS for 10 min at 37°C, washed in cold PBS, and permeabilized in 500 μl of 100% cold methanol for 10 min. After washing with cold PBS to remove the methanol, the cells were probed with phospho-Akt antibody in cold PBS containing 3% BSA, 1mM PMSF (Sigma), and 1X MS-SAFE protease and phosphatase inhibitor (Sigma) as described by manufacturers. The antibody used was mouse anti-phospho-Akt (Ser473), clone 6F5 IgGK (Millipore), which was labeled with Zenon One Alexfluor-647 mouse IgG (Molecular Probes) as described in the manufacturer’s instructions. Akt activation was quantified using FACS to acquire fluorescence.
2.9. In vitro inflammation assay
A total of 2.5 × 105 BMDCs per well in a 24-well plate were infected with subcultured bacteria (MOI = 1) to a final volume of 400 μl Roswell Park Memorial Institute 1640 (RPMI-1640) containing 10% Fetal Bovine Serum (FBS) (HyClone Laboratories, Logan, UT). Bacteria were incubated with the cells for 3 h at 37°C and 5%CO2. Cell-free supernatants were collected 3-h post infection (pi) and production of Interleukin-1β (IL-1β) and IL-6 cytokines was analyzed by ELISA. The DuoSet enzyme-linked immunosorbent assay (ELISA) kits for mouse IL-1β and IL-6 were acquired from R&D Systems (Minneapolis, MN).
2.10. LDH assay
Human neutrophils (healthy and from CF patients) were infected with subcultured bacteria at the indicated MOI in a total volume of 300 μl HBSS with 2% FBS for 2 h at 37°C and 5%CO2. The infections were carried out in 24-well plates using 2.5 × 105 neutrophils per well. The CytoTox kit (Promega) was used according to the manufacturer’s protocol to measure lactate dehydrogenase (LDH) release from cell-free supernatants, representing cytotoxicity.
2.11. Statistical analyses
Means ± standard deviations (SD) derived from multiple independent experiments with technical replicates are shown for each graph. Sample sizes for each experiment are noted in the figure legends. As indicated, unpaired Student’s t test with Welch’s correction or one-way ANOVA with Tukey’s post hoc analyses were performed using Prism 5.0a to determine statistical significance of the data. Statistical significance is represented in figures by asterisks.
3. RESULTS
3.1. Creation and validation of P. aeruginosa waaL mutants
To assess the functional roles of the waaL gene in P. aeruginosa, deletion mutants in the swimming-competent PA14 and PAK wildtype (wt) strains were generated using homologous recombination as described in the Methods section. Isogenic ΔwaaL (deletion mutants) were also generated in the PA14 motABmotCD and PAK motABmotCD backgrounds to assess the role of waaL in non-motile strains. The presence of the mutant gene in PA14 and PAK was verified by PCR screening of the double crossover products (Fig. 1A).
Validation of the ΔwaaL phenotype was confirmed by loss of O-antigen on the LPS of the mutant strains. LPS from wt PA14 and PA14 waaL was analyzed by Western blotting using polyclonal antisera raised against the serotype O-19 strain. As expected, we observed O antigen-positive LPS bands in wt PA14, while PA14 waaL was defective in the production of CPA and OSA LPS (Fig. 1B) [22].
3.2. ΔwaaL mutation in P. aeruginosa did not abrogate swimming motility nor ability of the bacteria to form biofilms
To determine the relationship between motility and O-antigen expression with regard to phagocytic susceptibility, we first sought to determine the effect of the ΔwaaL mutation on bacterial swimming motility. To test this, the strains were inoculated by stabbing puncture into 0.3% LB agar plates to monitor the zones surrounding the point of inoculation as an indication of swimming motility. In accordance with previous results, there was a complete lack of swimming motility in the PA14 and PAK motABmotCD stator-deficient mutants, compared to the swimming-competent wt strains (Fig. 2A). Importantly, PA14 waaL and PAK waaL exhibited swimming motility, while PA14 waaLmotABmotCD and PAK waaLmotABmotCD were completely non-motile. To functionally confirm the motility assay, the strains were evaluated for the capability to form biofilms: previous research has demonstrated that flagella-mediated motility in P. aeruginosa is required to initiate biofilm formation [35, 36]. The biofilm phenotypes corroborate the motility phenotypes observed with the mutants. PA14ΔwaaL and PAKΔwaaL were able to form biofilms, while PA14 waaLmotABmotCD and PAK waaLmotABmotCD were not (Fig. 2B). Notably, these results were reiterated in multiple strains (PA14 and PAK) suggesting that they may be broadly applicable in P. aeruginosa. Overall, these data indicate that mutants deficient for O-antigen expression are capable of swimming motility and biofilm formation.
Figure 2. P. aeruginosa deficient in O-antigen ligase expression exhibit swimming motility and biofilm formation.
