Abstract
The respiratory syncytial virus (RSV) is a leading cause of acute respiratory illnesses (ARI) in infants and young children. The objectives of this study were to investigate the RSV circulation among children aged <5 years in Bulgaria, to identify the RSV-A and RSV-B genotypes and to perform an amino acid sequence analysis of second hypervariable region (HVR2) of the G gene. During the 2014/15 and 2015/16 winter seasons, nasopharyngeal specimens of 610 children aged <5 years with ARI were tested using Real Time RT-PCR for influenza viruses, RSV, metapneumovirus, parainfluenza viruses, rhinoviruses and adenoviruses. Viral respiratory pathogens were detected in 429 (70%) out of 610 patients examined and RSV was the most frequently identified virus (26%) followed by influenza A(H1N1)pdm09 virus (14%) (p < .05). RSV was the most prevalent pathogen in patients with bronchiolitis (48%) and pneumonia (38%). In the 2014/15 season, RSV-A dominated slightly (53%), while in the next season RSV-B viruses prevailed more strongly (66%). The phylogenetic analysis based on the G gene indicated that all 21 studied RSV-A strains belonged to the ON1 genotype; the vast majority (96%) of the RSV-B strains were classified into BA9 genotype and only one - into BA10 genotype. All Bulgarian RSV-A and RSV-B sequences contained a 72-nt and a 60-nt duplication in the HVR2, respectively. The study showed the leading role of this pathogen as a causative agent of serious respiratory illnesses in early childhood, year-on-year fluctuations in RSV incidence, a shift from RSV-A to RSV-B subgroup dominance and relatively low genetic divergence in the circulating strains.
Keywords: Acute respiratory infection, respiratory syncytial virus, genetic characterization, amino acid substitution
Introduction
Acute respiratory illnesses (ARIs) are a major public health problem worldwide due to the associated high morbidity, large number of doctors’ visits, hospital admissions and significant mortality. Approximately 80% of the ARI cases with an identified aetiologic agent have viral aetiology [1]. Over 200 different virus types cause ARI, the main of which are influenza viruses, RSV, human metapneumovirus (hMPV), parainfluenza viruses (PIV), adenoviruses (AdV), rhinoviruses (RV), coronaviruses and bocaviruses.
RSV is one of the most common aetiologic agents of ARI and is a leading cause of serious lower respiratory tract infections (LRTI) (bronchiolitis and pneumonia) in infants and young children. Globally, in 2015, the virus caused 33.1 million episodes of acute LRTI, resulted in about 3.2 million hospital admissions, and 59 600 in-hospital deaths in children younger than 5 years [2]. Approximately 50–70% of children are infected with RSV during their first year of life, and by the age of 2, almost all children have had at least one RSV infection [3]. RSV is a member of the family Pneumoviridae, genus Orthopneumovirus [4]. It is an enveloped virus with a non-segmented, single-stranded, negative-sense RNA genome, and it is approximately 15.2 kb in length, containing 10 genes that encode 11 proteins [5]. The RSV envelope contains three surface glycoproteins: F and G are major antigens, and SH is a small hydrophobic protein. The G protein is responsible for the attachment of the virus to the sensitive cell; the F protein also participates in viral attachment and mediates virus and cell membrane fusion and viral entry. F and G are the only proteins that induce production of neutralizing antibodies. The F protein induces potent cross-protective antibodies against both antigenic subgroups and is highly conserved, while the G protein induces weak neutralizing subgroup-specific antibodies and has hypervariable regions [6]. The G protein is a type II integral membrane protein containing a N terminal cytoplasmic tail, transmembrane domain and ectodomain [7]. The first two regions are highly conserved, while the ectodomain contains two hypervariable regions (HVRs) with a mucin-like structure (a high percentage of serine, threonine and proline residues) separated by a conserved 13-residue segment comprising a receptor binding site [8–10]. The C-terminal region of the G gene (HVR2) contains multiple epitopes for genotypic and strain-specific antibodies and has the highest degree of variability, making it suitable for molecular-epidemiological analyses [11,12]. The G protein is heavily glycosylated with N-linked and O-linked sugars, which shield antigenic epitopes from immune recognition, thus contributing to the escape from pre-existing immunity [3].
RSV exists as a single serotype that is divided into two major subgroups (RSV-A and RSV-B) based on its antigenic and genetic variability [13]. In regard to the G gene, both subgroups show 67% homology at the nucleotide level and 53% homology at the amino acid level [8]. To date, based on nucleotide sequence analysis of the G gene, 14 RSV-A (GA1-GA7, SAA1, NA1-NA4, CB-A and ON1) and 25 RSV-B (GB1-GB4, SAB1-SAB4, URU1, URU2, BA1-BA12, BA-C, CB-B and CB1) genotypes have been identified [14–18]. Geographic and temporal variations are observed in the distribution of different RSV genotypes. The surveillance of circulating RSV genotypes in different countries is important in terms of the development of a globally effective vaccine. The analysis of nucleotide and amino acid sequences in detected RSV allows for characterization of the genetic variability of this pathogen, tracing of its evolution and clarification of the mechanisms required for immune escape.
