Abstract
There is increasing interest in discovering HDAC6 selective inhibitors as chemical probes to elucidate the biological functions of HDAC6 and ultimately as new therapeutic agents. Small-molecular fluorescent probes are widely used to detect target protein location and function, identify protein complex composition in biological processes of interest. In the present study, structural modification of the previously reported compound 4MS leads to two novel fluorescent HDAC inhibitors, 6a and 6b. Determination of IC50 values against the panel of Zn2+ dependent HDACs (HDAC1-11) reveals that 6b is a HDAC6 selective inhibitor, which can induce hyperacetylation of tubulin but not histone H4. Importantly, fluorescent and immunofluorescent analyses of cells treated with the proteasome inhibitor MG132 demonstrates that 6b can selectively target and image HDAC6 within the inclusion body, the aggresome. These results identify 6b not only as a HDAC6 selective inhibitor but also as a fluorescent probe for imaging HDAC6 and investigating the roles of HDAC6 in various physiological and pathological contexts.
Keywords: HDAC6, fluorescent probe, aggresome, neurodegenerative disease
Graphical abstract
Introduction
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl groups from the lysine residues located on histone and non-histone proteins. Such posttranslational modifications play various physiological and pathological roles [1–3]. HDACs family contains 18 isoforms, which can be categorized into four classes: class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9 and 10) and class IV (HDAC11) HDACs are Zn2+ dependent metalloproteases that are mechanistically distinct from NAD+ dependent class III HDACs (Sirtuins 1–7) [4,5].
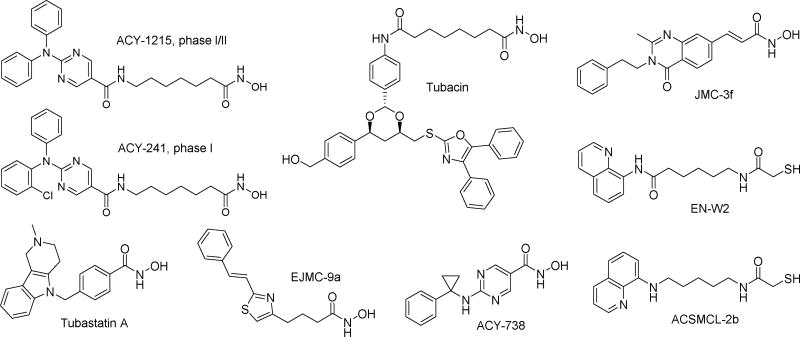
Compared with other Zn2+ dependent HDACs, HDAC6 has distinctive structural and functional profiles. HDAC6 is a cytoplasmic enzyme that uniquely features two catalytic domains [6]. Cellular substrates of HDAC6 include the cytoskeletal proteins α-tubulin [7], the chaperone Hsp90 [8], cortactin [9], peroxiredoxins I/II [10] and so on. In addition to two catalytic domains, HDAC6 also contains a zinc finger ubiquitin-binding domain and a dynein motor binding domain, through which polyubiquitinated misfolded protein cargo is recruited by HDAC6 to dynein motors for transport to aggresomes, then degraded by autophagy [11–13]. Through its direct deacetylation of various non-histone substrates, as well as its association with other interacting proteins, HDAC6 participates in multiple important biological processes, such as cell motility, cell survival, cell signaling, angiogenesis, transcription, inflammation and protein degradation [14]. Interestingly, genetic ablation or pharmacological inhibition of HDAC6 does not cause lethality or toxicity typically associated with pan-HDACs inhibition [15–18]. Therefore, modulation of HDAC6 with selective inhibitors is very attractive and holds great potential for treating numerous diseases ranging from cancer, autoimmunity to neurodegeneration [19]. In the oncology field, two moderately selective HDAC6 inhibitors ACY-1215 and ACY-241 (Fig. 1) have entered cancer clinical trials, alone or in combination with other antitumor agents [20]. In the immunology field, selective inhibition of HDAC6, using hydroxamate-based compounds such as tubacin [21,22], tubastatin A [22–25] and its analogues [26], EJMC-9a [27] or mercaptoacetamide-based compounds such as ACSMCL-2b [28] (Fig. 1), have exhibited preventive or therapeutic efficacy in various preclinical in vitro or in vivo studies. In the neurology field, recent preclinical advances of HDAC6 inhibitors focus on their therapeutic potential in Alzheimer's disease (AD). In diverse cellular or animal models, pleiotropic anti-AD effects and mechanisms have been observed for HDAC6 selective inhibitors tubacin [29–31], tubastatin A [32–34], ACY-1215 [34], ACY-738 [35,36], JMC-3f [37] and EN-W2 [38] (Fig. 1).
Figure 1. Chemical structures of representative HDAC6 selective inhibitors.
Despite these significant advances, the exact roles of HDAC6 in many pathological contexts are still obscure. In fact, HDAC6 can function as either tumor inducer or tumor suppressor depending on cancer type and stage [39]. With respect to neurodegenerative diseases, though HDAC6 inhibition is a promising therapy for AD, HDAC6 activity seems to be required to prevent progression of Parkinson's and Huntington's diseases [39]. Therefore, more selective and potent HDAC6 inhibitors are warranted to clarify the roles of HDAC6 in a specific disease and to verify the therapeutic potential of targeting HDAC6. In this regard, small-molecular fluorescent probes are powerful tools not only to visualize biological molecules of interest, but also to provide dynamic information regarding the localization and quantity of these target molecules [40,41].