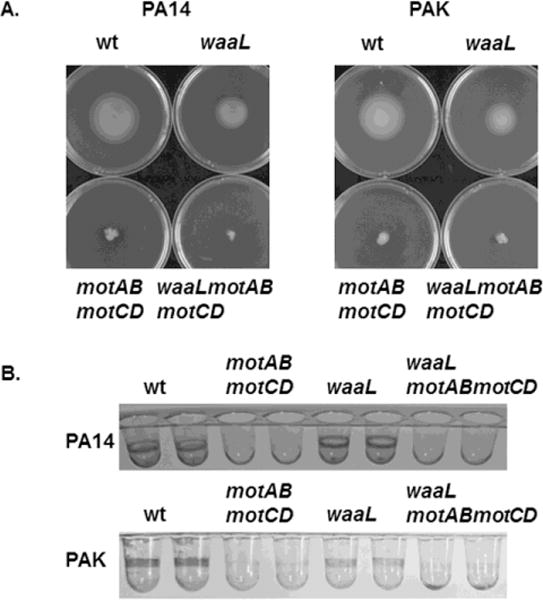
(A) wt and mutant P. aeruginosa strains were assayed for swimming motility in LB media containing 0.3% agar. wt PA14, wt PAK and single mutant waaL-deficient strains are fully competent at swimming motility, while the mutants deficient in the flagellar stator proteins (motABmotCD) and the double phenotypic mutants waaLmotABmotCD are completely impaired at swimming motility. The images shown are representative swimming plates of the indicated strains. (B) The P. aeruginosa strains described in (A) were assessed for biofilm formation following overnight culture in supplemented M63 minimal medium. Staining with 0.1% crystal violet was used to view the biofilms. Images of representative wells of a 96-well biofilm assay for each strain are shown.
3.3. Phagocytosis of P. aeruginosa by BMDCs is independent of O-antigen ligase expression in the bacteria
To evaluate the effect of loss of O-antigen ligase expression on phagocytic uptake of P. aeruginosa, we employed a quantitative gentamicin protection phagocytic assay with murine BMDCs. Interestingly, PA14 waaL and PAK waaL were phagocytosed by BMDCs at levels comparable or slightly higher than those for the wt strains. In comparison, and as a control, both motABmotCD strains exhibited a dramatic reduction in their phagocytic uptake compared to the respective parental strain (Fig. 3A and B) [20, 21, 27]. In order to account for the possibility that differences in intracellular killing of bacteria could be contributing to our phagocytosis results, we incubated the indicated bacteria with murine BMDCs for a shorter amount of time (20 minutes instead of 45 minutes) before addition of gentamicin. Indeed, similar to the previous results (Figs. 3A and B), the shorter incubation led to equivalent or greater phagocytosis of PA14 waaL and PAK waaL by BMDCs, compared to their wt strains (Fig. 3C and D). These data support that differential intracellular killing of bacteria is not responsible for the observed phagocytic phenotypes.
Figure 3. Phagocytic susceptibility of O-antigen -deficient P. aeruginosa is similar to that of bacteria with O-antigen.
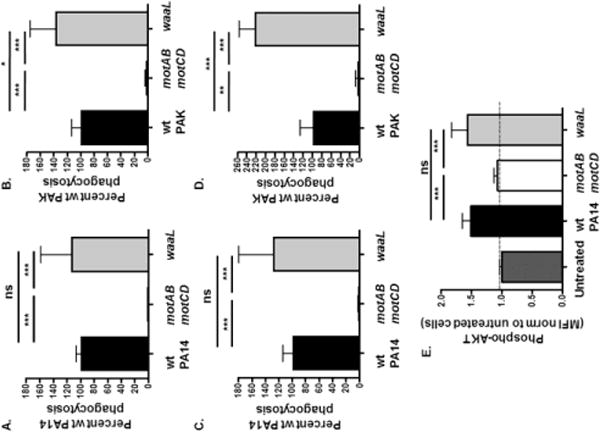
Murine bone marrow-derived dendritic cells (BMDCs) from C57BL/6 mice were assayed for relative in vitro phagocytosis of (A and C) wt PA14, motABmotCD, or waaL and (B and D) wt PAK, motABmotCD, or waaL bacteria by gentamicin protection assay incubated for 45 minutes (A and B) or 20 minutes (C and D) at MOI =10. Phagocytic uptake levels were normalized as percentages of respective mean wt phagocytosis. (E) Murine BMDCs were coincubated with wt PA14, motABmotCD, or waaL bacteria, and then fixed and probed for phospho-Akt. Akt activation (phospho-Akt) was quantified by FACS analysis and mean fluorescence intensity (MFI) was normalized to untreated cells (= 1 on Y-axis). All data are analyzed using one-way ANOVA with Tukey’s post hoc analyses and are representative of at least two independent biological experiments (n ≥ 4). ***, p ≤ 0.0005; *, p ≤ 0.05; ns, not significant.