Despite the important clinical significance of RSV infection, there are no data on the molecular epidemiology of RSV in Bulgaria. The main aims of the present study were to investigate the circulation pattern of RSV in Bulgaria, to determine the contribution RSV infection is making to the aetiological structure of ARI among children under the age of 5, to characterize the genotype diversity of RSV-A and RSV-B and to perform an amino acid sequence analysis of the C-terminal HVR2 region of the G gene.
Material and methods
Study population and specimen collection
Children under the age of 5 from different regions of Bulgaria treated for influenza like illness (ILI) or ARI in primary care settings or hospitals were enrolled within the National Influenza Sirveillance Programme. ILI and ARI were defined according to the European Centre for Disease Prevention and Control (ECDC) (https://ecdc.europa.eu/en/infectious-diseases-public-health/surveillance-and-disease-data/eu-case-definitions). The first 5–10 ILI/ARI patients attending a sentinel primary care/outpatient facility on a certain day of the week were selected for respiratory specimen collection. Samples were collected from hospital inpatients at various times based on a clinician’s judgement. The attending paediatrician diagnosed each patient based on standard clinical criteria. Clinical information, including demographic characteristics, history of illness, symptoms, initial diagnosis, and comorbid illnesses, was documented on case report forms. Combined nasal and pharyngeal specimens were collected from the enrolled patients by means of commercial polyester collection swabs (Deltalab, Spain). Following collection, swabs were stored at 4 °C for up to 72 hrs. Specimens were processed immediately or stored at −80 °C prior to analysis.
This study was performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans https://www.healthscience.net/resources/declaration-of-helsinki/ and approved by the local Ethics Committee of the National Centre of Infectious and Parasitic Diseases. All parents of sick children who participated in this study provided written informed consent before specimen collection and testing.
Extraction of nucleic acids and real time RT-PCR
Viral nucleic acids were extracted automatically from 500 μl of each respiratory specimen in a final eluate volume of 75 μl using a ExiPrep Dx Viral DNA/RNA kit (BioNeer, Korea) in accordance with the manufacturer’s instructions. Detection and typing/subtyping of influenza viruses were carried out by a real time RT-PCR method and the SuperScript III Platinum ® One-Step qRT-PCR System (Invitrogen, ThermoFisher Scientific, USA). Primers and probes were provided by the International Reagent Resource (IRR), USA. Amplification was performed using a Chromo 4 thermal cycler (Bio-Rad Laboratories, Inc., USA) according to the following protocol from CDC-Atlanta: reverse transcription (RT) at 50 °C for 30 min, Taq inhibitor inactivation at 95 °C for 2 min, 45 cycles of denaturation at 95 °C for 15 s and annealing at 55 °C for 30 s [19]. Samples with a cycle threshold (ct) value < 38 were considered positive.
The detection of RSV, hMPV, PIV 1/2/3, RV and AdV was performed using singleplex real time PCR assays and an AgPath-ID One Step RT-PCR kit (Applied Biosystems, ThermoFisher Scientific, USA). The primers, probes and PCR conditions used in the study were identical to those previously described [20]. Each 25 μL reaction mixture contained the following components: 12.5 μL of RT-PCR Buffer, 1.25 μL of detection enhancer, 0.5 μL of each primer and probe, 3.75 μL of RNAse-free water, 1 μL RT-PCR enzyme mix and 5 μL of RNA or DNA extracted from the specimen. Positive and negative controls were included in each run. The RNAase-P gene served as an internal positive control for human nucleic acid. Clinical specimens were tested in a separate real time RT-PCR reaction for the RNAase-P gene, which provided verification of RNA integrity and the absence of PCR inhibition [19]. For influenza type A and type B viruses, positive controls were provided by IRR, USA; for other targets, AmpliRun DNA/RNA Amplification Controls (Vircell, Spain) were used. Subgroup-specific primers and probes targeting the RSV F and N genes were used for Multiplex real time RT-PCR to distinguish RSV-A from RSV-B, respectively, as described previously [21]. Sequences of primers and probes used in this study, as well as thermocycling conditions, are shown in Supplementary Table 1.
PCR for analysis of the G Gene and DNA sequencing
Conventional RT-PCR was used to amplify fragments of the G and F gene open reading frames (ORFs) in the RSV-positive samples. We used RSV-A specific primers as follows: G513-F, corresponding to nucleotides 513–534 of the G gene of strain A2 (GenBank accession number U50362), and F131-R, corresponding to nucleotides 111–131 of the F gene [22]. RSV-B specific primers BGF and BGR corresponded to regions within the G gene (nt 4858–4881) and the intergenic G-F region (nt 5637–5658), respectively, in strain WV/B1/85 (GenBank accession number AF013254) [23] (Supplementary Table 1). Nucleic acid amplification was performed using an Eppendorf Mastercycler instrument (Eppendorf, UK) and a Qiagen One-Step RT-PCR kit (Qiagen, Germany). The 50-μL reaction mixture contained 30 pmol of forward and reverse primers and 10 μL of the extracted RNA. The amplified products (583 bp product for RSV-A and 800 bp product for RSV-B) were analysed by electrophoresis in 2% agarose gels stained with ethidium bromide. The PCR products were then extracted from the gels using a PureLink Quick Gel Extraction kit (Invitrogen, ThermoFisher Scientific, USA) according to the manufacturer’s instructions. The purified amplicons were sequenced commercially (BioNeer, Korea) in both directions using a BigDie Terminator v. 3.1 sequencing kit (Applied Biosystems, ThermoFisher Scientific, USA) and the same primer pairs previously used for PCR amplification. Sequences were analysed on an ABI 3130XL (Applied Biosystems, USA) automatic DNA Analyser.
