Abstract
The d, ash, and ln coat color mutations provide a unique model system for the study of vesicle transport in mammals. All three mutant loci encode genes that are required for the polarized transport of melanosomes, the specialized, pigment-containing organelles of melanocytes, to the neighboring keratinocytes and eventually into coat hairs. Genetic studies suggest that these genes function in the same or overlapping pathways and are supported by biochemical studies showing that d encodes an actin-based melanosome transport motor, MyoVa, whereas ash encodes Rab27a, a protein that localizes to the melanosome and is postulated to serve as the MyoVa receptor. Here we show that ln encodes melanophilin (Mlph), a previously undescribed protein with homology to Rab effectors such as granuphilin, Slp3-a, and rabphilin-3A. Like all of these effectors, Mlph possesses two Zn2+-binding CX2CX13,14CX2C motifs and a short aromatic-rich amino acid region that is critical for Rab binding. However, Mlph does not contain the two Ca2+-binding C2 domains found in these and other proteins involved in vesicle transport, suggesting that it represents a previously unrecognized class of Rab effectors. Collectively, our data show that Mlph is a critical component of the melanosome transport machinery and suggest that Mlph might function as part of a transport complex with Rab27a and MyoVa.
The pigmentation of mammalian hair and skin is a multistep process involving the synthesis of pigments (melanins) within melanosomes, the specialized, pigment-producing organelles of melanocytes; long-range, bidirectional, microtubule-dependent melanosome transport from the cell body to the melanocyte's extensive actin-rich dendritic arbor; capture and short-range actin-dependent melanosome transport in the cell periphery; and melanosome transfer from the dendritic tips to the neighboring keratinocytes and eventually into the hair shaft (1). More than 50 loci have been identified in the mouse that affect coat pigmentation (2), and these mutant loci provide a unique resource for the study of pigmentation in mammals.
In three pigment mutations, dilute (d), ashen (ash), and leaden (ln), melanin synthesis is normal but melanosome transport is impaired, resulting in the clumping of the melanosomes in the perinuclear region of the cell (3). Genetic evidence suggests that these loci encode proteins that function in the same or overlapping pathways. All three mutations have identical phenotypes. In addition, the doubly and triply homozygous mutant combinations on a nonagouti background have phenotypes that are indistinguishable from any of the single mutations alone (N.G.C. and N.A.J., unpublished observations). Furthermore, all three mutations are suppressed by the cell autonomous, semidominant dilute suppressor, dsu (4–6).
These genetic findings have been strengthened by the identification and biochemical analysis of the genes mutated in d and ash mice. Previous work has shown that dilute encodes unconventional myosin Va (MyoVa), a plus-end-directed, actin-based, myosin heavy-chain motor (7, 8), which colocalizes with end-stage melanosomes (9–11). MyoVa is a processive motor that moves in large steps approximating the 36-nm pseudorepeat of the actin filament (12). In the absence of MyoVa, long-range, bidirectional, microtubule-dependent movement of melanosomes along the length of dendrites is normal (1). However, the melanosomes are not captured in the actin-rich periphery of the cell and they thus become concentrated in the cell center, which is highly enriched in microtubules (9, 13).
ashen encodes Rab27a (14). Rab GTPases represent the largest branch of the p21 Ras superfamily and are recognized as key players in vesicular transport and organelle dynamics in eukaryotic cells (15). Like dilute melanocytes (9, 13), ashen melanocytes exhibit normal dendritic morphology and melanosome biogenesis, an abnormal accumulation of end-stage melanosomes in the cell center, and rapid, bidirectional, microtubule-dependent melanosome movements between the cell center and the periphery (16). This phenotype suggests that ashen melanocytes, like dilute melanocytes, are defective in peripheral melanosome capture and short-range melanosome transport. Consistent with this hypothesis, introduction of wild-type Rab27a cDNA into ashen melanocytes restores the peripheral accumulation of melanosomes in a microtubule-dependent manner (16). Conversely, introduction of a dominant interfering Rab27a cDNA into wild-type melanocytes causes the melanosomes to be redistributed to the perinuclear region of the cell (16, 17).
Rab27a colocalizes with MyoVa in end-stage melanosomes from wild-type cells (16–18). It can also be coimmunoprecipitated with MyoVa from wild-type melanosome extracts (17). However, MyoVa does not colocalize with melanosomes in ashen melanocytes, implying that Rab27a is, or is part of, the melanosome receptor that recruits MyoVa onto melanosomes (16).
Yeast two-hybrid studies have shown that MyoVa can also interact with a microtubule-based motor, ubiquitous kinesin heavy chain (Kif5b) (19). These data suggest that MyoVa, together with Kif5b and Rab27a, forms a transport complex that coordinates the long-range transport of melanosomes along the microtubules (Kif5b-dependent movement) and the short-range transport of melanosomes along actin filaments (MyoVa-dependent movement).
MyoVa has also been shown to bind a RING finger protein, BERP, in rat brain (20). BERP contains an N-terminal RING finger, a zinc-binding domain (B-box), a leucine coiled-coil region, and a C-terminal β-propeller through which it binds MyoVa (20). The multiple protein–protein interaction motifs carried by BERP make it a potential adaptor protein for mediating MyoVa cargo transport. Expression of a dominant interfering BERP mutant in rat PC12 cells has been shown to inhibit PC12 cell spreading and prevent neurite outgrowth in response to nerve growth factor (20). MyoVa also apparently plays a role in neurite extension and growth cone development (21), so it is tempting to speculate that the effects of dominant interfering mutants of BERP result from its interaction with MyoVa.
