Abstract
The cellular environment associated with coronary artery disease (CAD) can lead to mitochondrial DNA (mtDNA) damage. Mitochondrial variants in some copies of mtDNA (heteroplasmy) and mtDNA content are potential genetic biomarkers for CAD-associated disease states. Massively parallel sequencing and qRT-PCR techniques were used to measure heteroplasmic variants and mtDNA content in heart samples from donors with (n = 8) and without (n = 7) documented CAD. Both groups showed increased numbers of heteroplasmic mtDNA variants in the control region (CR) (p < 0.0010, ANOVA). The donors with CAD displayed a 41.07% increase in heteroplasmic mtDNA variant number in the CR (p = 0.043), an 87.50% increase in the number of heteroplasmic mtDNA deletions (p = 0.12), and a 48.76% increase in the number of heteroplasmic mtDNA single nucleotide variants (p = 0.029). This data suggests potential trends towards higher cardiac mtDNA heteroplasmy levels in heart samples from donors with CAD.
Keywords: coronary artery disease, mitochondrial DNA, heteroplasmy, mitochondrial variation, mitochondria
Introduction
Cardiovascular disease is the leading cause of death in the United States1. One of the most prevalent types of cardiovascular disease is coronary artery disease (CAD). CAD is characterized by the formation of a plaque (atheroma) in the coronary arteries (atherosclerosis), which can lead to ischemic cardiac conditions such as angina and myocardial infarction2. Many factors contribute to the manifestation and progression of CAD, one of which is oxidative stress. Increased levels of reactive oxygen species (ROS) can lead to vascular leukocyte infiltration, impairment of nitric oxide signaling, and other aberrations that result in the endothelial dysfunction typical of CAD. High cellular levels of ROS can also damage the mitochondrial DNA (mtDNA). The damaging pro-oxidative environment associated with CAD may contribute to the accumulation of mtDNA variants and mitochondrial dysfunction3. MtDNA damage alters the ability of the mitochondria to regulate the production of ROS, and leads to a destructive positive feedback loop that can culminate in cellular apoptosis4. The mitochondrion is being considered as a potential therapeutic target for heart diseases5,6.
It has been hypothesized that in the context of CAD, mtDNA damage would result in subpopulations of mitochondria carrying copies of mtDNA with insertions, deletions, or other mutations (heteroplasmy)6,7. In support of this notion, Botto et al., documented an increase in the incidence of the common mtDNA4977 deletion in peripheral blood cells from subjects with CAD (CAD group: 26.2% versus healthy control group: 4.5%, p = 0.03)8. Corral-Debrinski et al. reported a 7 – 220 fold increase in the levels of the mtDNA4977 deletion in heart tissue from individuals with CAD9. A recent study by Fetterman et al. showed significantly higher mtDNA lesion frequency in peripheral mononucleocytes (PMNs) in individuals with CAD versus those without CAD (p < 0.05) while noting that the mtDNA copy number did not differ between both groups10. In another study utilizing PMNs, subjects with CAD displayed a median mtDNA content of 0.65 versus a content value of 0.86 in the control group (p < 0.001); the authors concluded that depletion of mtDNA is associated with CAD11.