Based on the observed phagocytosis phenotype, we assessed the cellular Akt activation upon incubation of BMDCs with PA14ΔwaaL compared to wt PA14. Previous data demonstrated that the PI3K/Akt pathway, which is an important mammalian intracellular signaling pathway for phagocytosis of many bacteria by host cells, is activated in response to motile bacteria but not by non-motile bacteria [32, 37]. Relative Akt activation was quantitatively measured by FACS analysis following intracellular staining for phosphorylated Akt (phospho-Akt). The exposure of BMDCs with PA14 waaL elicited phosphorylation of Akt that was comparable to that of wt PA14. In comparison, and as a negative control, bacteria that lack flagellar motility (motABmotCD) did not elicit Akt activation, such that cellular phospho-Akt levels remained similar to untreated cells (Fig. 3E). Together, these data provided the evidence that leads to two initial insights. First, that phagocytosis of P. aeruginosa and the associated signaling are relatively unaffected by loss of O-antigen ligase expression and that loss of O-antigen is unlikely to correspond to selection for phagocytic resistance. Second, these data support that the P. aeruginosa O-antigen is not the predominant bacterial ligand recognized by the host cell phagocytic receptors.
3.4. P. aeruginosa defective in swimming motility are resistant to phagocytosis by BMDCs independent of rough LPS phenotype
Previous experiments have demonstrated that loss of bacterial motility enables phagocytic evasion by P. aeruginosa with the smooth LPS phenotype, however it is unknown whether O-antigen of LPS affects the phagocytosis of non-motile P. aeruginosa. We hypothesized that loss of motility would enable phagocytic evasion regardless of change in the LPS structure from smooth to rough. To address this, we tested the waaLmotABmotCD double phenotypic mutants in our BMDC phagocytic assay and compared the uptake to ΔwaaL single mutants in the PA14 and PAK backgrounds. When compared to the ΔwaaL mutant in wt PA14 and PAK, ΔwaaL in non-motile mutant backgrounds exhibited phagocytic resistance (Fig. 4A and B), similar to smooth non-motile bacteria as demonstrated in Fig 3. Moreover, analogous to results from Fig. 3, uptake of PA14 and PAK waaLmotABmotCD compared to the waaL mutants after 20 minutes incubation mirrored the data obtained after a a 45 minute co-incubation (Fig. 4C and D). This suggests that the phagocytic resistance exhibited by the non-motile bacteria with rough LPS is also independent of differential intracellular bacterial killing kinetics. To validate and extend these results, we assessed cellular Akt activation following infection of BMDCs with PA14 waaLmotABmotCD. The waaLmotABmotCD did not stimulate phosphorylation of Akt above the levels of untreated cells (Fig. 4E). These results suggest that loss of flagellar motility enables resistance of bacteria to uptake by phagocytic cells, independent of the presence or absence of O-antigen.
Figure 4. Non-motile P. aeruginosa with rough LPS architecture is resistant to phagocytosis by murine phagocytes.
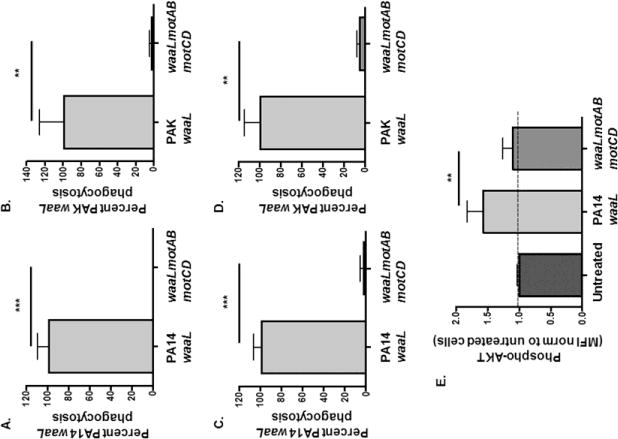
Murine BMDCs were assayed for relative in vitro phagocytosis of (A and C) PA14 waaL or waaLmotABmotCD and (B and D) PAK waaL or waaLmotABmotCD bacteria by gentamicin protection assay incubated for 45 minutes (A and B) or 20 minutes (C and D) at MOI=10. Phagocytic uptake levels were normalized as percentages of respective mean waaL phagocytosis. (E) Murine BMDCs were coincubated with PA14 waaL or waaLmotABmotCD bacteria, and then fixed and probed for phospho-Akt. Akt activation (p-Akt) was quantified by FACS analysis and MFI was normalized to untreated cells (= 1 on Y-axis). Data for panels A-D are analyzed using the unpaired t-test with Welch’s correction, while data for panel E are analyzed using one-way ANOVA with Tukey’s post hoc analyses. All data are representative of at least two independent biological experiments (n ≥ 4). ***, p ≤ 0.0005; **, p ≤ 0.005.