Here, we use positional cloning to identify the gene that is mutated in leaden mice. We show that leaden encodes melanophilin (Mlph), a previously unknown member of the Rab effector family. These results are consistent with previous experiments showing that melanosomes in leaden melanocytes clump in the perinuclear region of the cell in a manner similar to that of dilute and ash melanocytes (13). They are also consistent with transplantation studies, aggregation chimera and dermal–epidermal recombination graft experiments showing that leaden acts within, not on, the melanocyte (22, 23) and suggest that Mlph might function as part of a transport complex with MyoVa and Rab27a.
Materials and Methods
Mice.
C57BR/cdJ, C57L/J-ln/ln, SWXL-17/Ty-ln/ln, Mus castaneus (CAST/Ei), and In(1)1Rk-lnllRK3/+ mice were purchased from The Jackson Laboratory and maintained and propagated at the National Cancer Institute (Frederick, MD). The lnllRK3 mutation is lethal in the homozygous condition and all analyses of this allele were therefore done in In(1)1Rk-lnllRK3/+ or In(1)1Rk-lnllRK3/ln mice.
Genetic and Physical Mapping.
Genetic and yeast artificial chromosome (YAC)-based maps of the ln critical region were generated as previously described (24). Bacterial artificial chromosomes (BACs) from the contig were identified by screening two libraries with probes derived from simple sequence length polymorphism (SSLP) markers and YAC end clones from the ln critical region. BACs 63H24, 317P10, 341N13, and 172G11 were obtained from the 129/Sv BAC library (Research Genetics, Huntsville, AL), and 182B13 is a C57BL/6J-derived BAC from the RPCI-23 library (Roswell Park Cancer Institute, Buffalo, NY). A BAC contig was assembled as described (14).
BAC Complementation.
BAC DNAs were prepared and injected into (C57L/J-ln/ln × SWXL-17/Ty-ln/ln)F2 zygotes as described (25). Transgenic founders were identified on the basis of Southern blot analysis of genomic DNA isolated from tail biopsies (26), and complementation was assessed by coat color. Founder mice were mated to SWXL-17/Ty-ln/ln mice and their progeny were scored for the presence or absence of the BAC transgene and the ln coat color phenotype.
BAC Sample Sequencing.
Approximately 200 μg of DNA from BACs showing complementation was diluted to 2 ml in a solution that contained 20 mM Tris (pH = 7.5), 10 mM MgCl2, and 40% (vol/vol) glycerol final concentration. The DNA was sheared in a nebulizer by subjecting the mixture to 10 psi (1 psi = 6.89 kPa) of argon gas for 2.5 min. DNA in the 2- to 4-kb range was size-selected by gel electrophoresis on a 1% LE agarose (Roche Molecular Biochemicals) gel made in TAE (40 mM Tris-acetate/ 1 mM EDTA). DNA was gel-purified with the GeneClean III kit (Bio 101, Vista, CA), cloned into the pSTBlue-1 vector with the Perfectly Blunt Cloning Kit (Novagen), and electroporated into ElectroMAX DH10B Cells (GIBCO/BRL) under the following parameters: 1.75 kV, 25 μF, 200 Ω (Bio-Rad Gene Pulser). DNA from transformants containing inserts was prepared with the R.E.A.L. DNA prep kit (Qiagen, Valencia, CA) and sequenced with the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer). Data were acquired on an ABI model 377 DNA sequencer (Applied Biosystems). The sequence reads were assembled with phrap. After repetitive elements were masked, both the contigs and single reads were blast searched against the National Center for Biotechnology Information nucleotide and protein databases to look for similarities to known genes and expressed sequence tags (ESTs).
Cloning Mlph cDNA.
The mouse EST database at the National Center for Biotechnology Information was screened by using the human gene AK022207 (GenBank accession no.). On the basis of their similarities to this human gene, seven ESTs were ordered from Research Genetics, completely sequenced, and arranged into a complete unit of 4,229 bp (GenBank accession nos. BE290077, BE380367, BF160116, BE381191, BE916642, BE370113, and BF301891). BE290077 was also used to screen a phage library derived from melan-a cells (27). Sequence from the positive clones covers bp 62–2613 of the Mlph cDNA. The largest clone identified was 1,704 bp, extending from bp 904 to bp 2613 of the cDNA.
Expression Analysis of Mlph by Using Northern Blotting.
Expression patterns of Mlph in adult tissues were evaluated by using a mouse poly(A)+ Northern blot containing 12 major tissues from Swiss–Webster mice (OriGene Technologies, Rockville, MD). Embryonic expression was determined by hybridizing a 2.4-kb fragment of Mlph (bp 1–2400) to a poly(A)+ Northern blot derived from four stages of murine embryogenesis (CLONTECH). Mlph expression levels in mutant tissues were determined by first isolating total mouse RNA from 2- to 3-week-old shaved animals with STAT-60 RNAzol (Tel-Test, Friendswood, TX) and then single-selecting poly(A)+ RNA with the Poly(A) Pure Isolation kit from Ambion (Austin, TX). Two micrograms of poly(A)+ RNA from each tissue was loaded and separated on a 1.2% agarose and 6.2% formaldehyde gel and then transferred to a nylon membrane (Amersham Pharmacia). Hybridization was performed exactly as described for Southern blotting (26).