Phylogenetic analysis of the G-gene sequences
For phylogenetic analyses, the G gene nucleotide sequences of previously published strains representing different RSV-A and RSV-B genotypes, and sequences of recently circulating RSV strains, were retrieved from the BLAST (Basic Local Alignment Search Tool) [https://blast.ncbi.nlm.nih.gov/Blast.cgi] database (Supplementary Table 2). They were all aligned with the sequences determined from this study using the MUSCLE program embedded in Molecular Evolutionary Genetics Analysis software [MEGA, version 6.06; https://www.megasoftware.net/] [24]. Many representative sequences were based only on the second variable region (270 bp), so only the HVR2 sequences were retained in the alignment. The most appropriate nucleotide substitution models for phylogenetic analysis of RSV-A (Tamura–Nei, TN93) and RSV-B (Hasegawa-Kishino-Yano model with a gamma distribution, HKY+G) were determined using MEGA 6.06. Phylogenetic trees, based on the nucleotide sequences of the HVR2 regions of the G genes, were constructed using the Maximum Likelihood method within the MEGA 6.06. The reliability of the tree topology was assessed by bootstrapping with 1000 replications. The study strains were genotyped based on clustering with sequences representing known genotypes. Sequences were arbitrarily considered a genotype if they clustered together with bootstrap values of 70–100% [25].
Pairwise nucleotide distances (p-distances), the number of pairwise base differences divided by the total number of nucleotides in the sequenced segment, were calculated using the MEGA p-distance method to estimate the genetic variability among sequences within and between genotypes. The p-distances for RSV strains with the same genotype are <0.07 [25]. Strains with
100% nucleotide sequence identity were excluded from the analyses.
Deduced amino acid sequence analysis and prediction of N- and O-glycosylations
The amino acid sequences of the C-terminal HVR2 regions of the RSV G proteins were generated by translating nucleotide sequences with the standard genetic code using MEGA software. The deduced amino acid sequences of the RSV-A and RSV-B strains from this study were compared to those of the reference A2 and prototype BA1 strains (BA4128/99B), respectively, to identify amino acid substitutions.
Putative N-glycosylation sites were predicted using NetNGlyc 1.0 web server [https://www.cbs.dtu.dk/services/NetNGlyc] in order to identify the sequence motifs N–X–S/T (sequon), where X can be any amino acid except proline. Only the sites with scores higher than 0.5 were accepted as glycosylated [26]. Potential O-glycosylation sites were determined using the NetOGlyc 4.0 web server [https://www.cbs.dtu.dk/services/NetOGlyc]. O-glycosylation is based on the amino acids serine and threonine. Sites were predicted if the glycosylation potential was ≥0.5 [27].
Statistics
Patients’ age, gender, clinical features of illness and frequency of carrying each virus were compared using the χ2 or Fisher’s exact tests for categorical variables. A p value of < .05 was considered statistically significant. Non-normally distributed continuous or ordinal variables were compared using the Mann-Whitney U test. Data were analysed using SPSS version 16.0 for Windows.
Results
ARI Surveillance
This study covered two consecutive epidemic seasons in Bulgaria: week 40 of 2014 to week 20 of 2015 and week 40 of 2015 to week 20 of 2016. During these surveillance periods, the overall incidence of ARI ranged from 41.04 to 172.55 cases and from 41.04 to 158.74 cases per 10 000 people, respectively. The highest incidence rates for ARI were observed in children under the age of 4, with maximum values of 693.86 and 375.37 per 10 000 people, respectively [www.grippe.gateway.bg]. The study population consisted of 610 children under the age of 5 who demonstrated symptoms of ILI or ARI (217 in the first season and 393 in the second season). In total, 111 (18.2%) of these children attended outpatient healthcare centres and 499 (81.8%) were hospitalized. Patients’ ages ranged from 10 days to 59 months (median age 28 months). A total of 56.7% of the children were boys and 43.3% were girls. Most patients (56%) demonstrated symptoms of ILI or ARI without serious complications. Specific complications in the respiratory tract (laryngotracheitis, bronchiolitis, and pneumonia) and the central nervous system (CNS) (febrile seizures, cerebral edema, aseptic meningitis, encephalopathy, and encephalitis) were analysed.
Viral detection
Viral respiratory pathogens were identified in 429 (70%) of the 610 patients examined. Single infections were detected in 392 children (64.3%); 36 children (5.9%) were co-infected with two viruses, and one child (0.2%) was infected with 3 viruses. A total of 144 patients (23.6%) were positive for the following influenza viruses: A(H1N1)pdm09 (13.9%), A(H3N2) (6.1%) and type B (3.6%). Non-influenza respiratory viruses were detected in 285 children (46.7%), including RSV (25.7%), hMPV (4.9%), PIV-1 (0.7%), PIV-2 (1.3%), PIV-3 (1.6%), RV (7.9%) and AdV (4.6%). The most prevalent respiratory virus identified in this study was RSV, followed by influenza A(H1N1)pdm09 virus (p < .05). RSV was also the most commonly detected pathogen among patients with a viral coinfection (68%, 25/37), followed by RV and AdV. The detection rate of RSV did not vary significantly between the two study seasons (28.8% in the first season and 24.2% in the second, p = 0.2465).