Although the precise contribution of opsonins during P. aeruginosa chronic lung infections is unclear, we wanted to examine whether the presence of opsonin-containing human serum in the assay media would impact relative phagocytosis of the non-motile and O-antigen deficient mutants compared to wt strains. Therefore, we performed phagocytosis assays in media containing 10% human serum and compared them to media without serum. Results using serum-containing assay media recapitulated those obtained in serum-free conditions, such that waaL phagocytosis is comparable to wt, while phagocytosis of waaLmotABmotCD resembles motABmotCD (Fig. 5). This suggests that serum does not affect our relative phagocytic phenotypes.
Figure 5. Phagocytosis phenotypes of waaL mutants are recapitulated in the presence of serum.
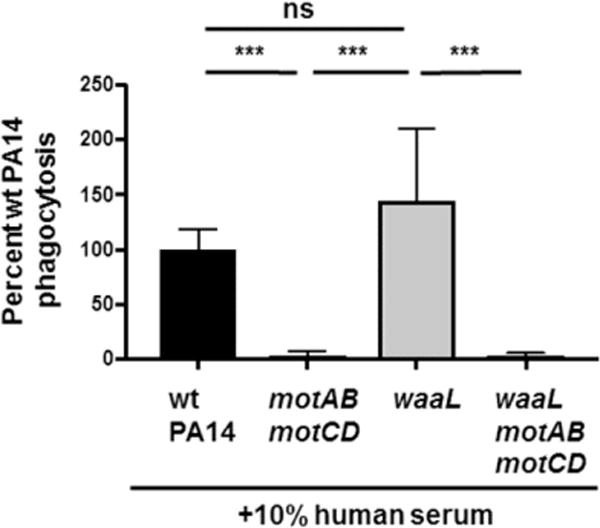
Murine BMDCs were assayed for relative in vitro phagocytosis of wt PA14, motABmotCD, waaL, or waaLmotABmotCD in the absence or presence of 10% human serum by gentamicin protection assay at MOI=10. Phagocytic uptake levels were normalized as percentages of mean wt phagocytosis. Data are accumulated from three independent biological experiments and analyzed using one-way ANOVA with Tukey’s post hoc analyses (n = 6). ***, p ≤ 0.0005; ns, not significant.
3.5. P. aeruginosa that lack O-antigen ligase elicit similar levels of pro-inflammatory cytokine production by BMDCs
We previously reported that loss of flagellar motility results in decreased cell-surface interactions with macrophages and thereby reduces bacterial type 3 secretion system (T3SS)-dependent inflammasome activation and IL-1β production in response to P. aeruginosa infection [27]. However, while TLR4 and TLR5 are integral to inflammatory cytokine responses to P. aeruginosa infection, neither is necessary for direct phagocytic recognition and uptake of P. aeruginosa in in vitro assays [21]. Therefore, we sought to evaluate whether waaL was required for IL-1β and IL-6 production in response to P. aeruginosa infection, the former dependent upon bacterial contact with the host cell and the latter dependent upon TLR signaling. BMDCs exhibited comparable elicited IL-1β responses following infection with wt PA14 and PA14 waaL. As a control, BMDCs infected with the motABmotCD mutant produced lower quantities of IL-1β, consistent with previous observations (Fig. 6A) [27]. Similar to the results observed for IL-1β, equivalent levels of IL-6 were produced by BMDCs infected with wt PA14 and ΔwaaL. Interestingly, infection with PA14 motABmotCD also elicited similar IL-6 production, consistent with the previous observation that induction of pro-inflammatory TLR signaling is independent of bacterial binding and phagocytosis (Fig. 6B) [21]. These findings demonstrate that cellular inflammatory cytokine responses to P. aeruginosa infection do not require bacterial O-antigen ligase expression nor does loss of O-antigen result in evasion of an inflammatory response.
Figure 6. Infection of BMDCs with waaL-deficient bacteria elicit comparable levels of inflammatory cytokines to wt bacteria.
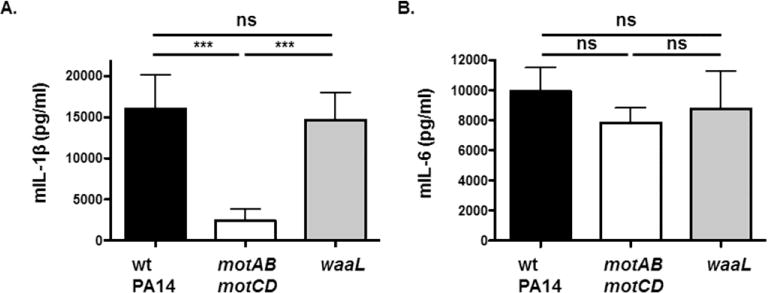
Murine BMDCs were infected with wt PA14, motABmotCD, waaL, or waaLmotABmotCD at a MOI of 1. Culture supernatants were collected 3h postinfection and analyzed by ELISA for (A) IL-1β and (B) IL-6 production. Data are accumulated from at least two independent biological experiments and analyzed using one-way ANOVA with Tukey’s post hoc analyses (n ≥ 4). ***, p ≤ 0.0005; ns, not significant.