Mutation Detection.
cDNA was synthesized from both poly(A)+ and total RNA by using the Superscript Preamplification System (GIBCO/BRL). These cDNAs were then amplified with a series of overlapping primer sets that spanned the entire 1773-bp ORF of Mlph with reagents from the High Fidelity amplification kit as directed (Roche Molecular Biochemicals). Amplification was performed with the following cycling conditions: 3 min at 94°C, 35 cycles of 1 min at 94°C, 2 min at 54°C, and 3 min at 72°C, followed by a 7-min extension at 72°C and a 4°C hold. Products were visualized on a TAE 4% NuSieve 3:1 gel (BioWhittaker). PCR products were gel-purified and subjected to direct sequencing. Primer pairs 187–206 (5′-AGATGGGGAAAAGGTTGGAC-3′) and 462–443 (5′-AGCTTTTGCAGACGAAGAGG-3′) yielded differently sized reverse transcription–PCR products. This result was confirmed by amplification, gel purification, and direct sequencing of genomic DNA by using primers that flank exon 2 (F = 5′-GTGGGTGGTTTTGCTGATCT-3′; R = 5′-CATAACGAGGAAGCCAAAGC-3′). Cycling conditions for this primer pair were as follows: 5 min at 94°C, 40 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by a 7-min extension at 72°C and a 4°C hold. All PCR was performed with a Perkin–Elmer 9700 machine.
Results and Discussion
A Genetic and Physical Map of the ln Critical Region.
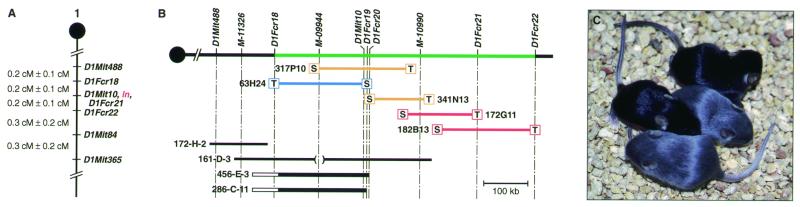
To initiate the positional cloning of ln we generated 1,031 (SWXL-17/Ty-ln/ln × M. castaneus) × SWXL-17/Ty-ln/ln N2 progeny, which were typed at weaning for ln by visual inspection. Tail DNAs from the mice were then typed for SSLP markers that are predicted to map near ln (28–30). These mapping studies defined a 0.9-centimorgan ln critical region that was flanked by D1Mit488 on the proximal side and D1Mit84 on the distal side (Fig. 1A). Next, a physical map of the ln critical region was generated. The mouse Massachusetts Institute of Technology Whitehead YAC map (31) contains four overlapping YAC clones that are located in the ln critical region (Fig. 1B). Three YACs (161-D-3, 456-E-3, 286-C-11) contain D1Mit10, which is nonrecombinant with ln (Fig. 1A). Three SSLP markers that had been localized to these YACs by the Whitehead group (M-11326, M-09944, M-10990) and two YAC end-sequence probes (D1Fcr19, D1Fcr20) were then used to isolate mouse BACs. The BACs were end-sequenced, and the end sequences were used to generate additional probes (D1Fcr18, D1Fcr21, D1Fcr22). These probes were, in turn, used to isolate additional BACs. Some of the BAC end-sequence probes were also mapped on the ln cross (Fig. 1A). Collectively, these studies defined a ln critical interval that was flanked by D1Fcr18 on the proximal side and D1Fcr22 on the distal side (Fig. 1B). This critical region is spanned by a minimum of five BACs and is estimated to represent ≈600 kb of genomic DNA.
Figure 1.
Meiotic and physical mapping of the ln critical region. (A) A meiotic linkage map showing the positions of SSLP markers (D1Mit followed by a number) and BAC end clones (D1Fcr followed by a number) that map within or near the ln critical region. The centromere is shown at the top (●). cM, centimorgan. (B) A physical map of the ln region of mouse chromosome 1 with the ln critical region shown in green, the centromere pictured on the left (●), and the position of various molecular markers shown on the top. YACs are shown in black at the bottom. YACs 456-E-3 and 286-C-11 are chimeric as indicated by the open boxes. BACs are shown in color: those that were used for BAC complementation and showed rescue are shown in red, and those that showed no complementation are depicted in orange. BACs in blue were not injected. (C) A litter from a BAC transgenic animal that had been bred to a SWXL-17/Ty-ln/ln animal contains two progeny that did not inherit the BAC and are ln in color, and two progeny that inherited the BAC and are nonagouti (i.e., wild type) in color.
BAC Complementation.