Currently, there is a paucity of information regarding the extent of mtDNA heteroplasmy and mtDNA depletion in cardiac tissue from individuals with and without CAD. It is reasonable to suspect that CAD may lead to an overall increase in heteroplasmic cardiac mtDNA variants and altered mtDNA content. In this pilot study, we aimed to characterize mtDNA content and heteroplasmic mtDNA variants in cardiac tissue from individuals with and without CAD using quantitative real time PCR techniques (qRT-PCR) and massively parallel sequencing, respectively. Similar qRT-PCR techniques have been used to reliably quantitate mtDNA content in various tissues12–16. Massively parallel sequencing is a robust, high throughput method that allows the identification of heteroplasmic mtDNA variants present at a frequency > 2.0%17,18. This study provides a first glance into the extent of mtDNA variability in heart tissue from donors with diagnosed CAD
Methods
Heart samples
This research was approved by the Institutional Review Board at the University at Buffalo. Heart samples from donors with (n = 8) and without CAD (n = 7) were procured from The National Disease Research Interchange and The Cooperative Human Tissue Network. The postmortem to tissue recovery interval was ≤ 10 hours. Samples (0.5 – 20 g, myocardium, left ventricle) were snap-frozen after recovery and stored in liquid nitrogen. Samples were binned into groups with and without CAD. The mean ages of both groups were not significantly different (mean ageCAD = 58.50 ± 22.08 versus mean agenon CAD = 60.86 ± 15.91 years, p = 0.64, Student’s t-test). The group of donors with documented CAD had an age range of 19 – 86 years of age while the group of donors without documented CAD had an age range of 34 – 80 years. The main demographics from donors with and without CAD are summarized in Table 1. Inclusion criteria for the group of donors with CAD included an explicit diagnosis of CAD, heart disease, or an ischemic cardiac event such as myocardial infarction in the medical history or pathology report. Inclusion criteria for the group of donors without CAD included a medical history or autopsy report that specifically noted the absence of CAD, congestive heart failure, and cardiac ischemia. Exclusion criteria from the study included absent or incomplete past medical history or pathology report, type 2 diabetes mellitus, documented chromosomal disorders (e.g. Down syndrome), documented non-cardiac ischemic events related to the presence of atherosclerosis (i.e. ischemic stroke), or documented history of cardiac transplant. Heart samples were processed following standardized procedures to isolate total cardiac DNA as previously described16.
Table 1.
Donor demographics.
| Donor Samples With CAD |
Age | Sex | Mitochondrial Haplogroup |
Cardiac Pathology |
|---|---|---|---|---|
| Sample 1 | 75 | M | HV-Group | History of myocardial infarction, CAD diagnosis |
| Sample 2 | 66 | F | L0, L1, L2 | Extensive CAD diagnosis, history of myocardial infarction |
| Sample 3 | 86 | F | L0, L1, L2 | CAD diagnosis, >50% stenosis noted in multiple coronary arteries |
| Sample 4 | 70 | M | L0, L1, L2 | History of myocardial infarction, atherosclerosis, and ischemic cardiomyopathy |
| Sample 5 | 19 | F | T | Diagnosed heart disease, increased cardiac enzymes at time off death |
| Sample 6 | 42 | M | UK | Myocardial infarct noted on autopsy, hypertensive cardiovascular disease diagnosis |
| Sample 7 | 42 | M | UK | Diagnosed severe CAD, up to 60% stenosis noted in multiple coronary arteries. |
| Sample 8 | 68 | M | H | Expired due to respiratory failure. Severe coronary artery disease noted. |
| Donor Samples Without CAD | ||||
| Sample 9 | 57 | M | H | Expired due to intracranial bleed. No history of CAD, thromboembolic disease, atherosclerosis was noted. |
| Sample 10 | 70 | M | I | Expired due to cancer. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
| Sample 11 | 67 | F | Y | Expired due to anoxia. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
| Sample 12 | 80 | F | UK | Expired due to hypoxia. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
| Sample 13 | 47 | F | V | Expired due to liver failure. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
| Sample 14 | 34 | F | V | Expired due to organ failure. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
| Sample 15 | 71 | M | H | Expired due to cancer. No history of CAD, thromboembolic disease, or atherosclerosis noted. |
Analysis of mtDNA content
Cardiac mtDNA content was measured as previously described16. Briefly, reaction mixtures for qRT-PCR were prepared using SYBR Advantage qPCR Premix (Clontech), forward and reverse primers (Table 2) for either MT-ND1 or 18S rRNA, and DNA template (10 ng). The conditions for amplification of both genes were: 95°C for 30 seconds, followed by 40 cycles at 95°C for 6 seconds and 60°C for 34 seconds. Each reaction run included negative controls and a 6-point calibration curve made with diluted control cardiac DNA (range: 0.0010 – 100 ng). Samples and standards for calibration curves were analyzed in triplicate in an iQ5 thermal cycler (Bio-Rad) and values were averaged. Cycle threshold values (Ct) for MT-ND1 and 18S rRNA were plotted versus DNA content to evaluate the calibration curves. Regression coefficient values of calibration curves (r2) were ≥ 0.99.
Table 2.
