Abstract
Pseudouridylation is the most abundant internal post-transcriptional modification of stable RNAs, with fundamental roles in the biogenesis and function of spliceosomal small nuclear RNAs (snRNAs) and ribosomal RNAs (rRNAs). Recently, the first transcriptome-wide maps of RNA pseudouridylation were published, greatly expanding the catalogue of known pseudouridylated RNAs. These data have further implicated RNA pseudouridylation in the cellular stress response and, moreover, have established that mRNAs are also targets of pseudouridine synthases, potentially representing a novel mechanism for expanding the complexity of the cellular proteome.
The field of RNA modifications was born in 1951 with the identification of an unknown nucleoside in total RNA hydrolysates of calf liver1. The structure of this compound was subsequently identified as 5-ribosyluracil, an isomer of 1-ribosyluracil (uridine)2. Because of its abundance, it gained the name ‘the fifth ribonucleoside’; shortly thereafter it was renamed pseudouridine (denoted by the Greek letter psi, ψ)3.
Pseudouridylation results from enzymatic isomerization (an internal transglycosylation) of a uridine in an RNA molecule. Structurally, two distinct features differentiate pseudouridine from uridine. First is the change of the canonical C-N glycosidic bond to a more inert C-C bond2. Second is the presence of an extra hydrogen-bond donor on the non-Watson–Crick edge of pseudouridine. As a result of these structural distinctions, the presence of pseudouridine within RNA increases both the rigidity of the phosphodiester backbone and the thermodynamic stability of ψ-A base pairs (compared with that of U-A base pairs) through effects on base stacking and water coordination, and by improving base pairing with adenosine4,5.
The function of pseudouridylation is best understood within the context of mRNA splicing and translation, as both the spliceosomal small nuclear RNAs (snRNAs; key components of the spliceosome) and the ribosomal RNAs (key components of the ribosome) are abundantly pseudouridylated6,7. In fact, pseudouridine residues are concentrated in evolutionarily conserved and functionally important regions of these RNAs, with implications for the primary, secondary and tertiary structures of the molecules. Indeed, experimental data have established the importance of pseudouridylation in rRNA and spliceosomal small nuclear ribonucleoprotein (snRNP) biogenesis, efficiency of pre-mRNA splicing and translation fidelity6.
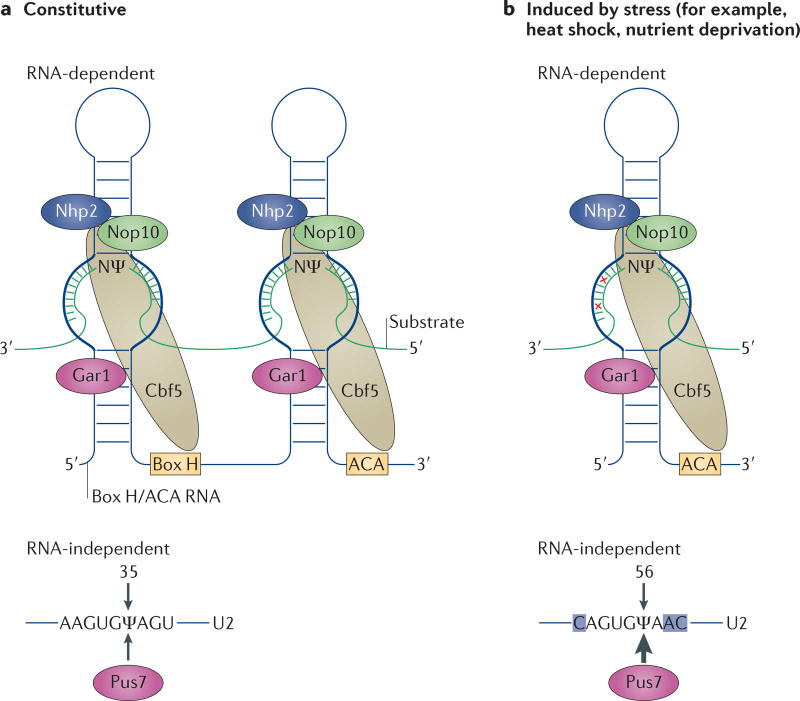
Pseudouridylation is catalysed by pseudouridine synthases (PUSs) and can be achieved through two distinct mechanisms, namely RNA-independent pseudouridylation and RNA-dependent pseudouridylation. Pseudouridylation by the RNA-independent, or stand-alone, mechanism (also referred to as the protein-only mechanism) is catalysed by a single PUS, which carries out both substrate recognition and catalysis. The RNA-dependent mechanism relies on RNA–protein complexes known as box H/ACA small RNPs, which