Because of the large size of the ln critical region, we sought to narrow the interval further by BAC complementation. Several BACs from the region were microinjected into ln zygotes, and the progeny were scored at weaning for their coat color. Among the seven transgenic founders (i.e., BAC-positive by Southern blot analysis) that resulted from injection of BAC 317P10, none showed rescue of the ln coat color. In contrast, two of three transgenic founders generated from the injection of BAC 182B13 showed partial (one founder) or complete (one founder) rescue. Both BACs were subsequently transmitted through the germ line and, in this case, both BACs generated a complete rescue of the ln coat color phenotype (Fig. 1C; data not shown). Likewise, one of two transgenic founders generated from the coinjection of BACs 341N13 and 172G11 showed partial rescue (data not shown). The most likely explanation for these results is that ln is located in the region of overlap between BACs 172G11 and 182B13 (Fig. 1B).
Identification of ln Candidate Genes.
To identify genes located in the region of overlap between BACs 172G11 and 182B13, both BACs were partially sequenced. Because this region was highly repetitive, 1,663,991 bp of raw data were generated, only 8% of which represented quality reads (i.e., not repetitive, Escherichia coli-derived, or vector sequence). These data were assembled into 165 contigs, the largest of which was 8,721 bp, constituting 194,474 bp of sequence. Analysis of these contigs and of the 819,216 bp contained in 1,092 single reads that did not assemble identified three genes in the overlapping region: preprolactin-releasing hormone, Rab17, and a human gene isolated from mammary tissue (AK022207). Preprolactin-releasing hormone was eliminated as a candidate for ln on the basis of its known function and expression pattern. On the other hand, Rab17 is an attractive candidate for ln because it is expressed in polarized epithelial cells (32–34) and because ash, a mutation with a phenotype identical to ln, encodes Rab27a (14). However, Northern blot and reverse transcription–PCR analysis showed that Rab17 is not expressed in wild-type melanocytes (i.e., melan-a cells; ref. 27) (data not shown). Sequence analysis also failed to identify any Rab17-coding region mutations in ln mice. Rab17 is thus unlikely to be mutated in leaden mice.
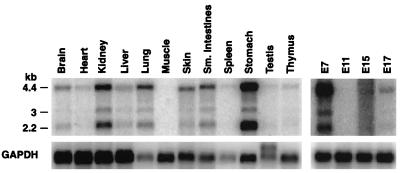
Subsequently, we identified mouse ESTs that corresponded to a human gene isolated from mammary tissue (AK022207), which we have renamed melanophilin (MLPH). Northern blot analysis identified three isoforms of Mlph, which are 2.2, 3.0, and 4.4 kb in size in mouse (Fig. 2). Mlph is expressed in most adult tissues, with the highest levels of expression observed in epithelial-enriched tissues such as kidney, lung, skin, small intestine, and stomach (Fig. 2). In the embryo, Mlph is highly expressed at E7. However, by E11 Mlph expression is undetectable. Later in development Mlph expression is up-regulated, albeit to low levels (Fig. 2). Mlph is also highly expressed in melan-a cells (27) and is thus a good candidate for leaden (Fig. 3A).
Figure 2.
Expression of Mlph in adult and embryonic tissues. Transcript sizes (in kb) are indicated on the left. A 2.4-kb Mlph EST was used as a probe and detected three isoforms of Mlph, which are predominantly expressed in epithelial-rich adult tissues. The highest embryonic Mlph expression was detected at embryonic day 7 (E7). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
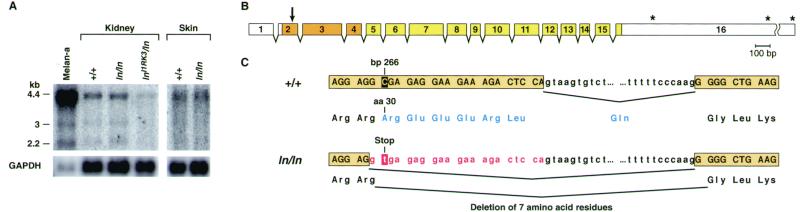
Figure 3.
The ln mutation results from a defect in Mlph. (A) Northern blot analysis of Mlph expression in wild-type (C57BR/cdJ), C57L/J-ln/ln, and In(1)1Rk-lnllRK3/ln tissues. Transcript sizes (in kb) are depicted on the left. The amount and size of Mlph transcripts seem unchanged in ln/ln mutants, but are reduced in lnllRK3/ln tissue. Also shown is Mlph expression in wild-type melan-a melanocytes (27). Note that Mlph is expressed at high levels in melan-a cells. (B) The intron/exon organization of the mouse Mlph gene. Coding regions are shown in color and untranslated region (UTR) sequences in white. The asterisks indicate the locations of three alternatively used polyadenylation sites, orange boxes represent the conserved Rab-effector domain, and the arrow denotes the site of the ln mutation. (C) A C-to-T transition at bp 266 (shaded) results in altered splicing between exons 2 and 3. The ln mutation generates a new splice donor site, so that the last 21 bp (shown in red) of exon 2 are spliced out of the mature message. If this message were translated, it would produce a protein that lacks amino acid residues 30–36 (REEERLQ), shown in blue. If there were some readthrough of the mutant message, a stop codon at amino acid 30 would severely truncate the protein.
Identification of a Mlph Defect in leaden Mice.