Here, we describe the discovery of a fluorescent HDAC6 selective inhibitor 6b. In HDACs isoform inhibition assays, 6b demonstrates high HDAC6 selectivity over the other Zn2+ dependent HDACs. Western blot analysis further confirms the intracellular HDAC6 selective inhibition of 6b, which is comparable with that of the well-known HDAC6 inhibitor tubastatin A. Fluorescent and immunofluorescent staining results show that the HDAC6 selectivity endows 6b with the distinctive ability of labelling and visualizing HDAC6 enriched inclusion body, aggresomes. To the best of our knowledge, 6b is the first reported fluorescent HDAC6 selective inhibitor, which is of great interest as chemical probe for cellular HDAC6 detection and inhibition.
Results
Compound design and synthesis
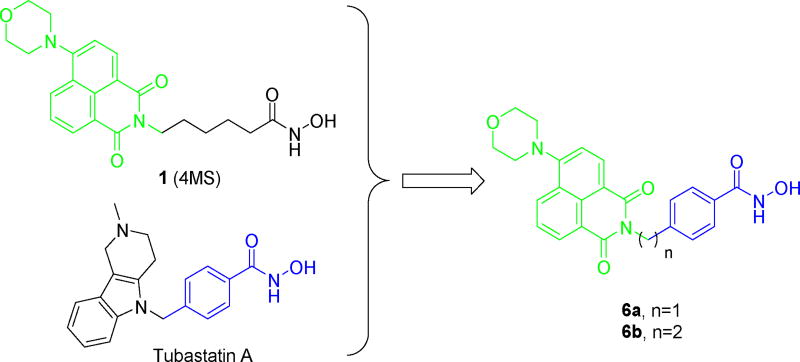
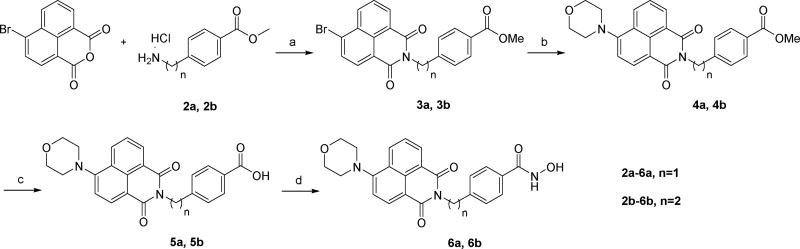
Recently, a fluorescent HDACs inhibitor 1 (4MS, Fig. 2) was developed for cellular imaging [42]. Based on the comprehensive understanding of the structure-selectivity relationships (SSRs) of HDAC6 inhibitor, we believe it was possible to create the fluorescent HDAC6 selective inhibitor by combination of the fluorophore of 1 and the para-N-hydroxybenzamide fragment of tubastatin A (Fig. 2). Compounds 6a and 6b were prepared according to the synthetic methods of 1 with minor modification (Scheme 1).
Figure 2. Compound design strategy.
The 1,8-naphthalimide-based fluorophore is indicated in green, the para-N-hydroxybenzamide fragment is indicated in blue.
Scheme 1.
Reagents and conditions: a) Et3N, EtOH, microwave, 100°C, 48–82%; b) morpholine, xantphos, Pd(dba)2, Cs2CO3, toluene, 65°C, 30–49%; c) LiOH, H2O/THF, 93–97%; d) (COCl)2, DMF, DCM, then NH2OH.HCl, Et3N, H2O/THF, 21–24%.
HDACs isoform selectivity profile
HDACs isoform inhibitory results (Table 1) showed that our compound 6a, as well as the previously reported fluorescent HDACs inhibitor 1, exhibited little discrimination among HDAC1/2/3/6. The HDACs inhibitory potency of 6a was comparable to that of the approved pan-HDACs inhibitor suberoylanilide hydroxamic acid (SAHA). In comparison with 6a and 1, 6b exhibited satisfying HDAC6 selectivity with the selective index of over 140, 700 and 210 against HDAC1/2/3, respectively. The IC50 values of 6b against HDAC1-11 in comparison with the reported data of tubastatin A were presented in Table S1 (Supporting Information), which confirmed the HDAC6 selectivity of 6b.
Table 1.
HDACs inhibitory activity and isoform selectivity
| Cpd | IC50 (nM) a | |||
|---|---|---|---|---|
|
| ||||
| class I | class IIb | |||
|
|
|
|||
| HDAC1 | HDAC2 | HDAC3 | HDAC6 | |
| 6a | 36.2±1.3 | 98.8±18.9 | 11.1±0.3 | 2.7±0.6 |
| 6b | >20000 | >100000 | >30000 | 139.0±1.4 |
| 1 | 5.25±0.65 | 25.6±0.45 | 2.65±0.15 | 0.65±0.15 |
| SAHA | 69.0±7.0 | 91.6±12.4 | 29.7±2.1 | 24.2±1.7 |
Assays were performed in replicate (n = 3); IC50 values are shown as mean ± SD.