consist of a box H/ACA non-coding RNA and four core proteins (in Saccharomyces cerevisiae, these are centromere-binding factor 5 (Cbf5), non-histone protein 2 (Nhp2), nucleolar protein 10 (Nop10) and glycine-arginine-rich protein 1 (Gar1); in mammals, the Cbf5 homologue is dyskerin (also known as NAP57))8,9 (FIG. 1a). Box H/ACA RNAs adopt a hairpin–hinge–hairpin–tail structure, in which the hinge and tail are single stranded and contain the box H (5′-ANANNA-3′, in which N is any nucleotide) and box ACA (5′-ACA-3′), respectively. Residing within each hairpin is a large internal loop referred to as the pseudouridylation pocket (FIG. 1). The pseudouridylation pocket is responsible for substrate recognition through complementary base-pairing interactions with the substrate RNA, and the catalytic activity is provided by Cbf5 (or dyskerin in mammals). It is worth noting that, at least in archaea, Cbf5 can also function as a stand-alone PUS, in addition to its pseudouridylation activity in archaeal box H/ACA RNPs.
Figure 1. Constitutive and inducible pseudouridylation.
a | Schematic of constitutive uridine-to-pseudouridine (ψ) conversions catalysed by box H/ACA ribonucleoproteins (RNPs) or by a stand-alone protein, pseudouridine synthase 7 (Pus7). Box H/ACA RNPs (top) consist of a small box H/ACA RNA (which has a typical hairpin–hinge–hairpin– tail structure) and four core proteins, namely centromere-binding factor 5 (Cbf5; known as dyskerin in mammals), glycine-arginine-rich protein 1 (Gar1), non-histone protein 2 (Nhp2) and nucleolar protein 10 (Nop10). Cbf5 is the PUS. The substrate RNA engages the box H/ACA RNP via complementary base-pair interactions with the pseudouridylation pocket (thick lines) of the box H/ACA RNA. The uridine targeted for modification and its 3′ adjacent nucleotide (N) are positioned at the base of the upper stem and remain unpaired throughout the reaction. Pus7 (bottom), a stand-alone PUS, recognizes and catalyses pseudouridylation of its substrate (the U2 small nuclear RNA (snRNA) substrate sequence is shown as an example). b | Schematic of a stress-induced uridine-to-pseudouridine conversion catalysed by a box H/ACA RNP or by Pus7. The 3′ hairpin complex (containing the 3′ pseudouridylation pocket) of a box H/ACA RNP is shown (top) with two mismatches between the guide sequence and its substrate (red crosses). Induced pseudouridylation of U2 by Pus7 is also shown (bottom). The nucleotides that differ from the constitutive Pus7 recognition sequence shown in part a are highlighted in blue.
In this Progress article, we review recent findings revealing that pseudouridylation is a dynamic and regulated process that is induced in response to cell state. In addition, we discuss how the application of novel ‘omics’ strategies to map and quantify pseudouridylation globally has revealed a greatly expanded catalogue of pseudouridylation substrates, implicating pseudouridylation in the control of various layers of gene expression regulation, including mRNA stability and proteome diversity.