The leaden mutation arose spontaneously in 1933 in the C57BR inbred strain (35). Northern blot analysis of wild-type (C57BR/cdJ) and C57L/J-ln/ln kidney and skin RNA did not identify any major alterations in Mlph transcript size or expression levels in ln RNA (Fig. 3A). However, reverse transcription–PCR analysis of Mlph transcripts expressed in ln mice by using primer pairs that spanned the entire Mlph ORF identified a region from bp 187 to bp 462 that was apparently altered in ln mice (data not shown). BAC and Mlph cDNA sequencing showed Mlph is encoded by 16 exons (Fig. 3B). The Mlph mRNA contains a relatively short (175 bp) 5′ untranslated region (UTR) and a long 3′UTR composed of 2,281 nucleotides. Two shorter Mlph transcripts were also identified that seemed to result from the use of alternative poly(A)+ addition sites, which accounts for the three differently sized Mlph transcripts identified on Northern blots. All three Mlph transcripts encode identical proteins. The longest Mlph transcript is 4,229 bp and contains a predicted 1,773-bp ORF. The Mlph protein is 590 aa.
Sequence analysis of the Mlph transcripts expressed in ln mice identified a 21-bp deletion that is not present in wild-type mRNA (Fig. 3C). Genomic sequence analysis showed that this 21-bp deletion results from a C-to-T transition at nucleotide position 266 of the mature message. This mutation creates a new splice donor site in exon 2. The resulting transcript has an in-frame 21-bp deletion that removes seven amino acids (REEERLQ) from the mature protein (Fig. 3C). Although sequence analysis failed to identify any normally spliced Mlph transcripts in ln mice, this C-to-T transition also introduces a stop codon in exon 2 (Fig. 3C). Thus, any normally spliced message expressed would produce a short, 30-aa, protein, which is unlikely to be functional.
We also analyzed the nature of the defect in another leaden allele, lnllRK3 (36). This mutation was identified in the descendants of a cross involving a triethylene melamine-treated male, which was homozygous for a chromosome 1 inversion [In(1)1Rk], and a female ln Sp/ln + animal. The lnllRK3 mutation is homozygous lethal, suggesting that it might be caused by a deletion. Consistent with this hypothesis, the Mlph-coding region is completely deleted in lnllRK3 DNA and Mlph transcripts are not expressed from the lnllRK3 chromosome (data not shown; Fig. 3A).
Three other radiation-induced leaden mutations have also been identified that are now extinct (37). Two of these mutations were homozygous viable, whereas one was homozygous lethal. No difference in the coat color phenotype of the two viable alleles and the original spontaneous ln allele was noted. In addition, the lnllRK3 allele, when balanced over ln, produces a phenotype that is similar to homozygous ln mice. These findings suggest that the lethality associated with lnllRK3 and one of the radiation-induced alleles results from the deletion of one or more flanking genes rather than from defects in Mlph itself. They also suggest that the original spontaneous leaden allele represents the null phenotype, although gene knockout studies will be needed to confirm this prediction.
Mlph Is a Member of the Rab Effector Gene Family.
blast and conserved domain searches against the nonredundant protein database showed that the N terminus of Mlph (amino acids 7–129) has homology to the Rab effector domains present in granuphilin-a and -b (38), synaptotagmin-like protein 3a (Slp3-a) (39), and rabphilin-3A (40) (Fig. 4). The Rab effector domain consensus sequence is ≈270 aa residues in length and contains two Zn2+-binding CX2CX13,14CX2C motifs and a short aromatic amino acid-rich region that is thought to be critical for Rab binding. Interactions between Rabs and their effectors are hypothesized to play many important roles in vesicle transport. First, Rabs may act in concert with their effectors to facilitate vesicle transport directly. The Rab effector, rabphilin-3A, has been shown in vitro to interact with α-actinin, an actin-bundling protein, but only when not bound to Rab3A (41). Actin bundling by rabphilin-3A/α-actinin complexes may help regulate vesicle interactions with the cytoskeleton and play an active role in vesicle transport. Second, these interactions may regulate membrane trafficking at the vesicle-docking step. A number of Rab effectors, including rabaptin-5, EEA1, rabphilin-3A, and Rim contain a Rab-binding domain and a linker region that is capable of interacting with a second Rab or the target membrane (reviewed in ref. 42). Third, Rab effectors may be required for Rabs to activate the SNARE fusion machinery. A Rab effector that interacts with a vacuole Rab (a Sec1p homolog) and a SNARE protein has recently been identified (43). This Rab effector could thus serve to connect Rab and SNARE function.
Figure 4.
Clustal W alignments of Mlph and three proteins that share a conserved Rab effector domain. The eight highly conserved Cys residues that make up the Zn2+ finger domain are shown in red. Identical and conserved amino acid residues are in shaded boxes. The seven amino acids deleted in leaden are shown in blue. The 6-aa aromatic-rich motif that binds to Rab is shown in yellow. Amino acids 1–129 of Mlph show 28% identity/48% similarity to granuphilin, 31% identity/49% similarity to Slp3-a, and 23% identity/41% similarity to rabphilin-3A.
As for the three Mlph-related proteins, granuphilin is thought to play a role in regulating exocytosis of insulin-containing dense-core granules from the pancreas (38). The function of Slp3a is not yet clear, but the related synaptotagmin I protein is an integral membrane protein that colocalizes to synaptic vesicles and is essential for the fusion of synaptic vesicles with the presynaptic plasma membrane (44). Rab3A recruits rabphilin-3A to synaptic vesicles before docking and neurotransmitter release (45). Rabphilin-3A has been postulated to regulate neurotransmitter release; however, mice that lack rabphilin-3A are viable and fertile without obvious physiological impairment (46). The function of rabphilin-3A thus remains to be determined.