Primers used for qRT and LD PCR amplification
| Amplicon | Primer Sequence |
|---|---|
| Fragment 1 forward | 5’-AACCAAACCCCAAAGACACC-3’ |
| Fragment 1 reverse | 5’-GCCAATAATGACGTGAAAGTCC-3’ |
| Fragment 2 forward | 5’-TCCCACTCCTAAACACATCC-3’ |
| Fragment 2 reverse | 5’-TTTATGGGGTGATGTGAGCC-3’ |
| MT-ND1 forward | 5’-CCCTAAAACCCGCCACATCT-3’ |
| MT-ND1 reverse | 5’-GAGCGATGGTGAGAGCTAAGGT-3’ |
| 18srRNA forward | 5’-TCAAGAACGAAAGTCGGAGG-3’ |
| 18s rRNA reverse | 5’-GGACATCTAAGGGCATCACA-3’ |
Cardiac mtDNA amplification for massively parallel sequencing
The cardiac mitochondrial genome was amplified in two fragments using high-fidelity long-distance PCR (LD-PCR) with a proof-reading polymerase (Takara LA) as described by Tang et al.17. The primers used to amplify cardiac mtDNA fragment 1 (9289 bp in length) and fragment 2 (7626 bp in length) are shown in Table 2. The LD-PCR amplification conditions were as follows: 25 cycles at 94°C for 25 seconds and 68°C for 16min. PCR amplification products were analyzed via electrophoresis using 0.80% agarose and purified with appropriate kits (Bio Basic, Ontario CA). A PicoGreen (Invitrogen) assay was used to determine DNA concentrations.
Massively parallel sequencing
cDNA libraries were prepared with Nextera sample preparation kits (Illumina). The cDNA libraries were quantified using a PicoGreen assay (Invitrogen) and Kapa qPCR library quantification kit (Kapa Biosystems). Size and quality of cDNA libraries were confirmed using an Agilent Bioanalyzer 2100 DNA high sensitivity chip. The cDNA libraries were then pooled based on qPCR values. cDNA products were sequenced in rapid mode using an Illumina HiSeq 2500 DNA sequencing system at the University at Buffalo Genomics and Bioinformatics Core Facility (Buffalo, NY). The sequence data were submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The accession number for this data is SRP074312. All heteroplasmic mitochondrial variants detected in the sample groups are shown on Supplementary Table 1.
Sequencing output analysis
Paired sequence reads were demultiplexed using bcl2fastq version 1.8.4 (Illumina). Reads were then mapped to the reference mitochondrial genome NC_012920.1 (version GI: 251831106) and underwent variant calling using the CLC Genomics Workbench version 8.0.1 alignment tool. This required a minimum coverage of 1,000× and a minimum frequency of 1.00 × 10−6. An average of 7.50 million reads were mapped per donor sample. Sequencing data (including variants) were then compiled into Microsoft Excel spreadsheet form for analysis.
Data analysis
Mitochondrial haplogroups were determined based on the presence of diagnostic single nucleotide polymorphisms (SNPs) in each sample (Table 1). Frequency values were used to classify heteroplasmic mtDNA variants after the application of a conservative 2.0% error rate to account for potential amplification and sequencing errors19. Cardiac mtDNA variants exhibiting frequency values between the 2.0% – 98.0% range were considered heteroplasmic. The mean number of cardiac heteroplasmic mtDNA variants was obtained from the average number of individual heteroplasmic variants present in the specified location from all samples within the group considered. The frequency represents the percentage of mitochondrial genomes that contain the specific variant. Mean frequency values were obtained from the average frequency of all heteroplasmic variants present in the specified location from all donor samples within the group being considered. MT-ND1/18S rRNA ratios were used to measure mtDNA content. MT-ND1/18S rRNA ratios were calculated based on values extrapolated from the calibration curves. The Shapiro-Wilk normality test was used to ascertain the normality of datasets. A two-tailed Student’s t-test was used to compare the means of normally distributed datasets and the Mann-Whitney U test was used to compare the means of non-normally distributed datasets. Analysis of variance (ANOVA) with post-hoc Tukey test was used to compare mean number of heteroplasmic variants in the control region, as well as protein-coding and RNA-coding genes. In all cases, differences between means were considered to be significant at p < 0.050.