3.6. O-antigen-deficient but motile P. aeruginosa exhibit phagocytic susceptibility to human blood-derived neutrophils
We extended our studies to human primary neutrophils in order to examine the phagocytic susceptibilities of the aforementioned bacterial strains and to validate the phagocytic phenotypes observed with murine BMDCs. Neutrophils play an essential role in clearing P. aeruginosa infections and neutropenic patients and mice are highly susceptible to those infections [17, 19]. Indeed, gentamicin protection assays employing purified human healthy neutrophils validated the mouse studies, such that motile rough LPS mutants (PA14ΔwaaL) were phagocytosed to a greater extent compared to wt PA14 in both serum-free and 10%human serum-containing assay media (Fig. 7A and B). On the other hand, non-motile rough LPS mutants (PA14 waaLmotABmotCD) were as resistant to phagocytosis by neutrophils as PA14 motABmotCD in serum-free as well as serum-containing media (Fig. 7A and B). Consistent with Fig. 5, addition of serum to the gentamicin assay media did not seem to alter the observed phagocytic phenotypes. Of specific interest, similar results were obtained with blood-derived neutrophils obtained from CF patients, although phagocytosis was not significantly higher for PA14 waaL compared to wt PA14 (Fig. 7C). Thus, the similar phagocytic outcomes comparing those observed with murine BMDC to those with human neutrophils indicate that this phenotype is consistent across multiple cell types and species and that it is likely to be relevant to interactions with human phagocytic cells.
Figure 7. Phagocytosis and cytotoxicity elicited by O-antigen ligase deficient P. aeruginosa in human neutrophils.
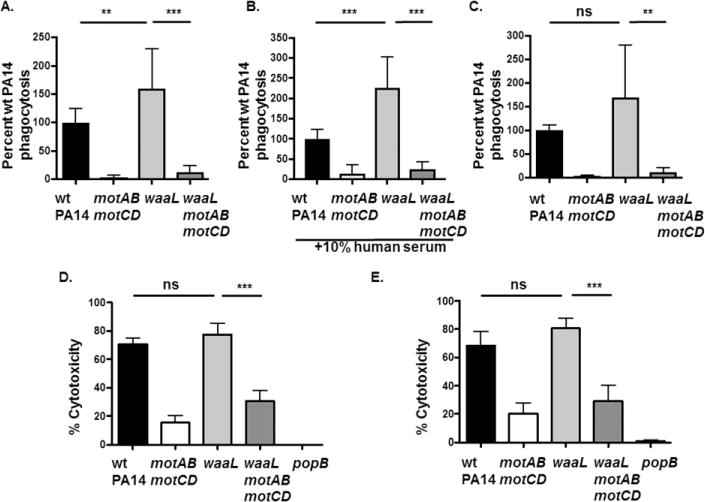
(A and B) Primary human blood neutrophils from healthy individuals or (C) from CF patients were assayed for relative in vitro phagocytosis of wt PA14, motABmotCD, waaL, or waaLmotABmotCD by gentamicin protection assay at MOI=25 in the (A) absence or (B) presence of 10% human serum. Phagocytic uptake levels were normalized as percentages of mean wt phagocytosis. (D) Cytotoxicity of neutrophils from healthy patients or (E) from CF patients following infection with wt PA14, motABmotCD, waaL, or waaLmotABmotCD (MOI=15) was assayed using the LDH assay. Release of LDH into the culture supernatants was measured at 2h post infection. All data are analyzed using one-way ANOVA with Tukey’s post hoc analyses and are representative of at least three independent biological experiments (n ≥ 5). ***, p ≤ 0.0005; **, p ≤ 0.005; ns, not significant.
Previous data have demonstrated that motility-deficient P. aeruginosa is less capable of inducing cell death of BMDCs in a process that is dependent upon bacterial interactions with the host cell and expression of the bacterial T3SS [27]. It is important to note that motABmotCD and wt bacteria have similar expression of T3SS-associated genes [20, 27]. However, the effect of O-antigen expression on bacterial-induced cytotoxicity is unknown. Therefore, we tested the effect of the ΔwaaL mutation, and therefore the loss of O-antigen, on human neutrophil viability following infection with P. aeruginosa. Human neutrophils from healthy (non-CF) individuals and CF patients were infected with PA14 and its mutants motABmotCD, waaL and waaLmotABmotCD at MOI = 15 and cytotoxicity was assessed by LDH release after 2 h. Robust, but comparable, cytotoxicity was observed in neutrophils infected with wt and waaL bacteria at the indicated MOIs, which contrasted with the lower cytotoxicity elicited by the non-motile mutants. The non-motile motABmotCD mutant triggered very low levels of cytotoxicity while cytotoxicity with waaLmotABmotCD was slightly higher (Fig. 7D and E). To confirm that cytotoxicity of neutrophils was dependent on bacterial T3SS activity, we also infected neutrophils with PA14 popB, which lacks a functional T3SS translocon. As expected, there was almost no cytotoxicity observed with the popB mutant (Fig. 7D and E). Overall, these results support that the interactions of O-antigen - deficient P. aeruginosa with host cells confers higher or similar phagocytic susceptibility and comparable induced cytotoxicity to that observed with O-antigen-expressing bacteria. However, loss of flagellar motility enables the bacteria, regardless of O-antigen expression, to evade phagocytosis.