Western blot analysis
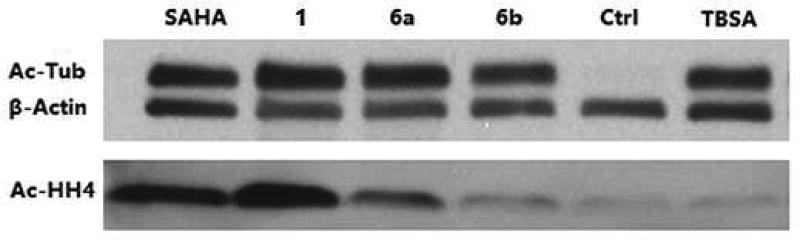
The intracellular HDACs inhibition and selectivity of compounds 6a, 6b and 1 were evaluated by western blot analysis, using a pan-HDACs inhibitor SAHA and a HDAC6 selective inhibitor tubastatin A as positive controls. Similar to SAHA, 1 simultaneously increased levels of the HDAC6 substrate acetylated α-tubulin (Ac-Tub) and the HDAC1/2/3 substrate acetylated histone H4 (Ac-HH4). Compared with 1, both 6a and 6b exhibited improved HDAC6 selectivity, validating the rationality of our compound design strategy. More importantly, at the concentration of 500 nM, 6b could potently induce α-tubulin acetylation without effecting histone H4, confirming the excellent intracellular HDAC6 selectivity of 6b, which was comparable to that of tubastatin A (Fig. 3).
Figure 3. Western blot results.
A549 cells were treated with 500 nM compounds or DMSO for 3h. The levels of α-tubulin acetylation and histone H4 acetylation were determined by immunoblotting. β-Actin was used as a loading control. TBSA represents tubastatin A.
Fluorescent and immunofluorescent staining results
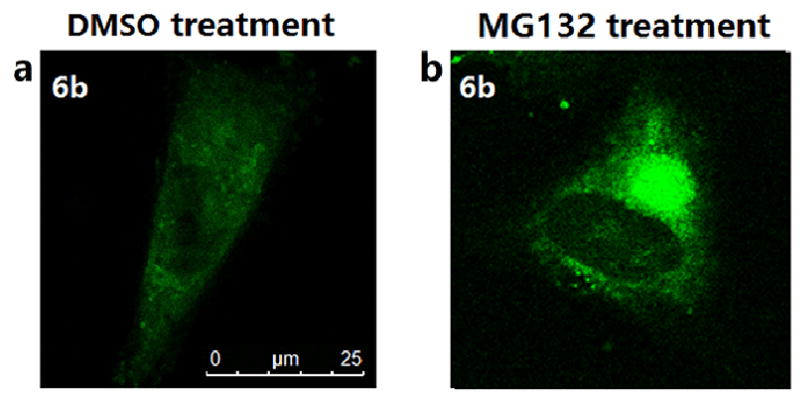
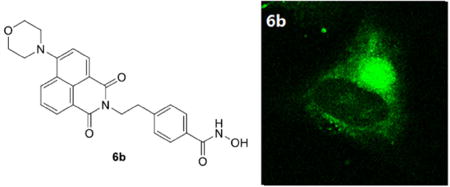
We next asked whether the HDAC6 selective inhibitor 6b could work as a fluorescent probe for cellular HDAC6 imaging. Considering HDAC6 is a component of aggresomes [13], which could be induced by proteasome inhibitor MG132 [43], we observed the intracellular distribution of 6b in A549 cells treated with DMSO or MG132 by confocal microscopy (Fig. 4). In DMSO-treated cells, 6b displayed a diffuse staining mainly in the cytoplasm (Fig. 4a), which was similar to the previously reported cellular imaging of 1 [42]. Strikingly, treatment of cells with MG132 resulted in a dramatic relocation of 6b to a single bright juxtanuclear structure (Fig. 4b), indicating that 6b could target HDAC6 concentrated in aggresomes.
Figure 4. Cellular imaging of A549 cells stained with 6b (green).
A549 cells were treated with DMSO or 5 µM MG132 for 24 h, then incubated with 2 µM 6b for 2 h.