Results
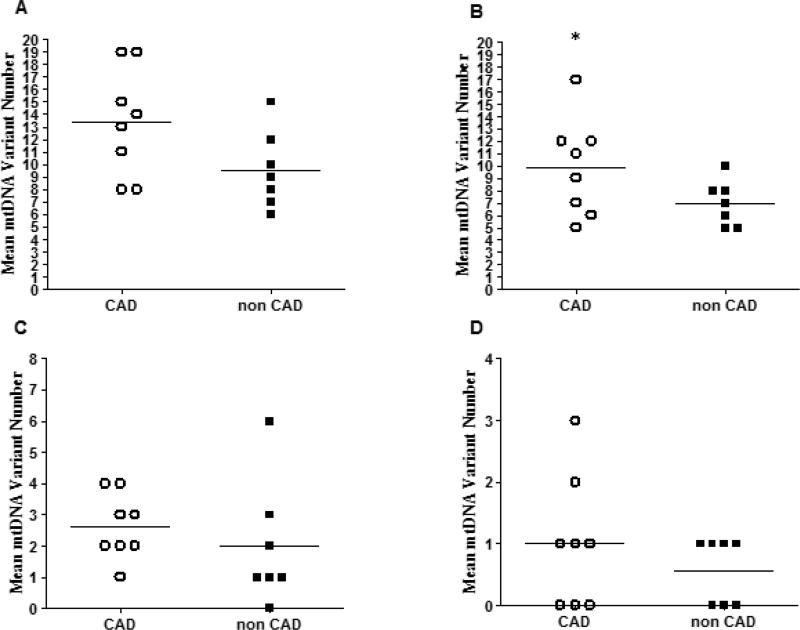
Massively parallel sequencing analysis showed that heart tissue samples from donors with (n = 8) and without (n = 7) CAD exhibited more heteroplasmic mtDNA variants in the CR in comparison to the number of variants in protein or RNA-coding regions (p < 0.0010, one-way ANOVA. Figure 1). The average number of total heteroplasmic mtDNA variants in heart samples from donors with CAD was 39.80% higher in comparison to the number of variants in samples from donors without CAD (mean variant numberCAD = 13.38 ± 4.31 versus mean variant numbernon CAD = 9.57 ± 2.31, p = 0.11. Figure 2A).
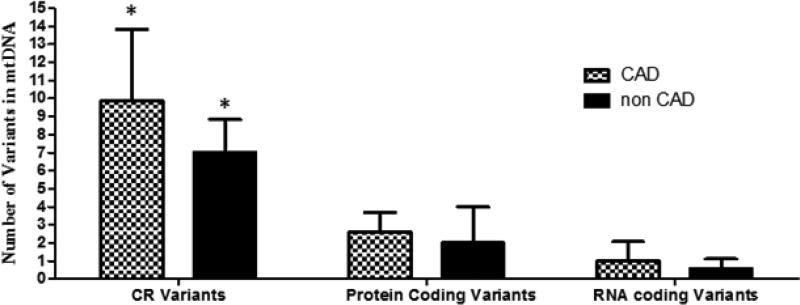
Figure 1.
Mean number of heteroplasmic mtDNA variants in CR, protein coding regions, and RNA coding regions in heart samples from donors with (n = 8) and without (n = 7) CAD. The bars represent mean ± SD. (*) indicates p < 0.0010 (one-way ANOVA with post-hoc Tukey test).
Figure 2.
Mean number of heteroplasmic mtDNA variants in: the complete mitochondrial genome (A), CR (B), protein coding regions (C), and RNA coding regions (D), in heart samples from donors with and without CAD. Horizontal bars indicate means, (*) indicates p < 0.050. See text for further details.
In regards to mtDNA variants in the CR, the average number of variants was 41.07% higher in samples from donors with CAD in comparison to samples from donors without CAD (mean CR variant numberCAD = 9.88 ± 3.94 versus mean CR variant numbernon CAD = 7.00 ± 1.83, p = 0.043. Figure 2B). On average, the number of mtDNA variants in protein and RNA coding regions in samples from donors with CAD were 31.25% and 75.44% higher than the number of variants in samples without CAD respectively; however, the differences did not reach statistical significance (Figures 2C and 2D).