4. DISCUSSION
Two of the established temporal trends observed from longitudinal samples of bacteria isolated from chronic pulmonary P. aeruginosa infection of patients with CF or COPD include loss of flagellar motility and alterations to the LPS structure [4, 6–8, 10]. We have previously determined that the loss of flagellar swimming motility, regardless of flagellar expression, is a critical factor that contributes to bacterial resistance to clearance through phagocytosis [20, 21, 27]. However, the contributions of other identified bacterial adaptations, with loss of LPS O-antigen being pertinent to these studies, to evasion of phagocytic recognition and clearance remain enigmatic. Therefore, based on the clinical observations that P. aeruginosa isolates acquire the rough LPS phenotype over the course of chronic infection [14], we have investigated whether LPS O-antigen loss confers an advantage to the bacteria in the context of phagocytic evasion of innate immune cells. Additionally, we have addressed independent and combined effects of losing flagellar swimming motility and O-antigen expression with regard to phagocytic resistance.
We employed a bacterial genetic approach to generate ΔwaaL constructs in both wild-type and non-swimming mutant backgrounds. Our results showed that the ΔwaaL mutants produce rough LPS lacking O-antigen, which is consistent with published literature [22]. Interestingly, deletion of the waaL gene in different bacterial species has been reported to have disparate effects on swimming motility [22, 38, 39]. For instance, a P. aeruginosa PAO1 waaL transposon-insertion mutant and a ΔwaaL mutant in an avian pathogenic Escherichia coli (APEC) were shown to exhibit partially-diminished swimming motility, while the ΔwaaL mutant in Proteus mirabilis had no impact on swimming motility in that species [22, 38, 39]. By generating independent ΔwaaL mutants in the PAK and PA14 strains of P. aeruginosa, we were able to show that these mutants only exhibited relatively minor decreases in flagellar motility as assessed by using swimming assays; however, we note that the assay could not discern whether this is due to alterations in flagellar rotation or, potentially, alterations to the exterior of the bacteria that diminish the ability of the bacteria to spread through the agar. However, consistent with the retention of flagellar motility, we showed that bacteria of ΔwaaL form robust biofilms. These data are supported by previous studies that showed that the P. aeruginosa PAO1 waaL strain forms thick biofilms [40] and waaL deletion in APEC actually increased the ability to form biofilms [38]. However, waaLmotABmotCD mutants are non-motile and are unable to form biofilms, which provides both a control for motility and an opportunity to assess whether there is an association between swimming motility and O-antigen phenotypes together. Our results indicate that flagella-mediated swimming motility and biofilm formation are largely independent of the presence or absence of O antigen on P. aeruginosa.
Importantly, our findings demonstrate that the level of phagocytosis of ΔwaaL mutant P. aeruginosa by dendritic cells and neutrophils is similar or slightly higher compared to phagocytosis of wt bacteria. Deletion of O-antigen ligase (and thus the presence of O-antigen, indirectly) in motile strains is dispensable for phagocytosis by neutrophils and dendritic cells, and may even provide a modest additional susceptibility. Thus, despite the clinical observation of selection for O antigen-deficiency in P. aeruginosa during chronic infection, the phenotype change to producing rough LPS did not confer evasion of phagocytosis by innate immune cells. Our results contrast with studies done using O antigen-deficient Burkholderia cenocepacia, which are more susceptible to phagocytic internalization [41]. However, it is noteworthy that Burkholderia has an intracellular lifecycle and is a non-obligate intracellular pathogen, while P. aeruginosa is largely regarded as an extracellular pathogen. Therefore, with regard to phagocytosis, loss of O-antigen may benefit the ability of Burkholderia to access an intracellular environment, while P. aeruginosa would not share similar benefits [41].