Inducible pseudouridylation
Pseudouridylation, and RNA modifications in general, were assumed to be constitutive. This assumption was recently challenged by the first evidence that pseudouridylation of yeast U2 snRNA can be induced when cells are subjected to either heat shock or nutrient deprivation10,11. U2 snRNA isolated from stressed cells contained, in addition to the three apparently constitutive pseudouridines (ψ35, ψ42 and ψ44), two novel pseudouridines (ψ56 and ψ93)10. Interestingly, the pseudouridylation of the novel sites was stress-specific: both U56 and U93 underwent pseudouridylation in response to nutrient deprivation, whereas during the heat-shock response only U56 was pseudouridylated. Both the stand-alone (RNA-independent) and the box H/ACA RNP-dependent modification machineries can engage in inducible pseudouridylation, as further analyses revealed that the RNA-independent PUS Pus7 catalyses ψ56 formation and the box H/ACA RNP complex catalyses ψ93 formation (using the 3′ pocket of small nucleolar RNA 81 (snR81))10 (FIG. 1b). Remarkably, the sites of inducible pseudouridylation deviate from the consensus target sites of both Pus7 and snR81, that is, the sequences surrounding positions U56 and U93 in U2 snRNA are similar but not identical to the sequences surrounding the constitutively pseudouridylated targets of Pus7 and snR81. For example, the 3′ pocket of snR81 pairs imperfectly (with two mismatches) with the sequence surrounding nucleotide U93 of U2 snRNA (FIG. 1b). Importantly, inducible pseudouridylation has functional implications, as demonstrated by the finding that ψ93 reduces the efficiency of pre-mRNA splicing10.
Inducible pseudouridylation of other RNAs has also been reported. For instance, during the yeast filamentous growth programme, U6 snRNA is pseudouridylated at U28 by Pus1 (an RNA-independent PUS)12. U6-ψ28 is functionally relevant, as targeted pseudouridylation of U28 within U6 snRNA by designer box H/ACA RNAs activates the filamentous growth programme, whereas blocking U6-ψ28 formation prevents filamentous growth. Interestingly, constitutive substrates of Pus1 lack a clearly identifiable consensus sequence, thus the role of non-consensus sequences in the pseudouridylation of U28 in U6 snRNA is unknown. Recently, using global pseudouridine-profiling techniques, a large number of inducible pseudouridylations were identified in S. cerevisiae and human mRNAs when cells were subjected to heat shock or nutrient deprivation13–15. Both stand-alone PUSs and box H/ACA RNPs were found to catalyse these inducible mRNA pseudouridylations. Interestingly, most of the box H/ACA RNP-catalysed pseudouridylations could not be associated with a known box H/ACA RNA, suggesting the existence of additional, unidentified box H/ACA RNAs. Furthermore, whereas the S. cerevisiae stand-alone PUS Pus7 was predominately nuclear under standard growth conditions, heat shock resulted in a pronounced relocalization of Pus7 to the cytoplasm15. The cytoplasmic relocalization may, in part, explain the increased repertoire of substrates during heat shock.
Pseudouridylation of mRNAs
Pseudouridine was traditionally thought to be restricted to various classes of non-coding RNAs. This notion was partially based on the fact that the modification machineries, particularly the box H/ACA RNPs, predominately colocalize with their substrate RNAs within the nucleolus and Cajal bodies9,16, the sites of rRNA and spliceosomal snRNA modification, respectively. However, some evidence suggests that the modification machinery is not functionally restricted to these subnuclear compartments, and that it is capable of carrying out the modification reaction elsewhere in the cell (for example, in the nucleoplasm). For instance, in Drosophila melanogaster knockouts of Coilin, which is an essential component of Cajal bodies, spliceosomal snRNAs are still pseudouridylated despite lacking detectable Cajal bodies17. In addition, the Xenopus laevis box H/ACA RNA pseudouridylation guide for U2 snRNA at positions U34 and U44 (pugU2-34/44) seems to localize to the nucleoplasm rather than to the nucleoli or Cajal bodies18. It is therefore possible that pre-mRNAs or mRNAs, which also localize to the nucleoplasm, are pseudouridylation substrates as well. In line with this, it has been reported that there are numerous small RNAs in mammalian cells that are predicted to fold into the typical box H/ACA RNA structure (hairpin–hinge–hairpin–tail) and assemble into box H/ACA RNPs19–21. Careful inspection of the guide sequences of these box H/ACA RNAs failed to identify complementarity to any of the known structurally stable non-coding RNAs, such as rRNAs, snRNAs and tRNAs. Thus, these RNAs have been dubbed ‘orphan box H/ACA RNAs’. Interestingly, many of these orphan box H/ACA RNAs exhibit tissue-specific expression and, although many of them are yet to be assigned a function, it is possible that some guide mRNA pseudouridylation in the nucleoplasm.