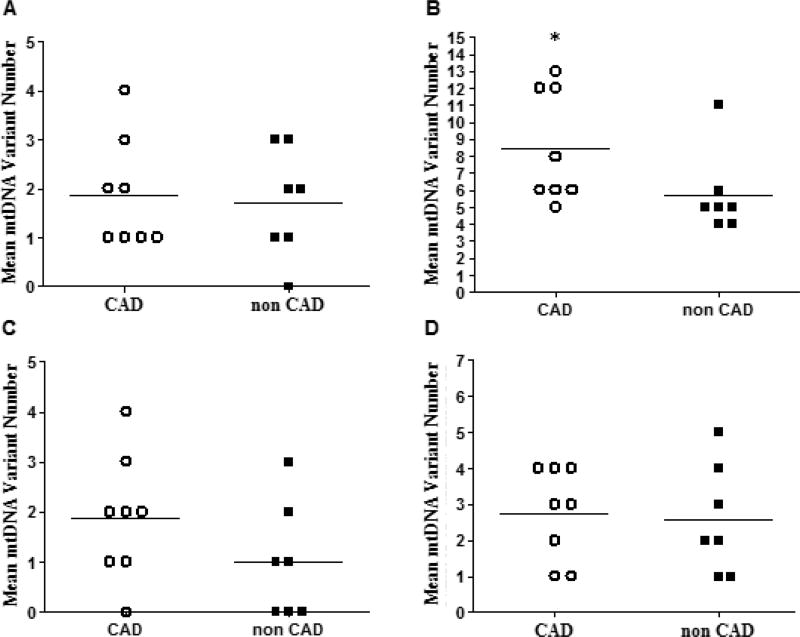
A breakdown of mtDNA variant types in heart samples with and without CAD showed that the total number of missense mtDNA variants in samples from donors with and without CAD were similar (Figure 3A and 3C). The total number of heteroplasmic deletions in samples from donors with CAD was 87.50% higher than those without CAD (mean deletion numberCAD = 1.88 ± 1.25 versus mean deletion numbernon CAD = 1.00 ± 1.16, p = 0.12. Figure 3D). The total number of heteroplasmic single nucleotide variants (SNVs) was significantly higher (48.76% increase) in hearts from donors with CAD in comparison to those without CAD (mean SNV numberCAD = 8.50 ± 3.30 versus mean SNV numbernon CAD = 5.71 ± 2.43, p = 0.029. Figure 3B).
Figure 3.
Mean number of heteroplasmic mtDNA missense variants (A), SNVs (B), deletions (C), and insertions (D), in heart samples from donors with and without CAD. Horizontal bars indicate means, (*) indicates p < 0.050. See text for further details.
Comparisons of variant frequency values (i.e. %) did not reveal any significant differences between heart samples from donors with and without CAD (Tables 3 and 4). Most heteroplasmic mtDNA variants in protein-coding genes were present in complexes I and IV (Table 5). There was one variant in complex V in the group of samples from donors with CAD, and one variant in complex III and two in complex V in the group of samples from donors without CAD.
Table 3.
Frequencies of heteroplasmic mtDNA variants in the CR, protein coding region, and RNA coding region in heart samples from donors with and without CAD.
| CAD(Mean ± SD) | nonCAD(Mean ± SD) | p value | |
|---|---|---|---|
| Mean total variant frequency | 31.90 ± 11.69 | 33.61 ± 10.89 | 0.95 |
| Mean CR variant frequency | 30.68 ± 9.11 | 36.34 ± 13.29 | 0.29 |
| Mean protein coding variant frequency | 26.52 ± 22.64 | 15.11 ± 15.27 | 0.54 |
| Mean RNA coding variant frequency | 37.37 ± 40.86 | 16.81 ± 34.19 | 0.63 |
Table 4.
Frequencies of heteroplasmic mtDNA single nucleotide, insertion, and deletion variants in heart samples from donors with and without CAD.
| CAD(Mean ± SD) | nonCAD(Mean ± SD) | p value | |
|---|---|---|---|
| Mean total missense variant frequency | 12.30 ± 15.59 | 10.74 ± 12.25 | 0.69 |
| Mean SNV frequency | 25.63 ± 13.80 | 33.08 ± 18.54 | 0.58 |
| Mean insertion variant frequency | 52.54 ± 17.68 | 49.75 ± 19.80 | 0.47 |
| Mean deletion variant frequency | 29.81 ± 30.77 | 10.57 ± 18.49 | 0.12 |
Table 5.