Among the 157 RSV-positive specimens, RSV-A was identified in 55 samples (35%) and RSV-B in 80 samples (51%). Coinfection with both subgroups of RSV was detected in 8 (5%) of the RSV-positive samples. For 14 specimens (9%), the subgroup could not be determined due to a low viral load. During the 2014/2015 season, RSV-A dominated slightly (53%, 33/62), while in the next season RSV-B prevailed (62%, 59/95) (p = 0.07). Influenza A(H1N1)pdm09, RSV-A and hMPV were more frequently detected in hospitalized children, while RV was more prevalent among outpatients (p < .05) (Table 1). The frequency of RSV and influenza in outpatients was 19.8% and 18%, respectively; in hospitalized children, these same frequencies were 27.1 and 24.8%, respectively.
Table 1.
Distribution of detected respiratory viruses in outpatients and hospitalized patients.
| Number (%) of patients infected with respiratory viruses | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A(H1N1) pdm | A(H3N2) | B/Yam* | B/Vic** | RSV (untyped) | RSVA | RSV B | RSV A+B | hMPV | PIV-1 | PIV-2 | PIV-3 | RV | AdV | Total | |
| Outpatients | 6 (5%) | 10 (9%) | 2 (2%) | 2 (2%) | 2 (2%) | 5 (5%) | 13 (12%) | 2 (2%) | 1 (1%) | 0 | 1 (1%) | 1 (1%) | 14 (13%) | 4 (4%) | 63/111 (57%) |
| Inpatients | 79 (16%) | 27 (5%) | 3 (0.6%) | 15 (3%) | 12 (2%) | 50 (10%) | 67 (13%) | 6 (1%) | 29 (6%) | 4 (1%) | 7 (1%) | 9 (2%) | 34 (7%) | 24 (5%) | 366/499 (73%) |
| Total | 85 (14%) | 37 (6%) | 5 (0.8%) | 17 (3%) | 14 (2%) | 55 (9%) | 80 (13%) | 8 (1%) | 30 (5%) | 4 (1%) | 8 (1%) | 10 (2%) | 48 (8%) | 28 (5%) | 429/610 (70%) |
B/Yamagata.
B/Victoria.
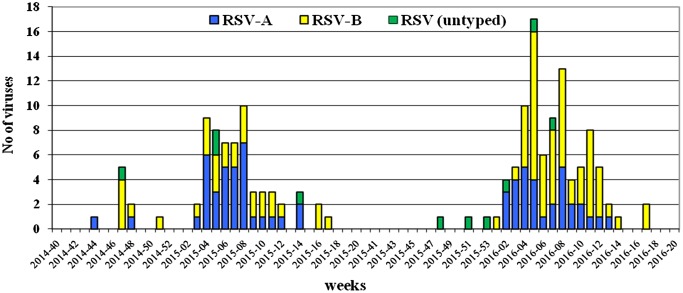
RSV was identified in specimens collected between November and April, and during both surveillance periods, the greatest number of respiratory viruses was detected in specimens obtained in February. Furthermore, the peak circulation of RSV occurred in week 8 of 2015 and week 5 of 2016 with detection rates of 48 and 43%, respectively (Figure 1).
Figure 1.
Weekly distribution of RSV detected during the 2014/15 and 2015/16 seasons in Bulgaria.
Age and gender distribution
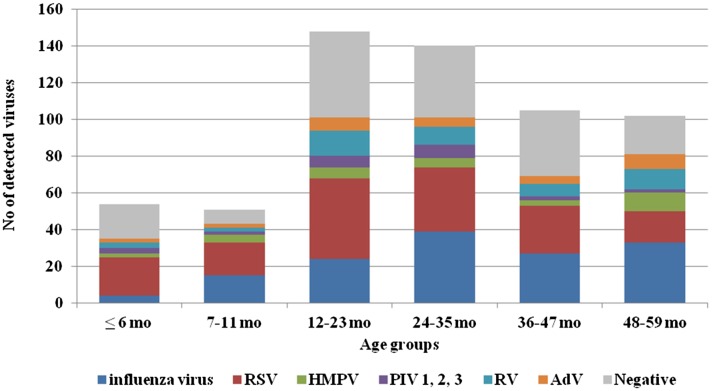
Viral respiratory infections occurred in all studied age groups (Figure 2). Co-infections were detected predominantly in children aged 24–37 months. The highest incidence rate for influenza viruses was found in children aged 48–59 months (32%), and the highest rates for non-influenza viruses were found in the youngest age groups (≤6 months (57.4%) and 7–11 months (54.9%)). Patients infected with RSV fell between the ages of 10 days and 59 months with a mean age of 23.5 ± 14.6 months and a median age of 25 months. The incidence of RSV infection was highest amongst the two youngest age groups (≤6 months (38.9%) and 7–11 months (35.3%)). During the 2014/2015 season, children ≤6 months of age (47.8%) were most affected by RSV, while in the subsequent season (2015/2016), the incidence of RSV infection was highest in both of the two youngest age groups (32.3% for both groups). RSV-B was detected in 4 out of 7 studied newborns at hospital ‘Maichin Dom’, Sofia, Bulgaria. In both seasons, RSV had the lowest frequency in children within the oldest age group (48–59 months) (14.6 and 18%, respectively). Among RSV-positive children for whom gender was recorded, 62% were boys and 38% were girls (p = 0.1265).