Recently, two studies provided evidence that the box H/ACA RNP machinery has access to mRNAs in two distinct experimental systems, namely in X. laevis oocytes and in S. cerevisiae. In one study, the X. laevis box H/ACA RNA pugU2-34/44 was altered to target the polypyrimidine tract (PPT) of a reporter adenovirus pre-mRNA22. Indeed, in vitro-transcribed adenovirus pre-mRNA, when injected into the oocytes, was susceptible to pseudouridylation. Moreover, targeted pseudouridylation of the PPT reduced RNA-backbone flexibility and prevented U2 auxiliary factor 65 kDa (U2AF65; a PPT-binding splicing factor) from binding the PPT. Thus, through targeted pseudouridylation of an important splicing regulatory element (the PPT), this study revealed a previously unknown role for RNA-backbone flexibility in U2AF65 binding.
In the other study23, the S. cerevisiae box H/ACA RNA snR81 guide’s sequence was modified to target the uridine residue of a premature translation termination codon (PTC) in the copper metallothionein 1 (CUP1)–PTC reporter system. The CUP1–PTC reporter system uses the CUP1 gene, which provides cells with resistance to copper; hence, the introduction of a PTC into the CUP1 gene renders cells sensitive to copper23. Remarkably, not only was CUP1–PTC mRNA pseudouridylated but the presence of a pseudouridine residue within the PTC promoted the incorporation of an amino acid at the pseudouridylated termination codon: under the experimental conditions used, ψAA and ψAG directed both serine and threonine incorporation, and ψGA directed tyrosine and phenylalanine incorporation, thus functionally converting the stop codons into sense codons23. Interestingly, these effects were mediated in part by unusual codon–anticodon interactions in the ribosome-decoding centre24. For example, during translational decoding, the tRNA–mRNA base-pair interaction is recognized by A1493 of the 18S rRNA in the ribosome-decoding centre. This nucleotide normally adopts the anti conformation; however, the decoding of ψAG by tRNASer is enabled by A1493 adopting the syn conformation. Further non-canonical interactions, including normally forbidden purine–purine base pairs at the second and third positions, were observed between ψAG and the anticodon of tRNASer.
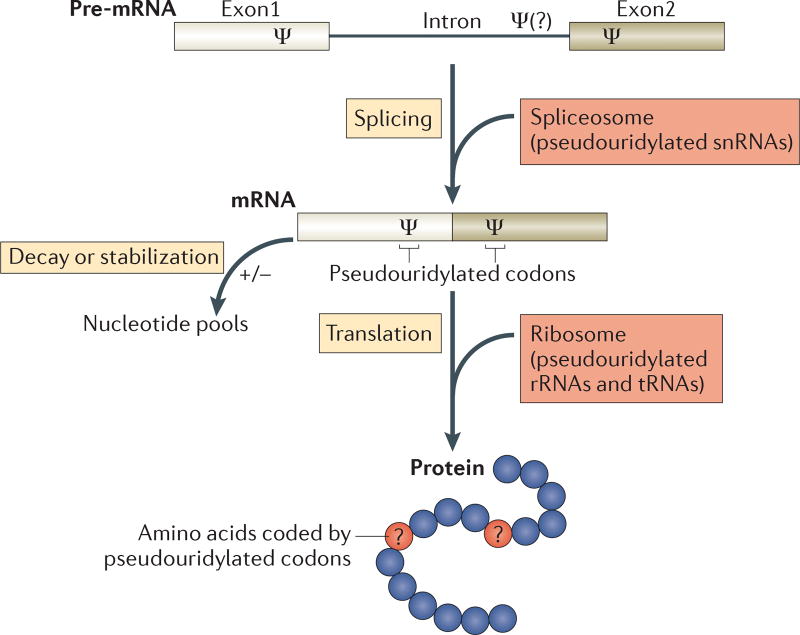
Together, these studies established that the box H/ACA RNA modification machinery has access to mRNAs and that mRNA pseudouridylation, along with the pseudouridylation of other RNAs that participate in the regulation of gene expression, can affect pre-mRNA splicing, translation fidelity and possibly mRNA stability and decay (FIG. 2 and see below). These studies further suggest that mRNA pseudouridylation may be yet another means to increase proteome diversity.