Heteroplasmic mtDNA variants (number and frequency) in complexes I and IV in heart samples from donors with and without CAD.
| CAD(Mean ± SD) | NonCAD(Mean ± SD) | p value | ||
|---|---|---|---|---|
| Complex I | Mean variant number | 1.38 ± 0.92 | 2.00 ± 1.16 | 0.33 |
| Mean variant frequency (freq%) | 5.67 ± 2.84 | 9.31 ± 13.31 | 0.82 | |
| Complex IV | Mean variant number | 0.38 ± 0.52 | 0.86 ± 1.07 | 0.40 |
| Mean variant frequency (freq%) | 14.74 ± 33.62 | 48.89 ± 46.61 | 0.32 |
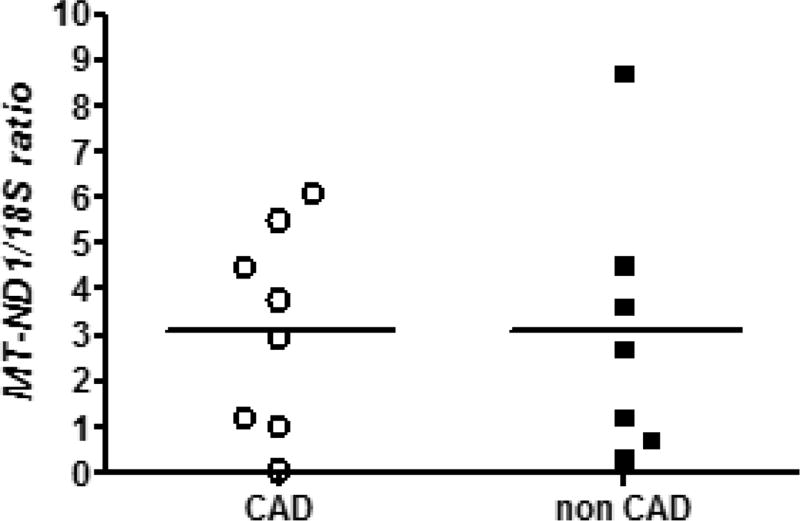
There were no apparent differences in mtDNA content between heart tissue samples from donors with and without CAD (Figure 4). The mean MT-ND1/18S ratio for samples from donors with CAD was 3.12 ± 2.22 versus 3.08 ± 2.91 for the donors without CAD (p = 0.48).
Figure 4.
MtDNA content in heart samples from donors with and without CAD. The horizontal bars represent mean (p = 0.48, Student’s t-test).
Discussion
In this pilot study, cardiac mtDNA heteroplasmy in samples from donors with documented CAD was investigated using massively parallel sequencing. In general, the data suggest potential trends towards higher levels of cardiac mtDNA heteroplasmy in heart tissue samples from subjects with diagnosed CAD. This observation is consistent with reports describing increased mitochondrial damage in subjects with CAD or ischemic heart disease9,10,20.
The cardiac mitochondrial genomes from donors with and without CAD exhibited relatively high concentrations of heteroplasmic mtDNA variants in the CR. The predominance of mtDNA variants in the CR has been noted in a variety of tissues, and this phenomenon has been attributed to various factors including the variable Displacement-loop within the CR and negative selective pressures associated with mtDNA variation in protein and RNA coding regions21–23. Since the CR is critical for mtDNA replication, it is possible that the increase in heteroplasmic variants could impact mtDNA replication24. Some evidence does suggest that lower mtDNA copy number in certain peripheral tissues (e.g. leukocytes) may be an independent risk factor and predictor of mortality in heart failure and cardiovascular disease25,26. In the present study, there were no apparent differences detected between cardiac mtDNA content in myocardial samples from donors with and without CAD. Based on the observed variability in cardiac mtDNA content and current sample size, the present study had a power of 0.8 at alpha 0.05 to detect differences between means in MT-ND1/18S ratios >4.0. Previous work by Chen et al. utilized similar methods and larger sample sizes to measure mtDNA content in peripheral blood lymphocytes in subjects with and without CAD (378 cases and 378 controls). The authors documented a ~24% decrease in mtDNA content in CAD cases in comparison to controls (i.e. a difference between means in MT-ND1/18S ratios of ~ 0.21)11. Based on this, it is reasonable to conclude that studies with larger samples sizes are needed to determine whether the presence of CAD affects mtDNA content in cardiac tissue.