The crystal structure of Rab3A complexed with the effector domain of rabphilin-3A has been determined (40). Rabphilin-3A contacts Rab3A in two distinct areas. The first interface involves the Rab3A switch I and switch II regions, which are sensitive to the nucleotide-binding state of Rab3A. The second interface consists of a deep pocket in Rab3A that interacts with a SGAWFF structural motif of rabphilin-3A. The last three amino acids of this structural motif are aromatic amino acids that adopt a perpendicular edge-to-plane configuration and are particularly important for this interaction. Although the regions in rabphilin-3A that contact switch regions I and II are not conserved in Mlph, the SGAWFF motif is conserved. Mlph encodes a related SLEWYY motif with the last three amino acids being aromatic residues (Fig. 4). The divergence in the first part of this motif likely reflects the fact that Mlph binds a different Rab.
Like granuphilin, Slp3-a, and rabphilin-3A, Mlph also has a series of eight conserved Cys residues that form a Zn2+ finger-binding domain (Fig. 4). In rabphilin-3A, the Zn2+-binding domain is far removed from the sites of interaction with Rab3A. However, mutations that disrupt Zn2+ binding abolish Rab binding, suggesting that the Zn2+-binding domain is essential and perhaps serves as a structural scaffold, which is necessary for Rab binding.
Outside the Rab effector domain, there is little similarity between Mlph and granuphilin, Slp3-a, or rabphilin-3A, or to any other protein in current databases. The lack of similarity exists partly because the C terminus of granuphilin, Slp3-a, and rabphilin-3A contain Ca2+-sensing, membrane-targeting C2 domains that are not present in Mlph (47). Mlph is thus unlikely to be regulated by Ca2+. In this respect, Mlph is similar to another rabphilin family member, Noc2, which also lacks C2 domains (48).
Human Mutations.
Griscelli syndrome is a rare autosomal recessive disorder that results in hypopigmentation of the skin and hair, the presence of large clumps of pigment granules in hair shafts, and an accumulation of melanosomes in melanocytes. Most patients also develop an uncontrolled T lymphocyte and macrophage activation syndrome (known as hemophagocytic syndrome). In contrast, some patients develop a severe neurological impairment without apparent immune abnormalities. Recently, de Saint Basile and her colleagues (49) showed that Griscelli patients with immune defects carry mutations in RAB27A, whereas those with neurological impairments carry mutations in MYOVA (50). These Griscelli's phenotypes are consistent with what has been observed in mutant mice. For example, dilute mice have smooth endoplasmic reticulum transport defects that result in ataxia (51, 52), in addition to melanosome transport defects, whereas ashen mice have T lymphocyte granule transport defects that produce immune abnormalities (53, 54).
MLPH maps to human 2q37 (55), a region of conserved synteny with mouse chromosome 1 where leaden maps. In leaden mice, the only defect that has been observed to date is in melanosome transport. So far, all characterized Griscelli's syndrome patients carry mutations in MYOVA or RAB27A. However, it would not be surprising to find patients with Griscelli's syndrome, or patients with other hypopigmentation disorders, that have mutations in MLPH.
Mlph Function in Melanosome Transport.
Although the function of Mlph in melanosome transport remains to be firmly established, one likely possibility is that Mlph is a Rab27a effector that is recruited onto melanosomes by Rab27a. This hypothesis is consistent with mutant data showing that ashen and leaden mice have identical melanocyte defects and with Rab3A/rabphilin-3A knockout studies showing that rabphilin-3A is recruited onto synaptic vesicles by Rab3A (46, 56). According to this hypothesis, Mlph would not be recruited onto melanosomes in leaden melanocytes, the MyoVa-Rab27a transport complex would not function properly, and melanosomes would not be captured and transported in the melanocyte periphery. Instead, the melanosomes would become clumped in the perinuclear region of the cell.
Mlph could also play additional roles in later steps of melanosome transport, such as vesicle targeting and vesicle fusion. For example, binding of Mlph to activated Rab27a could inhibit melanosome fusion with target membranes and release of pigment granules to neighboring keratinocytes. In this case, vesicle fusion could be induced by the activation of MyoVa-binding proteins such as BERP, which contains a RING finger motif. Many, if not all, RING finger proteins function as E3 ubiquitin protein ligases (57), although BERP has not yet been demonstrated formally to be an E3 ligase. Activation of BERP could, in turn, lead to the degradation of Mlph by the 26S proteasome. The loss of any such function in ln mice would not be evident because these pathways are downstream of the primary site of action of Mlph.
Finally, Mlph could have Rab-independent functions. As discussed previously, rabphilin-3A, a Mlph-related protein, binds α-actinin, an actin-bundling protein (41). Binding to α-actinin occurs only after rabphilin-3A is released from Rab3A. Mlph might also bind α-actinin or proteins that bind to the cytoskeleton and thereby play an active role in targeting vesicles to their appropriate destination. BERP has also been shown to bind to α-actinin-4, a Ca2+-independent actin-bundling protein implicated in cell motility and carcinogenesis (58). Therefore, it is also possible that BERP, not Mlph, supplies the functions provided by rabphilin-3A in actin bundling. Furthur biochemical and cell biological studies will be needed to clarify these important issues.