Figure 2. Possible roles of pseudouridines in gene regulation.
Pseudouridine (Ψ) nucleosides are introduced into pre-mRNAs at coding and non-coding exons and presumably also at introns. Given that they are present in pre-mRNAs and mRNAs, as well as in non-coding RNAs (such as spliceosomal small nuclear RNAs (snRNAs), ribosomal RNAs (rRNAs) and tRNAs) that are involved in every step in the pathway (splicing, translation and mRNA decay), pseudouridylation is likely to have a complex role in the regulation of gene expression. In fact, the functions of some pseudouridine residues in snRNAs, rRNAs and tRNAs have already been well characterized. In the schematic diagram of a protein (bottom), the blue circles represent amino acids coded by unmodified codons and the red circles (with question marks) represent amino acids coded by pseudouridylated codons. Although it is known that the pseudouridylation of nonsense (stop) codons can result in the suppression of translation termination, it is not clear whether pseudouridylation of sense codons will lead to changes in coding specificity.
The pseudouridylation of stop codons has been shown to promote a decrease in the efficiency of translation termination, whereas the effect of pseudouridylation on the decoding of sense codons is unclear. Molecular modelling data suggest a possibility that the pseudouridylation of specific codons may lead to altered tRNA–mRNA interactions; for example, pseudouridylation of the phenylalanine codon UUU (generating ψUU) could result in the incorporation of either cysteine or tyrosine25. Interestingly, when transfected into mammalian cells, in vitro-transcribed pseudouridine-containing mRNA is capable of generating a functional protein, suggesting that the decoding of sense codons is perhaps not grossly affected26. By contrast, the incorporation of N6-methyladenosine (m6A), a naturally occurring and abundant mRNA modification, into an in vitro-transcribed mRNA prevented translation and resulted in no protein being produced26.
Pseudouridylation goes global
The indications that mRNAs and potentially other non-coding RNAs are pseudouridylated sparked great interest in characterizing the global landscape of RNA pseudouridylation27. Identifying new pseudouridine residues within RNA has historically relied on targeted approaches, particularly the production of chemical derivatives of pseudouridines within total RNA followed by transcript-specific reverse transcription. This approach is based on the preferential reaction of CMCT (N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate) with uracil, guanine and pseudouridine residues28. Incubation of the CMCT-modified RNA at alkaline pH (pH 10.3) results in the hydrolysis of U-CMCT and G-CMCT adducts, which are less stable than ψ-CMCT adducts. The remaining ψ-CMCT adducts, which block the passage of reverse transcriptase, are then revealed as pauses or premature stops detected by primer extension.
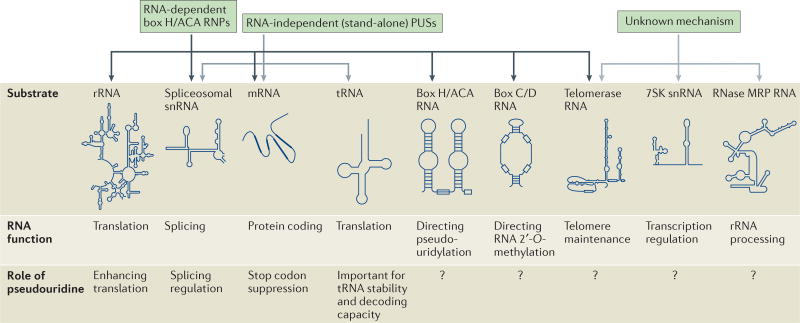
Recently, by coupling the production of CMCT derivatives with deep-sequencing and bioinformatics analyses, three groups have reported transcriptome-wide maps of pseudouridylation in S. cerevisiae and human cells13–15. Together, these studies have revealed remarkable complexity in the global landscape of RNA pseudouridylation (FIG. 3). For example, numerous pseudouridine residues were identified in box H/ACA RNAs, box C/D RNAs (which guide site-specific RNA 2′-O-methylation), telomerase RNA, ribonuclease mitochondrial RNA processing RNA (RNase MRP RNA; the catalytic component of an RNP complex involved in 5.8S rRNA processing and mitochondrial DNA replication) and 7SK snRNA (an abundant, non-coding RNA that regulates the activity of positive transcription elongation factor b (P-TEFb)).