While the data did show a significant increase in the number of SNVs in the group of heart tissue samples from subjects with CAD, this group of samples also exhibited a relatively large non-significant increase (87.50%) in the number of heteroplasmic deletions. This is of interest since increases in heteroplasmic mtDNA deletion frequency may have deleterious effects on mitochondrial function and have been associated with various disease states27. The number of missense mtDNA variants, which may also be damaging to mitochondrial functions, did not show trends of increase in the group of samples from subjects with a documented history of CAD.
High variability precluded comparisons between cardiac mtDNA variant frequency data. The frequency of cardiac mtDNA heteroplasmic variants may change throughout the life of the individual. Some potentially pathogenic variants are phenotypically silent until they reach a certain tissular threshold value28. MtDNA variants may have different threshold values for pathogenesis; however, a frequency of 60% has been reported as a general threshold value at which pathogenic heteroplasmic variants maybe phenotypically expressed21.
There are limitations in this study. This is a retrospective analysis of a small number of cardiac tissue samples. There were no maternal DNA samples to control for potential inherited mtDNA variants. Limited sample sizes only allowed for reliable detection of large differences between groups (Supplementary Table 2). Although the study included high quality samples with detailed pathology reports, it is possible that some donors were either incorrectly diagnosed with CAD or that a CAD diagnosis was missed. Some subjects with documented CAD had a history of myocardial infarction, and there is a possibility that this could further exacerbate mtDNA damage beyond what would be present with CAD alone. These subjects may have also experienced reperfusion injury, which could increase damage to cardiomyocytes and cardiac mtDNA29.
The data from this pilot study suggest that individuals with CAD display increased numbers of heteroplasmic mtDNA variants in cardiomyocytes compared to those without documented CAD. Further investigation into the potential role of cardiac mtDNA heteroplasmy on CAD progression is warranted.
Supplementary Material
Acknowledgments
Special thanks to the University at Buffalo Genomics and Bioinformatics Core, New York State Center of Excellence in Bioinformatics and Life Sciences.
Funding Sources:
This work was supported by the National Institute of General Medical Sciences under award R01GM073646. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography
Erik Hefti (ORCiD: 0000-003-0358-2539) is a PhD candidate at the the School of Pharmacy and Pharmaceutical Sciences, University at Buffalo. . Javier G. Blanco is an associate professor at the School of Pharmacy and Pharmaceutical Sciences, University at Buffalo.
Footnotes
Disclosures:
The authors have no conflicts of interest to disclose.
References
- 1.Mensah GA, Brown DW. An overview of cardiovascular disease burden in the United States. Health Aff (Millwood) 2007;26(1):38–48. doi: 10.1377/hlthaff.26.1.38. [DOI] [PubMed] [Google Scholar]
- 2.Hansson GK. Inflammation, Atherosclerosis, and Coronary Artery Disease. New England Journal of Medicine. 2005;352(16):1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 3.Madamanchi NR, Runge MS. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100(4):460–73. doi: 10.1161/01.RES.0000258450.44413.96. [DOI] [PubMed] [Google Scholar]
- 4.Mikhed Y, Daiber A, Steven S. Mitochondrial oxidative stress, mitochondrial DNA damage and their role in age-related vascular dysfunction. International journal of molecular sciences. 2015;16(7):15918–15953. doi: 10.3390/ijms160715918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayeva M, Gheorghiade M, Ardehali H. Mitochondria as a therapeutic target in heart failure. J Am Coll Cardiol. 2013;61(6):599–610. doi: 10.1016/j.jacc.2012.08.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A Sobenin I, V Zhelankin A, Y Mitrofanov K, V Sinyov V, A Sazonova M, Y Postnov A, N Orekhov A. Mutations of mitochondrial DNA in atherosclerosis and atherosclerosis-related diseases. Current pharmaceutical design. 2015;21(9):1158–1163. doi: 10.2174/1381612820666141013133000. [DOI] [PubMed] [Google Scholar]