Figure 2.
Age distribution of children infected with respiratory viruses.
Clinical characteristics
In children under the age of 5, respiratory viruses can cause complications in many organs and regions of the body including the lower respiratory tract, CNS and heart, to name a few. In this study, the contributions of influenza viruses, RSV, hMPV, PIV 1/2/3, RV and AdV in the development of the most common complications (laryngotracheitis, bronchiolitis, pneumonia, CNS complications) were analysed (Table 2). Patients with different clinical diagnoses did not differ significantly in terms of age. The median age of children with bronchiolitis was the lowest, and the median age of children with pneumonia was the highest (p = .0048). Male predominance was observed in patients with bronchiolitis, pneumonia and CNS complications, but the association was not statistically significant. The proportion of detected influenza viruses among patients with laryngotracheitis was 12 out of 45 (27%), the proportion among patients with bronchiolitis was 12 out of 62 (19%), the proportion among patients with pneumonia was 14 out of 81 (17%), and the proportion among patients with neurological complications was 9 out of 55 (16%). With regards to non-influenza viruses, these proportions were 27/45 (60%), 42/62 (68%), 46/81 (57%), and 20/55 (36%), respectively. RSV was the most common virus identified in patients with bronchiolitis (48%) and pneumonia (38%) (p < .05). Among RSV-positive children with lower respiratory tract complications, 54% were under 2 years of age. Influenza viruses, especially the A(H1N1)pdm09 virus, were detected more frequently (16%) among patients with neurological symptoms. Co-infections were detected in children from all four studied clinical groups, more often in those patients with pneumonia and laryngotracheitis. RSV was the most common virus found in mixed infections.
Table 2.
Distribution of detected respiratory viruses according to patients’ clinical diagnoses.
| Clinical diagnoses (No of cases, %) | ||||
|---|---|---|---|---|
| Laryngotracheitis (n = 45) | Bronchiolitis (n = 62) | Pneumonia (n = 81) | CNS complications (n = 55) | |
| Mean age (months) | 26.9 ± 16.2 | 20.6 ± 12.6 | 29.3 ± 15.7 | 25.2 ± 13.4 |
| Median age (months) | 22.5 | 22 | 29 | 23 |
| Male/female ratio | 22/23 | 37/25 | 46/35 | 31/24 |
| Total influenza viruses | 12 (27%) | 12 (19%) | 14 (17%) | 9 (16%) |
| RSV (untyped) | 1 | 1 | 1 | 1 |
| RSV-A | 5 | 14 | 10 | 2 |
| RSV-B | 4 | 15 | 16 | 4 |
| hMPV | 5 | 6 | 8 | – |
| PIV1 | 1 | – | – | – |
| PIV2 | 1 | – | – | 2 |
| PIV3 | – | 1 | 1 | 2 |
| RV | 2 | 4 | 2 | 3 |
| AdV | 2 | – | 3 | 3 |
| A(H1N1)pdm09+RSV | 1 | – | – | – |
| A(H1N1)pdm09+RSV-A | 1 | – | – | – |
| A(H1N1)pdm09+RSV-B | 1 | – | – | – |
| RSV-A+ RSV-B | 1 | – | 3 | – |
| RSV-A+RV | – | – | – | 1 |
| RSV-B+AdV | – | – | 1 | – |
| hMPV+PIV1 | – | 1 | – | – |
| hMPV+AdV | 1 | – | 1 | – |
| PIV3+RV | 1 | – | – | – |
| RV+AdV | – | – | – | 2 |
| Total non-influenza | 27 (60%) | 42 (68%) | 46 (57%) | 20 (36%) |
Phylogenetic analysis of RSV
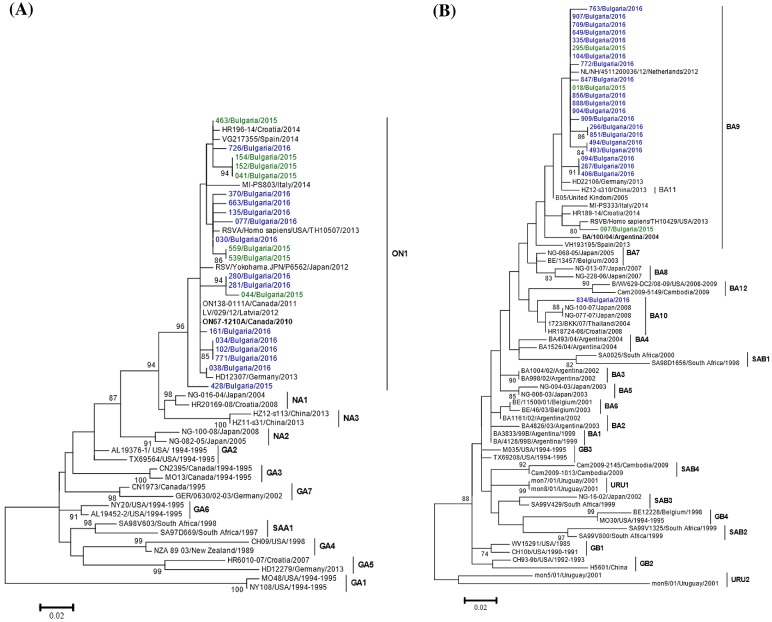
Phylogenetic trees based on the HVR2 regions of the G genes of 52 RSV-A and 74 RSV-B strains were constructed, including study sequences and representative sequences of all known genotypes that had been downloaded from GenBank. The phylogenetic analysis showed that all 21 sequenced Bulgarian RSV-A strains belonged to the ON1 genotype (Figure 3(a)). The vast majority (96%, 24/25) of the studied RSV-B strains were classified as genotype BA9, but one was classified as BA10 (Figure 3(b)). The Bulgarian sequences clustered with sequences of RSV circulating in the same period in other European countries (Croatia, Germany, Italy, Latvia, Spain, The Netherlands).