Figure 3. The occurrence and function of pseudouridines in various eukaryotic RNAs.
Pseudouridines have been identified in a range of eukaryotic RNAs. Shown are typical secondary structures of the substrates (dark arrows) and potential substrates (light arrows) of the two types of pseudouridylation machineries — box H/ACA ribonucleoproteins (RNPs) and RNA-independent (stand-alone) pseudouridine synthases (PUSs). Also indicated are the known functions of the RNA substrates and of some pseudouridines in certain RNAs (unknown functions are denoted by question marks). rRNA, ribosomal RNA; RNase MRP RNA, ribonuclease mitochondrial RNA-processing RNA; snRNA, small nuclear RNA.
In addition to novel pseudouridine residues within non-coding RNA species previously suggested to be devoid of any modifications, a large number of pseudouridines were detected within mRNAs. Specifically, 50–100 pseudouridine residues were identified in yeast mRNAs and 100–400 pseudouridine residues were found in human mRNAs. The variable number of pseudouridine residues identified in the different studies is a reflection of sequencing depth and methods. Although all three studies concluded that the majority of the newly identified pseudouridines are catalysed by the stand-alone PUSs (PUS1–PUS4, PUS6, PUS7 and PUS9), several of the new pseudouridylations were catalysed by box H/ACA RNPs13–15. Remarkably, mRNA pseudouridylation was also found to be highly inducible, and both the stand-alone PUSs and box H/ACA RNAPs catalysed inducible pseudouridylation.
In mRNAs, pseudouridine residues were detected within 5′ untranslated regions (5′ UTRs), coding sequences and 3′ UTRs, with no clear positional bias. This is in stark contrast to m6A, which shows clear positional biases towards long internal exons, the vicinity of stop codons and 3′ UTRs29,30. Furthermore, whereas m6A seems to accelerate mRNA decay, it was noted that mRNAs containing heat-shock-induced, Pus7-dependent pseudouridines were 25% more highly expressed in wild-type S. cerevisiae as compared with in Pus7-deficient cells, raising the possibility that pseudouridine enhances mRNA stability15,31. This is in line with a previous study demonstrating that in vitro-transcribed mRNAs containing pseudouridines that were transfected into mammalian cells or administered intravenously into mice displayed enhanced stability relative to uridine-containing in vitro-transcribed mRNA26.
Interestingly, evolutionary conservation of mRNA pseudouridylation was also found. For instance, pseudouridylation of 60S ribosomal protein L11-A mRNA was conserved in Saccharomyces mikatae, and tef1 mRNA pseudouridylation was conserved in both S. mikatae and Schizosaccharomyces pombe14. These results suggest that mRNA pseudouridylation may provide an evolutionary advantage, as the last common ancestor of S. cerevisiae and S. pombe is estimated to have lived 600 million years ago.
Transcriptome-wide quantitative mapping of pseudouridine (Psi-seq) was also used to monitor pseudouridine levels in fibroblasts from patients suffering from X-linked dyskeratosis congenital (X-DC), a rare bone-marrow-failure disorder resulting from mutations in components of the box H/ACA RNA modification or biogenesis machinery15,32. Although X-DC has primarily been considered a result of telomere dysfunction, recent studies have suggested that rRNA pseudouridylation contributes to disease pathology and severity33–35. Indeed, Psi-seq independently confirmed that rRNA pseudouridylation is indeed reduced, on average, by 10% per modified site in X-DC patients15. Interestingly, the study further revealed that pseudouridylation of telomerase RNA is also reduced (the mechanism of telomerase RNA pseudouridylation is unknown). These findings further support the notion that RNA pseudouridylation is disrupted in X-DC, and they suggest that both RNA pseudouridylation and telomerase dysfunction contribute to X-DC pathology, perhaps in a mechanistically coupled manner through the pseudouridylation of telomerase RNA.