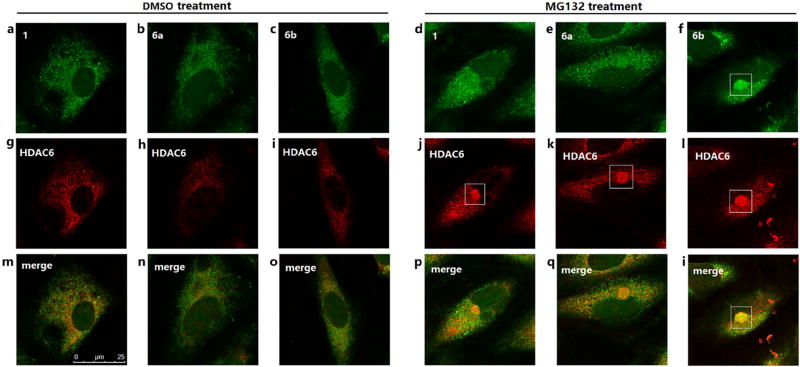
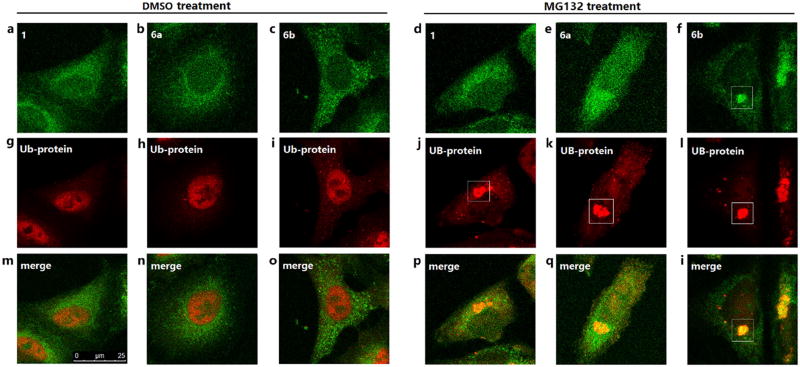
We also compared the cellular imaging behaviors of 1, 6a and 6b. In DMSO-treated cells, no significant difference was observed for these three fluorescent HDACs inhibitors (Fig. 5a–5c, Fig. 6a–6c). However, in MG132-treated cells, 6b could form obvious aggresome-like structures (Fig. 5f, Fig. 6f), which were not observed in cells stained with 1 (Fig. 5d, Fig. 6d) or 6a (Fig. 5e, Fig. 6e). We further characterized the observed 6b-enriched aggresome-like structure for the presence of HDAC6 and another component of aggresomes: polyubiquitin. As shown by staining results, the 6b-enriched structure colocalized with HDAC6 (Fig. 5l) and polyubiquitin (Fig. 6l), verifying that 6b could label and image aggresomes by selectively targeting HDAC6. These results indicated the higher selectivity but not the higher inhibition towards HDAC6 should be the main determinant of intracellular imaging, probably because the higher HDAC6 selectivity generated lower fluorescent background and higher contrast.
Figure 5. Cellular imaging of A549 cells double stained with fluorescent compounds (green) and HDAC6 antibody (red).
A549 cells were treated with DMSO or 5 µM MG132 for 24 h, then sequentially incubated with HDAC6 antibody overnight and 2 µM fluorescent compounds for 2 h.
Figure 6. Cellular imaging of A549 cells double stained with fluorescent compounds (green) and polyubiquitin antibody (red).
A549 cells were treated with DMSO or 5 µM MG132 for 24 h, then sequentially incubated with polyubiquitin antibody overnight and 2 µM fluorescent compounds for 2 h.
Conclusions and Discussion
Inspired by the previously reported fluorescent HDACs inhibitor 1, two new fluorescent HDACs inhibitors 6a and 6b have been discovered. General design strategy for fluorescent probe uses a linker to connect a recognition moiety and an additional fluorophore, which often generates a large probe with high molecular weight and poor physicochemical property. In our case, 6a and 6b possess low molecular weights of less than 400 daltons because their fluorophore is also a part of the recognition moiety. Most importantly and different from 1 and 6a, 6b could label and image the proteasome inhibitor MG132-induced aggresomes due to its high HDAC6 selectivity. Considering aggresomes are similar to inclusion bodies, such as Lewy bodies, commonly found in many neurodegenerative diseases [13,43,44], our study provides a proof-of-concept for discovering effective imaging probes for diagnosis of Parkinson’s and other neurodegenerative diseases.
As a fluorescent probe, 6b could also be useful to visualize HDAC6 and other HDAC6 associated protein complex in various physiological and pathological contexts. In comparison to immunofluorescent staining using antibodies, HDAC6 targeting fluorescent probe 6b is more convenient, more economical and less time-consuming. 6b will also facilitate dynamic imaging of HDAC6 in live cells because HDAC6 selective inhibitors show extremely low cytotoxicity. It is worth noting that if one biological process imaged by HDAC6 immunofluorescent staining can also be detected by 6b in live cells, it means this biological process is not HDAC6 catalytic activity dependent; if one biological process can only be imaged by HDAC6 immunofluorescent staining but not 6b, it indicates this biological process is probably HDAC6 catalytic activity dependent. Therefore, simultaneous HDAC6 imaging and HDAC6 inhibition by 6b in live cells will help to determine one biological process is HDAC6 catalytic activity dependent or not, which will facilitate the investigation of HDAC6 biological function and subsequent discovery of HDAC6 inhibitors as therapeutic agents.
Experimental Section
Chemistry
All starting materials, reagents and solvents were commercially available. All reactions except those in aqueous media were carried out by standard techniques for the exclusion of moisture. All reactions were monitored by thin-layer chromatography on 0.25mm silica gel plates (60GF-254) and visualized with UV light, ferric chloride or iodine vapor. NMR spectrums were determined on Bruker DRX spectrometer, δ in parts per million and J in Hertz. ESI-MS spectrums were determined on an API 4000 spectrometer. HRMS spectrums were conducted by Shandong Analysis and Test Center. Silica gel was used for column chromatography purification. All tested compounds are >95% pure by HPLC analysis.
N-hydroxy-6-(6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)hexanamide (1) was synthesized according to the previously reported methods [42].