Figure 3.
Phylogenetic trees based on the G gene sequences of RS-A (a) and RSV-B (b) strains. Phylogenetic trees were constructed using maximum-likelihood method with 1000 bootstrap replicates running within MEGA 6.06 software. Only bootstrap values ≥70% are displayed at the branch nodes. The scale bar indicates the number of nucleotide substitutions per site. The GenBank name of the strains, the country and the year of isolation are shown in the phylogenetic trees. The genotypes are denoted by lines on the right side. The ON67–1210A and BA/100/04 strains, prototype for the ON1 and BA9 genotypes, respectively, are indicated in bold. Bulgarian RSV strains detected during the 2014/15 and 2015/16 seasons are indicated in green and blue, respectively.
At the nucleotide level, the mean pairwise distance amongst Bulgarian ON1 sequences was 0.027 ± 0.0025 (mean ± standard deviation, SD). The mean p distance amongst Bulgarian BA9 sequences was 0.020 ± 0.013, while the mean distance between BA9 and BA10 sequences was 0.072 ± 0.0015.
Deduced Amino Acid Sequence Analysis
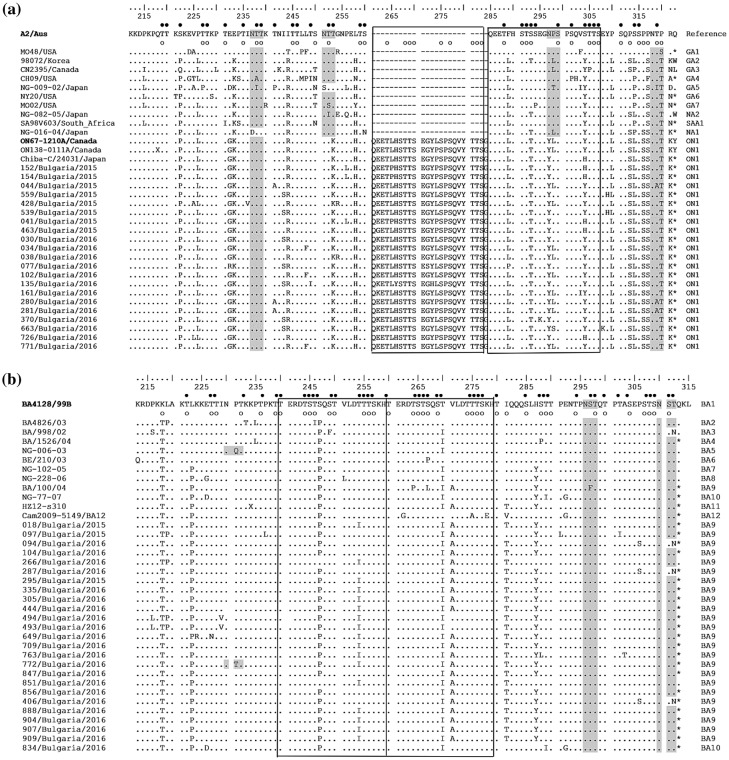
The deduced amino acid sequence of the HVR2 region of Bulgarian RSV-A was compared to those of the A2 reference strain (accession number: M11486), prototype ON1 strain ON67–1210A (accession number: JN257693) and other representative sequences of known genotype (Figure 4(a)). The prototype ON1 virus was characterized by a unique nucleotide insertion (72 nt duplication) in the C-terminal end of the G gene that starts after residue 850 (A2 numbering) and leads to duplication of 23 amino acids and lengthening of the polypeptide to 321 amino acids (298 amino acids in the prototype RSV-A subgroup virus A2). The stop codon in the ON1 viruses is at position 322, while in the A2 virus it is at position 299 [28]. All Bulgarian RSV-A ON1 genotype viruses were found to contain the indicated duplication of 23 amino acids (QEETLHSTTSEGYLSPSQVYTTS) (positions 261–283 and 285–307). In comparison to A2, ON1 viruses also contained E232G, T253 K, N298Y and S314L substitutions. Relative to the prototype strain ON67–1210A, amino acid substitutions at 24 positions were found in the Bulgarian ON1 viruses; I243S, E262 K, L274P, L298P and Y304H were the most common substitutions. Substitutions at 12 positions were identified either in the insertion or in its homologous site.