Future perspectives
Although great progress has been made in recent years in identifying pseudouridylation in various RNAs (FIG. 3), the functions of pseudouridylation in the vast majority of RNAs remain unknown. Perhaps the most interesting question to be addressed pertains to the function of mRNA pseudouridylation. Given that pseudouridines are detected in various parts of mRNAs, it remains to be seen whether these pseudouridines play a part in pre-mRNA splicing, mRNA stability and/or protein coding (FIG. 2). In addition, the mechanism by which stress induces pseudouridylation deserves closer attention. A possible explanation is that, under conditions of stress, PUS proteins are rapidly modified post-translationally, resulting in reduced substrate specificity and pseudouridylation of uridine residues that are not normally modified. Finally, the role of pseudouridylation and the pseudouridylation machinery in human disease is certainly deserving of more attention. For instance, besides X-DC, mutations in the pseudouridylation machinery (both in stand-alone PUSs and box H/ACA RNPs) are associated with a range of diseases, including mitochondrial myopathy and sideroblastic anaemia, and pituitary adenoma36,37. Furthermore, various box H/ACA RNAs have been shown to exert an effect on viral infection38. Revealing the role of pseudouridylation in the pathology of diseases may open the door to novel therapeutic strategies. Although pseudouridylation is a long-established modification, its cellular repertoire and functions continue to amass, and future studies are sure to reveal more surprises.
Acknowledgments
The authors would like to thank members of the Yu laboratory for insightful discussions.
Footnotes
Competing interests
The authors declare no competing interests.
Contributor Information
John Karijolich, Department of Plant and Microbial Biology, University of California, 565 Li Ka Shing Center #3370, Berkeley, California 94720-337, USA.
Chengqi Yi, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Synthetic and Functional Biomolecules Center and Peking-Tsinghua Center for Life Sciences, Peking University, 5 Summer Palace Road, Haidian District, Beijing 100871, China.
Yi–Tao Yu, Department of Biochemistry and Biophysics, University of Rochester Medical Center, School of Medicine and Dentistry, 601 Elmwood Avenue, Box 712 Rochester, New York 14642, USA.
References
- 1.Cohn WE. Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J. Cell. Physiol. Suppl. 1951;38:21–40. doi: 10.1002/jcp.1030380405. [DOI] [PubMed] [Google Scholar]
- 2.Cohn WE. 5-ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim. Biophys. Acta. 1959;32:569–571. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 3.Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem. 1960;235:1488–1498. [PubMed] [Google Scholar]
- 4.Arnez JG, Steitz TA. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 5.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 6.Karijolich J, Kantartzis A, Yu Y-T. RNA modifications: a mechanism that modulates gene expression. Methods Mol. Biol. 2010;629:1–19. doi: 10.1007/978-1-60761-657-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine) RNA Biol. 2014;11:1540–1554. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 9.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, Xiao M, Yang C, Yu Y-T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier UT. Pseudouridylation goes regulatory. EMBO J. 2011;30:3–4. doi: 10.1038/emboj.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 2014;8:966–973. doi: 10.1016/j.celrep.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in. S. cerevisiae. PLoS ONE. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darzacq X, et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell. 2009;20:5250–5259. doi: 10.1091/mbc.E09-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Li Z-H, Terns RM, Terns MP, Yu Y-T. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA. 2002;8:1515–1525. [PMC free article] [PubMed] [Google Scholar]
- 19.Huttenhofer A, et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitali P, et al. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Zhao X, Kierzek R, Yu Y-T. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol. Cell. Biol. 2010;30:4108–4119. doi: 10.1128/MCB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karijolich J, Yu Y-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez IS, et al. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA. 2012;18:355–367. doi: 10.1261/rna.031351.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kariko K, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishore S, et al. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013;14:R45. doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 29.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 30.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier UT. Dissecting dyskeratosis. Nat. Genet. 2003;33:116–117. doi: 10.1038/ng0203-116. [DOI] [PubMed] [Google Scholar]
- 33.Ruggero D, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 34.Bellodi C, et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013;3:1493–1502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 36.Bellodi C, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray JL, Sheng J, Rubin DH. A role for H/ ACA and C/D small nucleolar RNAs in viral replication. Mol. Biotechnol. 2014;56:429–437. doi: 10.1007/s12033-013-9730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]