Acknowledgments
Sequence assembly and blast analysis were greatly facilitated by excellent computer assistance provided by Bob Stevens. This research was supported by the National Cancer Institute, Department of Health and Human Services.
Abbreviations
- Mlph
melanophilin
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- SSLP
simple sequence length polymorphism
- EST
expressed sequence tag
- En
embryonic day n
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF384098 (mouse Mlph)].
References
- 1.Wu X, Kocher B, Wei Q, Hammer J A., III Cell Motil Cytoskeleton. 1998;40:286–303. doi: 10.1002/(SICI)1097-0169(1998)40:3<286::AID-CM7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 2.Jackson I J. BioEssays. 1991;13:439–446. doi: 10.1002/bies.950130903. [DOI] [PubMed] [Google Scholar]
- 3.Silvers W K. The Coat Colors of Mice. New York: Springer; 1979. [Google Scholar]
- 4.Moore K J, Seperack P K, Strobel M C, Swing D A, Copeland N G, Jenkins N A. Proc Natl Acad Sci USA. 1988;85:8131–8135. doi: 10.1073/pnas.85.21.8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore K J, Swing D A, Rinchik E M, Mucenski M L, Buchberg A M, Copeland N G, Jenkins N A. Genetics. 1988;119:933–941. doi: 10.1093/genetics/119.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sweet H O. J Hered. 1983;74:305–306. doi: 10.1093/oxfordjournals.jhered.a109794. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Nature (London) 1981;293:370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- 8.Mercer J A, Seperack P K, Strobel M C, Copeland N G, Jenkins N A. Nature (London) 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Bowers B, Wei Q, Kocher B, Hammer J A., III J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- 10.Nascimento A A, Amaral R G, Bizario J C, Larson R E, Espreafico E M. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert J, Onderwater J, Vander Haeghen Y, Vancoillie G, Koerten H K, Mommaas A M, Naeyaert J M. J Invest Dermatol. 1998;111:835–840. doi: 10.1046/j.1523-1747.1998.00395.x. [DOI] [PubMed] [Google Scholar]
- 12.Mehta A D, Rock R S, Rief M, Spudich J A, Mooseker M S, Cheney R E. Nature (London) 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 13.Provance D W, Jr, Wei M, Ipe V, Mercer J A. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson S M, Yip R, Swing D A, O'Sullivan T N, Zhang Y, Novak E K, Swank R T, Russell L B, Copeland N G, Jenkins N A. Proc Natl Acad Sci USA. 2000;97:7933–7938. doi: 10.1073/pnas.140212797. . (First Published June 20, 2000; 10.1073/pnas.140212797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick P, Zerial M. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Rao K, Bowers M B, Copeland N G, Jenkins N A, Hammer J A. J Cell Sci. 2001;114:1091–1100. doi: 10.1242/jcs.114.6.1091. [DOI] [PubMed] [Google Scholar]
- 17.Hume A N, Collinson L M, Rapak A, Gomes A Q, Hopkins C R, Seabra M C. J Cell Biol. 2001;152:795–808. doi: 10.1083/jcb.152.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahadoran P, Aberdam E, Mantoux F, Busca R, Bille K, Yalman N, de Saint-Basile G, Casaroli-Marano R, Ortonne J P, Ballotti R. J Cell Biol. 2001;152:843–850. doi: 10.1083/jcb.152.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J D, Brady S T, Richards B W, Stenolen D, Resau J H, Copeland N G, Jenkins N A. Nature (London) 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 20.El-Husseini A E, Vincent S R. J Biol Chem. 1999;274:19771–19777. doi: 10.1074/jbc.274.28.19771. [DOI] [PubMed] [Google Scholar]
- 21.Wang F S, Wolenski J S, Cheney R E, Mooseker M S, Jay D G. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 22.Markert C L, Silvers W K. Genetics. 1956;41:429–450. doi: 10.1093/genetics/41.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson D A, Glenister P H, Hornby J E. Genet Res. 1985;46:193–205. doi: 10.1017/s0016672300022655. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher C F, Lutz C M, O'Sullivan T N, Shaughnessy J D, Jr, Hawkes R, Frankel W N, Copeland N G, Jenkins N A. Cell. 1996;87:607–617. doi: 10.1016/s0092-8674(00)81381-1. [DOI] [PubMed] [Google Scholar]
- 25.Antoch M P, Song E J, Chang A M, Vitaterna M H, Zhao Y, Wilsbacher L D, Sangoram A M, King D P, Pinto L H, Takahashi J S. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins N A, Copeland N G, Taylor B A, Lee B K. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett D C, Cooper P J, Hart I R. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 28.Copeland N G, Gilbert D J, Jenkins N A, Nadeau J H, Eppig J T, Maltais L J, Miller J C, Dietrich W F, Steen R G, Lincoln S E, et al. Science. 1993;262:67–82. doi: 10.1126/science.8211131. [DOI] [PubMed] [Google Scholar]