Figure 4.
Deduced amino acid alignments of the second variable region of the G-protein from RSV-A (a) and RSV-B (b) strains. The alignment (a) is shown relative to the sequence of reference RSV-A strain A2 (GenBank accession number M11486) and the alignment (b) is shown relative to the sequence of prototype BA strain BA4128/99B (GenBank accession number AY333364). The amino acid numbers correspond to G protein positions 212 to 298 of the strain A2 (a) and to G protein positions 213 to 315 of the strain BA4128/99B (b). Identical residues are identified as dots. Dashes indicate gaps corresponding to the nucleotide insertions; asterisks indicate the stop codons. The outlined rectangles represent the two copies of the duplicated 23-amino acid region in RSV-A strains (a) and the two copies of the duplicated 20-amino acid region in RSV-B strains (b). Genotype names are shown on the right. Light grey shading highlights the predicted N-glycosylation sites. Black circles denote the predicted O-glycosylation sites of the A2 reference strain (a) and BA prototype strain BA4128/99B (b); predicted O-glycosylation sites of Bulgarian strains are indicated by unfilled circles.
The sequences of the Bulgarian and other RSV-A ON1 genotype viruses have two potential N-glycosylation motifs in the HVR2 region at positions 237 and 318. The first glycosylation site was conserved, while at the second site, an amino acid substitution (NTT → NAT) was observed in three strains. The amino acid substitutions at positions 253 and 298 resulted in the loss of two potential glycosylation sites, which are present in the A2 strain and other RSV-A genotypes. In the HVR2 region of Bulgarian RSV-A, 27–38 serine and threonine residues were predicted to be O-glycosylated with a G score ≥0.5. The area of duplication contained a maximum of 10 O-glycosylation sites.
The deduced amino acid sequence of the HVR2 region of Bulgarian RSV-B was analysed in comparison to that of the prototype BA1 strain BA4128/99B (accession number: AY333364). The analysis also included sequences of the prototype BA9 strain BA/100/04 (accession number: DQ227395), prototype BA10 strain NG-077–07 (accession number: HM459886) and other representative sequences of known genotype strains (Figure 4(b)). A feature characteristic of BA viruses is a duplication of 60 nucleotides, resulting in a duplication of 20 amino acids (TERDTSTSQSTVLDTTTSKE) starting at position 240. As a result, protein G is extended to 312 amino acids (the stop codon in BA viruses is set to 313 amino acids). Amino acid substitutions V271A, I281T, S267L and S297F are genotype-specific for the different BA9 viruses, but Bulgarian BA9 strains possessed only these first two substitutions. In the studied RSV-B BA10 genotype virus, genotype-specific substitutions E226D, T289I and E292G were found, but substitution S269P was not [29,30]. In comparison to the prototype BA4128/99B strain, the HVR2 regions of Bulgarian BA9 and BA10 viruses contained amino acid substitutions at 22 positions. Substitutions K218T, L223P, S247P, T254I, T270I, V271A, I281T and H287Y were identified in most Bulgarian RSV-B viruses, while other substitutions were unique to individual viruses.
The RSV-B BA genotype sequences have two putative N-glycosylation motifs in the HVR2 region at positions 296 and 310. The amino acid substitution T312 N resulted in the loss of the second potential N-glycosylation site in three Bulgarian BA9 strains. In other strains, both N-glycosylation motifs were conserved, without changes to the amino acids therein. In the HVR2 region of the RSV-B strains studied here, 29–40 serine and threonine residues had the potential for O-glycosylation with a G score ≥0.5; 10 of those sites were found in the duplication region.
Discussion
Here, we describe the circulation of 12 respiratory viruses among children under the age of 5 in Bulgaria, a country in Southeast Europe. This study highlighted the circulation of RSV in particular, in addition to its genetic analysis, during two consecutive seasons. During the studied surveillance periods, the percentage of positive cases for influenza and non-influenza respiratory viruses were 23.6 and 46.7%, respectively. Among all studied respiratory viruses, RSV was the most frequently identified aetiologic agent of ARI. The incidence of RSV infections can vary from country-to-country and from year-to-year depending on the population, detection methods, climate and other factors [31]. In this study, the overall incidence rate of RSV was 26%: 28.8% in the first season with a dominance by RSV-A and 24.2% in the second season when RSV-B prevailed. Year-to-year fluctuations in the incidence of RSV and a shift from RSV-A to RSV-B dominance were observed in Bulgaria, in agreement with other studies [30,32]. The change in the circulation of the two RSV subgroups is likely due to the formation of a specific herd immunity against the RSV subgroup that was prevalent in the country in the preceding year. Consistent shifts in RSV subgroup dominance at different time intervals (1–3 years) have been reported worldwide [21,30,32,33]. Like other countries, the incidence of RSV infections was highest in the youngest age groups (≤11 months) and decreased with increasing age due to the development of immunity after repeated infections [5]. The early onset of RSV infection points to the need for the administration of a RSV vaccine soon after birth or during the last months of pregnancy [34]. No statistical significance in gender predominance was found, which contradicts other studies [31]. In countries with a temperate climate, such as Bulgaria, RSV episodes generally occur during the winter and spring months, although there are considerable variations [31]. In the present study, RSV epidemics continued from November to late April, which is consistent with the data from other European countries [35]. Information about the monthly distribution of detected RSV is important for enacting prophylactic measures among high risk children and for strengthening control measures to prevent nosocomial infections.