- 7.Emma P, Bennett MR. Mitochondrial DNA damage and atherosclerosis. Trends in Endocrinology & Metabolism. 2014;25(9):481–487. doi: 10.1016/j.tem.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Botto N, Berti S, Manfredi S, Al-Jabri A, Federici C, Clerico A, Ciofini E, Biagini A, Andreassi MG. Detection of mtDNA with 4977bp deletion in blood cells and atherosclerotic lesions of patients with coronary artery disease. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2005;570(1):81–88. doi: 10.1016/j.mrfmmm.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Corral-Debrinski M, Shoffner J, Lott M, Wallace D. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutation Research/DNAging. 1992;275(3):169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 10.Fetterman JL, Holbrook M, Westbrook DG, Brown JA, Feeley KP, Bretón-Romero R, Linder EA, Berk BD, Weisbrod RM, Widlansky ME. Mitochondrial DNA damage and vascular function in patients with diabetes mellitus and atherosclerotic cardiovascular disease. Cardiovascular diabetology. 2016;15(1):1. doi: 10.1186/s12933-016-0372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Xie X, Wang Y, Gao Y, Xie X, Yang J, Ye J. Association between leukocyte mitochondrial DNA content and risk of coronary heart disease: a case-control study. Atherosclerosis. 2014;237(1):220–226. doi: 10.1016/j.atherosclerosis.2014.08.051. [DOI] [PubMed] [Google Scholar]
- 12.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100(15):1104–12. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grady JP, Murphy JL, Blakely EL, Haller RG, Taylor RW, Turnbull DM, Tuppen HAL. Accurate Measurement of Mitochondrial DNA Deletion Level and Copy Number Differences in Human Skeletal Muscle. PLOS ONE. 2014;9(12):e114462. doi: 10.1371/journal.pone.0114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venegas V, Wang J, Dimmock D, Wong LJ. Real-Time Quantitative PCR Analysis of Mitochondrial DNA Content. Current Protocols in Human Genetics. 2011:19.7. 1–19.7. 12. doi: 10.1002/0471142905.hg1907s68. [DOI] [PubMed] [Google Scholar]
- 15.Bai R-K, Perng C-L, Hsu C-H, Wong L-JC. Quantitative PCR Analysis of Mitochondrial DNA Content in Patients with Mitochondrial Disease. Annals of the New York Academy of Sciences. 2004;1011(1):304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- 16.Hefti E, Quiñones-Lombraña A, Redzematovic A, Hui J, Blanco JG. Analysis of mtDNA, miR-155 and BACH1 expression in hearts from donors with and without Down syndrome. Mitochondrial DNA Part A. 2016;27(2):896–903. doi: 10.3109/19401736.2014.926477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 2010;48(4):287–96. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- 18.Hefti E, Bard J, Blanco JG. Analysis of Heteroplasmic Variants in the Cardiac Mitochondrial Genome of Individuals with Down Syndrome. Human Mutation. 2017;38(1):48–54. doi: 10.1002/humu.23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corral-Debrinski M, Stepien G, Shoffner JM, Lott MT, Kanter K, Wallace DC. Hypoxemia is associated with mitochondrial DNA damage and gene induction. Implications for cardiac disease. JAMA. 1991;266(13):1812–6. [PubMed] [Google Scholar]
- 21.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci U S A. 2014;111(29):10654–9. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. The American Journal of Human Genetics. 2000;67(4):1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma H, Singh A, Sharma C, Jain SK, Singh N. Mutations in the mitochondrial DNA D-loop region are frequent in cervical cancer. Cancer Cell International. 2005;5:34–34. doi: 10.1186/1475-2867-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nature Reviews Genetics. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashar FN, Zhang Y, Moes A, Grove ML, Wilsdon AG, Chaves PH, Coresh J, Newman AB, Bandeen-Roche K, Boerwinkle E, et al. Abstract 19318: Mitochondrial DNA Copy Number as a Predictor of Cardiovascular Disease. Circulation. 2014;130(Suppl 2):A19318–A19318. [Google Scholar]
- 26.Huang J, Tan L, Shen R, Zhang L, Zuo H, Wang DW. Decreased Peripheral Mitochondrial DNA Copy Number is Associated with the Risk of Heart Failure and Long-term Outcomes. Medicine. 2016;95(15) doi: 10.1097/MD.0000000000003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 28.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16(9):530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 29.Lucas DT, Szweda LI. Cardiac reperfusion injury: aging, lipid peroxidation, and mitochondrial dysfunction. Proceedings of the National Academy of Sciences. 1998;95(2):510–514. doi: 10.1073/pnas.95.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.