General Procedure for the Preparation of Compounds 3a and 3b
Methyl 4-((6-bromo-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)methyl)benzoate (3a)
Starting materials 6-bromo-1H,3H-benzo[de] isochromene-1,3-dione (1.35 g, 4.87 mmol), 2a (982 mg, 4.87 mmol) and Et3N (493 mg, 4.87 mmol) were taken up into a microwave tube in EtOH (20 mL). The sealed tube was heated at 100°C for 45 min under microwave. The resulting mixture was diluted with H2O (100 mL) and extracted with EtOAc (3 × 20 mL). The combined organic layers were washed with 0.1 M HCl (10 mL), saturated NaHCO3 (10 mL) and brine (10 mL), then dried over Na2SO4, filtered and the solvent was evaporated to afford compound 3a (1.00 g, 2.36 mmol, 48% yield) as yellow solid, which was used directly in next step without any purification. 1H NMR (400 MHz, DMSO-d6) δ 8.58–8.62 (m, 2H), 8.37 (d, J = 8.0 Hz, 1H), 8.25 (d, J = 8.0 Hz, 1H), 8.02 (t, J = 7.6 Hz, 1H), 7.89 (d, J = 8.0 Hz, 2H), 7.49 (d, J = 8.0 Hz, 2H), 5.31 (s, 2H), 3.82 (s, 3H). ESI-MS m/z: 424.1 [M+H]+.
Methyl 4-(2-(6-bromo-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)ethyl)benzoate (3b)
Yellow solid (yield: 82%). 1H NMR (400 MHz, DMSO-d6) δ 8.57–8.60 (m, 2H), 8.35 (d, J = 7.6 Hz, 1H), 8.24 (d, J = 8.0 Hz, 1H), 8.02 (t, J = 8.4 Hz, 1H), 7.89 (d, J = 8.4 Hz, 2H), 7.43 (d, J = 8.0 Hz, 2H), 4.27–4.31 (m, 2H), 3.84(s, 3H), 3.01–3.06 (m, 2H). ESI-MS m/z: 438.2 [M+H]+.
General Procedure for the Preparation of Compounds 4a and 4b
Methyl 4-((6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)methyl)benzoate (4a)
To a mixture of compound 3a (1.00 g, 2.36 mmol) and morpholine (617 mg, 7.08 mmol) in toluene (10 mL) was added Xantphos (54.6 mg, 94.4 µmol), Pd(dba)2 (54.3 mg, 94.4 µmol) and Cs2CO3 (2.31 g, 7.09 mmol) under N2. The mixture was stirred at 65°C for 12 h. The solid was filtered. The solvent was evaporated and the residue was purified by column (petroleum ether: EtOAc=20:1 to 3:1) to give compound 4a (500 mg, 1.16 mmol, 49% yield) as yellow solid. 1H NMR (400 MHz, CDCl3) δ 8.55–8.63 (m, 2H), 8.45 (d, J = 8.4 Hz, 1H), 7.97 (d, J = 8.4 Hz, 2H), 7.70–7.74 (m, 1H), 7.57 (d, J = 8.8 Hz, 2H), 7.25 (d, J = 8.4 Hz, 1H), 5.47 (s, 2H), 4.02 (t, J = 4.4 Hz, 4H), 3.89 (s, 3H), 3.28 (t, J = 4.4 Hz, 4H). ESI-MS m/z: 431.2 [M+H]+.
Methyl 4-(2-(6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)ethyl)benzoate (4b)
Yellow solid (yield: 30%). 1H NMR (400 MHz, CDCl3) δ 8.53–8.61 (m, 2H), 8.43–8.46 (m, 1H), 7.98 (d, J = 8.0 Hz, 2H), 7.72–7.75 (m, 1H), 7.43 (d, J = 8.4 Hz, 2H), 7.25 (d, J = 7.6 Hz, 1H), 4.39–4.44 (m, 2H), 4.04 (t, J = 4.4 Hz, 4H), 3.91 (s, 3H), 3.29 (t, J = 4.4 Hz, 4H), 3.07–3.11 (m, 2H). ESI-MS m/z: 445.2 [M+H]+.
General Procedure for the Preparation of Compounds 5a and 5b
4-((6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)methyl)benzoic acid (5a)
To a solution of compound 4a (500 mg, 1.16 mmol) in H2O (5 mL) and THF (5 mL) was added LiOH.H2O (486 mg, 11.6 mmol). The mixture was stirred at room temperature for 12 h. The THF was evaporated. 2M HCl was added to the mixture until pH = 3. The water phase was extracted with DCM (20 mL × 3). The combined organic phase was washed with brine (20 mL), dried over Na2SO4 and evaporated to give compound 5a (470 mg, 1.13 mmol, 97% yield) as yellow solid, which was used directly in next step without any purification. 1H NMR (400 MHz, CDCl3) δ 8.55–8.63 (m, 2H), 8.45 (d, J = 8.4 Hz, 1H), 8.02 (d, J = 8.4 Hz, 2H), 7.70–7.75 (m, 1H), 7.60 (d, J = 8.4 Hz, 2H), 7.25 (d, J = 8.4 Hz, 1H), 5.44 (s, 2H), 4.03 (t, J = 4.4 Hz, 4H), 3.28 (t, J = 4.4 Hz, 4H). ESI-MS m/z: 417.2 [M+H]+.
4-(2-(6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)ethyl)benzoic acid (5b)
Yellow solid (yield: 93%). 1H NMR (400 MHz, CDCl3) δ 8.54–8.62 (m, 2H), 8.44–8.47 (m, 1H), 8.03 (d, J = 8.0 Hz, 2H), 7.71–7.76 (m, 1H), 7.46 (d, J = 8.4 Hz, 2H), 7.25 (d, J = 6.4 Hz, 1H), 4.41–4.45 (m, 2H), 4.03 (t, J = 4.4 Hz, 4H), 3.28–3.34 (m, 4H), 3.09–3.14 (m, 2H). ESI-MS m/z: 431.2 [M+H]+.