- 29.Dietrich W F, Miller J C, Steen R G, Merchant M, Damron D, Nahf R, Gross A, Joyce D C, Wessel M, Dredge R D, et al. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 30.Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O'Connor T J, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 31.Haldi M L, Strickland C, Lim P, VanBerkel V, Chen X, Noya D, Korenberg J R, Husain Z, Miller J, Lander E S. Mamm Genome. 1996;7:767–769. doi: 10.1007/s003359900228. [DOI] [PubMed] [Google Scholar]
- 32.Hunziker W, Peters P J. J Biol Chem. 1998;273:15734–15741. doi: 10.1074/jbc.273.25.15734. [DOI] [PubMed] [Google Scholar]
- 33.Lutcke A, Jansson S, Parton R G, Chavrier P, Valencia A, Huber L A, Lehtonen E, Zerial M. J Cell Biol. 1993;121:553–564. doi: 10.1083/jcb.121.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zacchi P, Stenmark H, Parton R G, Orioli D, Lim F, Giner A, Mellman I, Zerial M, Murphy C. J Cell Biol. 1998;140:1039–1053. doi: 10.1083/jcb.140.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray J M. Am Nat. 1933;67:278–283. [Google Scholar]
- 36.Roderick T H. In: Utilization of Mammalian Specific Locus Studies in Hazard Evaluation and Estimation of Genetic Risk. de Serres F J, Sheridan W, editors. Vol. 28. New York: Plenum; 1983. pp. 135–167. [Google Scholar]
- 37.Lyon M F, Morris T. Mutat Res. 1969;8:191–198. doi: 10.1016/0027-5107(69)90153-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Takeuchi T, Yokota H, Izumi T. J Biol Chem. 1999;274:28542–28548. doi: 10.1074/jbc.274.40.28542. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda M, Mikoshiba K. Biochem Biophys Res Commun. 2001;281:1226–1233. doi: 10.1006/bbrc.2001.4512. [DOI] [PubMed] [Google Scholar]
- 40.Ostermeier C, Brunger A T. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 41.Kato M, Sasaki T, Ohya T, Nakanishi H, Nishioka H, Imamura M, Takai Y. J Biol Chem. 1996;271:31775–31778. doi: 10.1074/jbc.271.50.31775. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez L, Jr, Scheller R H. Cell. 1999;96:755–758. doi: 10.1016/s0092-8674(00)80585-1. [DOI] [PubMed] [Google Scholar]
- 43.Peterson M R, Burd C G, Emr S D. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- 44.Bommert K, Charlton M P, DeBello W M, Chin G J, Betz H, Augustine G J. Nature (London) 1993;363:163–165. doi: 10.1038/363163a0. [DOI] [PubMed] [Google Scholar]
- 45.Stahl B, Chou J H, Li C, Sudhof T C, Jahn R. EMBO J. 1996;15:1799–1809. [PMC free article] [PubMed] [Google Scholar]
- 46.Schluter O M, Schnell E, Verhage M, Tzonopoulos T, Nicoll R A, Janz R, Malenka R C, Geppert M, Sudhof T C. J Neurosci. 1999;19:5834–5846. doi: 10.1523/JNEUROSCI.19-14-05834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukuda M, Saegusa C, Mikoshiba K. Biochem Biophys Res Commun. 2001;283:513–519. doi: 10.1006/bbrc.2001.4803. [DOI] [PubMed] [Google Scholar]
- 48.Kotake K, Ozaki N, Mizuta M, Sekiya S, Inagaki N, Seino S. J Biol Chem. 1997;272:29407–29410. doi: 10.1074/jbc.272.47.29407. [DOI] [PubMed] [Google Scholar]
- 49.Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffraat N, Bianchi D, Fischer A, Le Deist F, et al. Nat Genet. 2000;25:173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 50.Pastural E, Barrat F J, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, Seger R, Griscelli C, Fischer A, de Saint Basile G. Nat Genet. 1997;16:289–292. doi: 10.1038/ng0797-289. [DOI] [PubMed] [Google Scholar]
- 51.Dekker-Ohno K, Hayasaka S, Takagishi Y, Oda S, Wakasugi N, Mikoshiba K, Inouye M, Yamamura H. Brain Res. 1996;714:226–230. doi: 10.1016/0006-8993(95)01560-4. [DOI] [PubMed] [Google Scholar]
- 52.Takagishi Y, Oda S, Hayasaka S, Dekker-Ohno K, Shikata T, Inouye M, Yamamura H. Neurosci Lett. 1996;215:169–172. doi: 10.1016/0304-3940(96)12967-0. [DOI] [PubMed] [Google Scholar]
- 53.Stinchcombe J C, Barral D C, Mules E H, Booth S, Hume A N, Machesky L M, Seabra M C, Griffiths G M. J Cell Biol. 2001;152:825–834. doi: 10.1083/jcb.152.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haddad E K, Wu X, Hammer J A, III, Henkart P A. J Cell Biol. 2001;152:835–842. doi: 10.1083/jcb.152.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 56.Li C, Takei K, Geppert M, Daniell L, Stenius K, Chapman E R, Jahn R, De Camilli P, Sudhof T C. Neuron. 1994;13:885–898. doi: 10.1016/0896-6273(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 57.Freemont P S. Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 58.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]