While the studies presented in this manuscript test one aspect of the host innate immune response to P. aeruginosa lacking O-antigen, it is reasonable to suggest that WaaL (O-antigen ligase) likely plays an important role in the regulation of other factors that contribute to P. aeruginosa virulence. For instance, it has been previously shown that P. aeruginosa O-antigen-deficient isolates were more sensitive to in vitro killing by serum complement and oxidative stress (H2O2) [14, 42]. Moreover, there are data to support that some adaptations associated with modifications of LPS of P. aeruginosa that favor persistence in the CF airways confers reduced virulence and pathogenicity, and these studies are corroborated in experiments that tested waaL mutants of several bacterial species within a variety of in vivo infection models [38, 42–45]. However, it is noteworthy that many of the reports that describe adaptations of P. aeruginosa resulting in modification in the LPS were focused on changes to the lipid-A portion rather than the O-antigen, and observed changes in lipid A likely reflect an adaptation or selection to evade inflammatory responses [9]. Importantly, we also demonstrate that P. aeruginosa strains that are defective in swimming motility are resistant to phagocytosis regardless of whether smooth LPS or rough LPS is produced on the cell surface of the bacteria. These data are the first test of the contribution of O-antigen to susceptibility to phagocytic activities in swimming competent and swimming-defective P. aeruginosa backgrounds. We conclude that loss of swimming motility confers phagocytic evasion regardless of the presence or absence of O-antigen on the bacteria, and in some cases, may even overcome additional phagocytic susceptibility conferred by loss of O-antigen.
In summary, there is a need to understand how these adaptive or selective changes to the bacteria affect immune evasion in the hopes of developing strategies to effectively eliminate the notoriously difficult to eradicate chronic infections. Using P. aeruginosa bacteria with genetic deletions of the waaL gene, we demonstrate that the loss of O-antigen (ligase) does not confer phagocytic evasion of the motile bacteria by murine and human phagocytic cells. Importantly, we also identify that the resistance to phagocytosis exhibited by non-swimming mutants is independent of LPS O-antigen composition. Overall, these findings provide key insights into how P. aeruginosa adaptations observed during chronic infections of the CF lung environment contribute to host phagocytic evasion.
Highlights.
New bacterial mutants enable combinatorial test of motility and O-antigen loss on phagocytosis.
Phagocytosis of P. aeruginosa is independent of presence of O-antigen.
Non-motile P. aeruginosa, regardless of LPS architecture, are resistant to phagocytosis.
Outcomes validated with primary human neutrophils.
Acknowledgments
We thank George O’Toole, Deborah Hogan, Alix Ashare, William Rigby, Harini Natarajan, Sladjana Skopelja-Gardner, Lynn Theprungsirikul, Haley Hazlett, and Amanda Nymon (Geisel School of Medicine at Dartmouth) for reagents and discussion. This work was facilitated by the Dartmouth Lung Biology Translational Research Core. This work was supported by grants from the National Institutes of Health (NIH) (P30 RR032136-01, P30 GM106394, R21 AI121820) and the Cystic Fibrosis Foundation Research Development Program (STANTO19R0 and STANTO11R0) (BB), and NIH RO1 Al103003 (MJW). Work performed in the lab of JSL is funded by an operating grant from the Canadian Institutes of Health Research (MOP-14687). JSL is a recipient of a Canada Research Chair award funded by the Canadian Foundation of Innovation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES. All authors: No conflicts.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser AR, Jain M, Bar-Meir M, McColley SA. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev. 2011;24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 5.Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzar MA, Thomassen MJ, Montie TC. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray TS, Egan M, Kazmierczak BI. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr. 2007;19:83–88. doi: 10.1097/MOP.0b013e3280123a5d. [DOI] [PubMed] [Google Scholar]
- 9.Maldonado RF, Sa-Correia I, Valvano MA. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev. 2016;40:480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B, Haagensen JA, Ciofu O, Andersen JB, Hoiby N, Molin S. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Clin Microbiol. 2005;43:5247–5255. doi: 10.1128/JCM.43.10.5247-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 12.Cigana C, Curcuru L, Leone MR, Ierano T, Lore NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML, Bragonzi A. Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One. 2009;4:e8439. doi: 10.1371/journal.pone.0008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tummler B. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol. 2011;13:1690–1704. doi: 10.1111/j.1462-2920.2011.02483.x. [DOI] [PubMed] [Google Scholar]