General Procedure for the Preparation of Compounds 6a and 6b
N-hydroxy-4-((6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)methyl)benza mide (6a)
To a solution of compound 5a (470 mg, 1.13 mmol) in DCM (10 mL) was added (COCl)2 (301 mg, 2.37 mmol) and DMF (100 µL). The mixture was stirred at room temperature for 0.5 h. Then the mixture was poured into the solution of NH2OH.HCl (314 mg, 4.52 mmol) and Et3N (686 mg, 6.78 mmol) in THF (5 mL) and H2O (2.5 mL). The mixture was stirred at room temperature for 1 h. DCM and THF was evaporated. The residue was taken up with H2O and extracted with DCM (20 mL × 3). The combined organic phase was washed with brine (20 mL) and dried over Na2SO4. The solvent was evaporated to give 6a (100 mg, 232 µmol, 21% yield) as yellow solid. 1H NMR (400 MHz, DMSO-d6) δ 11.19 (s, 1H), 9.01 (s, 1H), 8.50 (t, J = 8.0 Hz, 2H), 8.42 (d, J = 8.0 Hz, 1H), 7.82 (t, J = 8.0 Hz, 1H), 7.68 (d, J = 8.0 Hz, 2H), 7.35–7.40 (m, 3H), 5.27 (s, 2H), 3.92 (s, 4H), 3.23 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 164.51, 164.10, 163.55, 156.20, 141.16, 133.00, 132.02, 131.46, 131.35, 129.73, 127.69, 127.49, 126.65, 125.76, 122.89, 116.04, 115.61, 66.65, 53.50, 43.06. HRMS (AP-ESI) m/z calcd for C24H22N3O5 [M+H]+ 432.1559, found 432.1567.
N-hydroxy-4-(2-(6-morpholino-1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)ethyl)benza mide (6b)
Yellow solid (yield: 24%). 1H NMR (400 MHz, DMSO-d6) δ 11.18 (s, 1H), 9.01 (s, 1H), 8.47 (t, J = 8.0 Hz, 2H), 8.39 (d, J = 8.0 Hz, 1H), 7.81 (t, J = 8.0 Hz, 1H), 7.69 (d, J = 8.0 Hz, 2H), 7.33–7.35 (m, 3H), 4.25 (t, J = 8.0 Hz, 2H), 3.91 (s, 4H), 3.22 (s, 4H), 2.96 (t, J = 8.0 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 164.65, 163.89, 163.36, 155.97, 142.65, 132.71, 131.29, 131.18, 131.10, 129.57, 129.12, 127.52, 126.60, 125.73, 122.96, 116.20, 115.58, 66.66, 53.49, 41.10, 33.85. HRMS (AP-ESI) m/z calcd for C25H24N3O5 [M+H]+ 446.1716, found 446.1736.
Biology
HDAC1 and HDAC6 were purchased from Abcam (AB101661 and AB42632), HDAC2 and HDAC3 were purchased from SignalChem (H84-30G and H85-30G). HDACs substrate Boc-Lys(acetyl)-AMC was purchased from Bachem. Pan-HDACs inhibitor SAHA, HDAC6 selective inhibitor tubastatin A and proteasome inhibitor carbobenzoxy-Leu-Leu-leucinal (MG132) were purchased from Selleckchem. The following antibodies were used for western blot: acetylated α-tubulin antibody (Sigma: T6793, 1:1000), β-actin antibody (Sigma: A1978, 1:5000), acetylated histone H4 antibody (Abcam: ab15823, 1:1000), anti-mouse IgG HRP conjugate (Promega: W402B, 1:10000) and anti-rabbit IgG HRP conjugate (Promega: W401B, 1:10000). The following antibodies were used for immunofluorescent staining: HDAC6 antibody (Santa Cruz: sc-11420, 1:200), ubiquitin antibody (Millipore: 05-1307, 1:500) and Alexa Fluor® 647 goat anti-rabbit IgG (Invitrogen: A21245, 1:200).
HDACs inhibition assays
In vitro HDACs inhibition assays were conducted as previously [45]. In brief, the compounds were diluted in 10% DMSO, and 5 µL of the dilution was added to a 50 µL reaction mixture containing HDAC isoform and substrate. The deacetylation reactions were conducted at 37 °C for 30 min, then stopped by addition of 100 µL of developer containing trypsin and Trichostatin A (TSA). 20 min later, fluorescence was then analyzed with excitation at 350–360 nm and emission at 450–460 nm using a SpectraMax M5 microplate reader. The IC50 values were calculated based on the amounts of fluorescent product.
Cell culture
The human lung carcinoma cell line A549 (ATCC) was cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum, penicillin-streptomycin 100U/mL, and maintained in a humidified incubator at 37 °C with 5% CO2.
Western blotting analysis
After treatment, cells were washed twice with cold PBS and then lysed in ice-cold RIPA buffer (10 mM Tris pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 1 mM Na3VO4, 1 mM NaF, and a protease inhibitor cocktail). Lysates were cleared by centrifugation (10,000 r.p.m. for 20 min). Protein concentrations were determined using the BCA assay (Pierce). Equal amounts of cell extracts were then resolved by SDS-PAGE, transferred to nitrocellulose membranes and probed with antibodies. Blots were detected using an ECL system (Thermo).
Fluorescent and immunofluorescent staining
For single staining using compound 6b, A549 cells were treated with 5 µM MG132 for 24 h, fixed with 4% formaldehyde in PBS for 20 min, and then incubated with 2 µM 6b in PBS for 2 h. For double staining using fluorescent compounds and antibodies, A549 cells were treated with MG132 for 24 h, fixed with 4% formaldehyde in PBS for 20 min and then permeabilized with 0.1% Tritonx-100 in PBS for 5 min at room temperature. Cells were blocked with 5% bovine serum albumin in PBS and incubated with primary antibodies overnight at 4 °C, followed by Alexa Fluor® 647 goat anti-rabbit IgG for 30 min and fluorescent compounds for 2 h. Using a Leica SP5 inverted confocal microscope, staining images of compounds 1, 6a and 6b were obtained with excitation at 405 nm and emission at 500–600 nm, while staining images of HDAC6 and ubiquitin were obtained with excitation at 633 nm and emission at 650–750 nm.
Supplementary Material
Two novel fluorescent HDAC inhibitors 6a and 6b were designed and synthesized.
6b is a HDAC6 selective inhibitor which can induce hyperacetylation of tubulin but not histone H4.
6b can selectively target and image HDAC6 within the inclusion body.
6b might also be useful to visualize other HDAC6 associated protein complex in various physiological and pathological contexts.
Acknowledgments
This work was supported by National High-Tech R&D Program of China (863 Program, 2014AA020523 to Y. Z.), Major Project of Science and Technology of Shandong Province (2015ZDJS04001, 2017CXGC1401), Young Scholars Program of Shandong University (YSPSDU, 2016WLJH33 to Y. Z.), and 2R01-NS054022 (National Institutes of Health) to T.-P.Y.
Abbreviations Used
- HDAC
histone deacetylase
- AD
Alzheimer's disease
- SSRs
structure-selectivity relationships
- SAHA
suberoylanilide hydroxamic acid
- Ac-Tub
acetylated α-tubulin
- Ac-HH4
acetylated histone H4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
HDACs isoform selectivity comparison of 6b and tubastatin A. 1H NMR, 13C NMR spectra and HPLC analysis of compounds 6a and 6b.
References
- 1.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 3.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 6.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl Acad. Sci. USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, Yao TP, Lane WS, Seto E. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parmigiani RB. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc. Natl Acad. Sci. USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hook SS, Orian A, Cowley SM, Eisenman RN. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl Acad. Sci. USA. 2002;99:13425–13430. doi: 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Shin D, Kwon SH. Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J. 2013;280:775–793. doi: 10.1111/febs.12079. [DOI] [PubMed] [Google Scholar]
- 15.Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schlüter OM, Bradke F, Lu J, Fischer A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 2013;5:52–63. doi: 10.1002/emmm.201201923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J. Am. Chem. Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivieccio MA, Brochier C, Willis DE, Walker BA, D'Annibale MA, McLaughlin K, Siddiq A, Kozikowski AP, Jaffrey SR, Twiss JL, Ratan RR, Langley B. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc. Natl Acad. Sci. USA. 2009;106:19599–19604. doi: 10.1073/pnas.0907935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalin JH, Bergman JA. Development and therapeutic implications of selective histone deacetylase 6 inhibitors. J. Med. Chem. 2013;56:6297–6313. doi: 10.1021/jm4001659. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues DA, Thota S, Fraga CA. Beyond the selective inhibition of histone deacetylase 6. Mini. Rev. Med. Chem. 2016;16:1175–1184. doi: 10.2174/1389557516666160428115959. [DOI] [PubMed] [Google Scholar]
- 21.Akimova T, Ge G, Golovina T, Mikheeva T, Wang L, Riley JL, Hancock WW. Histone/protein deacetylase inhibitors increase suppressive functions of human FOXP3+ Tregs. Clin. Immunol. 2010;136:348–363. doi: 10.1016/j.clim.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, Bradner JE, Mazitschek R, Kozikowski AP, Matthias P, Hancock WW. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol. Cell. Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vishwakarma S, Iyer LR, Muley M, Singh PK, Shastry A, Saxena A, Kulathingal J, Vijaykanth G, Raghul J, Rajesh N, Rathinasamy S, Kachhadia V, Kilambi N, Rajgopal S, Balasubramanian G, Narayanan S. Tubastatin, a selective histone deacetylase 6 inhibitor shows anti-inflammatory and anti-rheumatic effects. Int. Immunopharmacol. 2013;16:72–78. doi: 10.1016/j.intimp.