- 14.Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infection and Immunity. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King JD, Kocincova D, Westman EL, Lam JS. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 2009;15:261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- 16.Lore NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, Eberl L, Bragonzi A. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One. 2012;7:e35648. doi: 10.1371/journal.pone.0035648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh AY, Priebe GP, Ray C, Van Rooijen N, Pier GB. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun. 2009;77:5300–5310. doi: 10.1128/IAI.00501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurahashi K, Sawa T, Ota M, Kajikawa O, Hong K, Martin TR, Wiener-Kronish JP. Depletion of phagocytes in the reticuloendothelial system causes increased inflammation and mortality in rabbits with Pseudomonas aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol. 2009;296:L198–209. doi: 10.1152/ajplung.90472.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews T, Sullivan KE. Infections in patients with inherited defects in phagocytic function. Clin Microbiol Rev. 2003;16:597–621. doi: 10.1128/CMR.16.4.597-621.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovewell RR, Collins RM, Acker JL, O’Toole GA, Wargo MJ, Berwin B. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog. 2011;7:e1002253. doi: 10.1371/journal.ppat.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amiel E, Lovewell RR, O’Toole GA, Hogan DA, Berwin B. Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect Immun. 2010;78:2937–2945. doi: 10.1128/IAI.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abeyrathne PD, Daniels C, Poon KK, Matewish MJ, Lam JS. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J Bacteriol. 2005;187:3002–3012. doi: 10.1128/JB.187.9.3002-3012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren AE, Boulianne-Larsen CM, Chandler CB, Chiotti K, Kroll E, Miller SR, Taddei F, Sermet-Gaudelus I, Ferroni A, McInnerney K, Franklin MJ, Rosenzweig F. Genotypic and phenotypic variation in Pseudomonas aeruginosa reveals signatures of secondary infection and mutator activity in certain cystic fibrosis patients with chronic lung infections. Infect Immun. 2011;79:4802–4818. doi: 10.1128/IAI.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dettman JR, Rodrigue N, Aaron SD, Kassen R. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2013;110:21065–21070. doi: 10.1073/pnas.1307862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toutain CM, Zegans ME, O’Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patankar YR, Lovewell RR, Poynter ME, Jyot J, Kazmierczak BI, Berwin B. Flagellar motility is a key determinant of the magnitude of the inflammasome response to Pseudomonas aeruginosa. Infect Immun. 2013;81:2043–2052. doi: 10.1128/IAI.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amiel E, Alonso A, Uematsu S, Akira S, Poynter ME, Berwin B. Pivotal Advance: Toll-like receptor regulation of scavenger receptor-A-mediated phagocytosis. J Leukoc Biol. 2009;85:595–605. doi: 10.1189/jlb.1008631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhns DB, Long Priel DA, Chu J, Zarember KA. Isolation and Functional Analysis of Human Neutrophils. Curr Protoc Immunol. 2015;111:7 23 21–16. doi: 10.1002/0471142735.im0723s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalivoda EJ, Horzempa J, Stella NA, Sadaf A, Kowalski RP, Nau GJ, Shanks RM. New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol Biotechnol. 2011;48:7–14. doi: 10.1007/s12033-010-9342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. Journal of Bacteriology. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovewell RR, Hayes SM, O’Toole GA, Berwin B. Pseudomonas aeruginosa flagellar motility activates the phagocyte PI3K/Akt pathway to induce phagocytic engulfment. Am J Physiol Lung Cell Mol Physiol. 2014;306:L698–707. doi: 10.1152/ajplung.00319.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol, Chapter. 2005;1 doi: 10.1002/9780471729259.mc01b01s00. Unit 1B 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernier SP, Ha DG, Khan W, Merritt JH, O’Toole GA. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res Microbiol. 2011;162:680–688. doi: 10.1016/j.resmic.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 36.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 37.Lovewell RR, Patankar YR, Berwin B. Mechanisms of phagocytosis and host clearance of Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2014;306:L591–603. doi: 10.1152/ajplung.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y, Han X, Wang S, Meng Q, Zhang Y, Ding C, Yu S. The waaL gene is involved in lipopolysaccharide synthesis and plays a role on the bacterial pathogenesis of avian pathogenic Escherichia coli. Vet Microbiol. 2014;172:486–491. doi: 10.1016/j.vetmic.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 39.Morgenstein RM, Rather PN. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol. 2012;194:669–676. doi: 10.1128/JB.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol. 2012;14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saldias MS, Ortega X, Valvano MA. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J Med Microbiol. 2009;58:1542–1548. doi: 10.1099/jmm.0.013235-0. [DOI] [PubMed] [Google Scholar]
- 42.Berry MC, McGhee GC, Zhao Y, Sundin GW. Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora. FEMS Microbiol Lett. 2009;291:80–87. doi: 10.1111/j.1574-6968.2008.01438.x. [DOI] [PubMed] [Google Scholar]
- 43.Miyashita A, Iyoda S, Ishii K, Hamamoto H, Sekimizu K, Kaito C. Lipopolysaccharide O-antigen of enterohemorrhagic Escherichia coli O157:H7 is required for killing both insects and mammals. FEMS Microbiol Lett. 2012;333:59–68. doi: 10.1111/j.1574-6968.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 44.Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R. 3rd, Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun. 2011;79:4227–4239. doi: 10.1128/IAI.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Billips BK, Yaggie RE, Cashy JP, Schaeffer AJ, Klumpp DJ. A live-attenuated vaccine for the treatment of urinary tract infection by uropathogenic Escherichia coli. J Infect Dis. 2009;200:263–272. doi: 10.1086/599839. [DOI] [PubMed] [Google Scholar]