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Rao YH, Inoue M, Hao R, Lai CH, Chen D, McDonald SL, Choi MC, Wang Q, Shinohara ML, Yao TP. Microtubule acetylation amplifies p38 kinase signalling and anti-inflammatory IL-10 production. Nat. Commun. 2014;5:3479. doi: 10.1038/ncomms4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhao T, Liu B, Halaweish I, Mazitschek R, Duan X, Alam HB. Inhibition of histone deacetylase 6 improves long-term survival in a lethal septic model. J. Trauma Acute Care Surg. 2015;78:378–385. doi: 10.1097/TA.0000000000000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalin JH, Butler KV, Akimova T, Hancock WW, Kozikowski AP. Second-generation histone deacetylase 6 inhibitors enhance the immunosuppressive effects of Foxp3+ T-regulatory cells. J. Med. Chem. 2012;55:639–651. doi: 10.1021/jm200773h. [DOI] [PubMed] [Google Scholar]
- 27.Yoo J, Kim SJ, Son D, Seo H, Baek SY, Maeng CY, Lee C, Kim IS, Jung YH, Lee SM, Park HJ. Computer-aided identification of new histone deacetylase 6 selective inhibitor with anti-sepsis activity. Eur. J. Med. Chem. 2016;116:126–135. doi: 10.1016/j.ejmech.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Segretti MC, Vallerini GP, Brochier C, Langley B, Wang L, Hancock WW, Kozikowski AP. Thiol-based potent and selective HDAC6 inhibitors promote tubulin acetylation and T-regulatory cell suppressive function. ACS Med. Chem. Lett. 2015;6:1156–1161. doi: 10.1021/acsmedchemlett.5b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding H, Dolan PJ, Johnson GV. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J. Neurochem. 2008;106:2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Owens GC, Makarenkova H, Edelman DB. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS One. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Y, Zhao K, Wu J, Xu Z, Jin S, Zhang YQ. HDAC6 mutations rescue human tau-induced microtubule defects in Drosophila. Proc. Natl Acad. Sci. USA. 2013;110:4604–4609. doi: 10.1073/pnas.1207586110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selenica ML, Benner L, Housley SB, Manchec B, Lee DC, Nash KR, Kalin J, Bergman JA, Kozikowski A, Gordon MN, Morgan D. Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimers Res. Ther. 2014;6:12. doi: 10.1186/alzrt241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim C, Choi H, Jung ES, Lee W, Oh S, Jeon NL, Mook-Jung I. HDAC6 inhibitor blocks amyloid beta-induced impairment of mitochondrial transport in hippocampal neurons. PLoS One. 2012;7:e42983. doi: 10.1371/journal.pone.0042983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Liu C, Wu J, Tao JJ, Sui XL, Yao ZG, Xu YF, Huang L, Zhu H, Sheng SL, Qin C. Tubastatin A/ACY-1215 improves cognition in Alzheimer's disease transgenic mice. J. Alzheimers Dis. 2014;41:1193–1205. doi: 10.3233/JAD-140066. [DOI] [PubMed] [Google Scholar]
- 35.Cook C, Carlomagno Y, Gendron TF, Dunmore J, Scheffel K, Stetler C, Davis M, Dickson D, Jarpe M, DeTure M, Petrucelli L. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014;23:104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majid T, Griffin D, Criss Z, II, Jarpe M, Pautler RG. Pharmocologic treatment with histone deacetylase 6 inhibitor (ACY-738) recovers Alzheimer’s disease phenotype in amyloid precursor protein/presenilin 1 (APP/PS1) mice. Alzheime's & Dementia: Translat. Res. Clin. Intervent. 2015;1:170–181. doi: 10.1016/j.trci.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu CW, Chang PT, Hsin LW, Chern JW. Quinazolin-4-one derivatives as selective histone deacetylase-6 inhibitors for the treatment of Alzheimer's disease. J. Med. Chem. 2013;56:6775–6791. doi: 10.1021/jm400564j. [DOI] [PubMed] [Google Scholar]
- 38.You MS, Lee T, Yoon H, Dibattista AM, Song JM, Sohn Y, Moffat EI, Turner RS, Jung M, Kim J, Hoe HS. Mercaptoacetamide-based class II HDAC inhibitor lowers Aβ levels and improves learning and memory in a mouse model of Alzheimer's disease. Exp. Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase 6 in health and disease. Epigenomics. 2015;7:103–118. doi: 10.2217/epi.14.69. [DOI] [PubMed] [Google Scholar]
- 40.Xu W, Zeng Z, Jiang J, Chang YT, Yuang L. Discerning the chemistry in individual organelles with small-molecule fluorescent probes. Angew. Chem. Int. Ed. Engl. 2016;54:13658–13699. doi: 10.1002/anie.201510721. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Gao X, Shi W, Ma H. Design strategies for water-soluble small molecular chromogenic and fluorogenic probes. Chem. Rev. 2014;114:590–659. doi: 10.1021/cr300508p. [DOI] [PubMed] [Google Scholar]
- 42.Fleming CL, Ashton TD, Nowell C, Devlin M, Natoli A, Schreuders J, Pfeffer FM. A fluorescent histone deacetylase (HDAC) inhibitor for cellular imaging. Chem. Commun. 2015;51:7827–7830. doi: 10.1039/c5cc02059j. [DOI] [PubMed] [Google Scholar]
- 43.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J. Cell. Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell. Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 45.Duan W, Li J, Inks ES, Chou CJ, Jia Y, Chu X, Li X, Xu W, Zhang Y. Design, synthesis, and antitumor evaluation of novel histone deacetylase inhibitors equipped with a phenylsulfonylfuroxan module as a nitric oxide donor. J. Med. Chem. 2015;58:4325–4338. doi: 10.1021/acs.jmedchem.5b00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.