As in previous seasons, positive respiratory virus tests were higher among inpatients (74%) compared to outpatients (60%) (p < .05) [36]. RSV was also a major pathogen associated with hospitalization in children; 86% (135/157) of all RSV infections detected occurred in hospitalized children. Not surprisingly, RSV is the most important pathogen of the lower respiratory tract in infants and young children and is the major cause of bronchiolitis and pneumonia [37]. In our study, the highest percentage of bronchiolitis (48%) and pneumonia (38%) cases were associated with an RSV infection. No statistically significant differences in the incidence of RSV-A vs. RSV-B were found in these clinical groups. RSV-associated cases of LRTI were observed more frequently in children under the age of 2, which indicates higher susceptibility of this age group to RSV infection.
Phylogenetic analysis showed that all 21 Bulgarian RSV-A strains were of the ON1 genotype initially detected in Ontario, Canada in 2010 [28]. The ON1 genotype then became the predominant genotype among recently circulating RSV-A strains in many countries [15,17,35,38–51], and it was suggested that ON1 evolved from the ancestral genotype NA1 [15]. The 24 studied RSV-B strains belonged to the BA9 genotype; only one sequence belonged to the BA10 genotype. The BA genotype was first detected in Buenos Aires, Argentina in 1999 [52], and since then has rapidly disseminated across the world, evolving into at least 12 branches (BA1-BA12 genotypes) [35,39,46,53,54]. By 2006, BA had become the globally dominant RSV-B genotype and almost completely replaced other RSV-B genotypes [55]. BA9 and B10 genotypes were described for the first time in Niigata, Japan in 2007 [29] and were recently the prevalent genotype in many countries [35,56–62]. It has been reported that several genotypes within each subgroup can co-circulate in a single population during one epidemic season [30]. The small genotypic diversity of Bulgarian RSV could be explained by the short study period (two seasons) and the small territory of the country. All studied Bulgarian RSV-A and RSV-B viruses contained a 72-nt and a 60-nt duplication in the HVR2 region, respectively, which is characteristic of ON1 and BA genotypes. The rapid and widespread distribution throughout the world of viruses harbouring these unique tandem repeat insertions suggests that the duplications provide selective advantages to the viruses [55]. Hotard et al. (2015) demonstrated that the duplicated region in the BA strain augments virus attachment and fitness [63]. In the HVR2 of Bulgarian RSV-A and RSV-B, 24 and 22 amino-acid substitutions were detected compared to prototype ON67–1210A and BA4128/99B strains, respectively, which confirms the high genetic variability of the RSV G gene. The duplication of 23 and 20 amino acids in the studied RSV-A and RSV-B strains, respectively, resulted in the acquisition of additional potential glycosylation sites in the G protein. The continuous accumulation of amino acid changes and the extensive glycosylation of the RSV G protein with N- and O-linked sugars are important viral mechanisms for circumventing neutralization by pre-existing antibodies. The high degree of genetic variability allows RSV the ability to cause repeated infections during the same individual and annual epidemics.
In summary, this study provides important information concerning the major viral respiratory infections, in particular RSV, detected among children under the age of 5 in Bulgaria. It pinpoints RSV and influenza viruses as the causative agents of serious respiratory diseases that occur during early childhood. This is the first study that has identified the genotype of the RSV strains circulating in Bulgaria. In addition, this study highlights the necessity of routine laboratory-based surveillance of this pathogen in order to determine RSV prevalence and genotype distribution throughout the country. Phylogenetic and molecular analyses of RSV are essential for identifying new epidemic strains, understanding the clinical impact of genetic changes and facilitating the development of effective strategies for prevention and therapy. In paediatric practice, diagnostic testing for RSV and other respiratory viruses using molecular methods may lead to the reduced use of antibiotics and may assist in measures to control infection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by Ministry of Health in Bulgaria (National plan of Republic of Bulgaria for influenza pandemic preparedness).
Supplemental data
The supplemental material for this paper is available online at https://doi.org/10.1080/20477724.2017.1375708
Supplementary Material
Acknowledgements
The authors are grateful to all the study’s nurses and paediatricians for their submission of patient clinical samples and clinical information.
References
- [1].Mäkelä MJ, Puhakka T, Ruuskanen O, et al. . Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36(2):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shi T, McAllister DA, O’Brien KL, et al.. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390(10098):946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cane PA. 2001. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol. 2001;11(2):103–116. 10.1002/(ISSN)1099-1654 [DOI] [PubMed] [Google Scholar]
- [4].Afonso CL, Amarasinghe GK, Banyai K, et al. . Taxonomy of the order Mononegavirales: update 2016. Adv Virol. 2016;161:2351–2360. 10.1007/s00705-016-2880-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Collins PL, Karron RA. Respiratory syncytial virus and metapneumovirus In: Knipe DM, Howley PM, editors. Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia: 2013, vol. 1, p. 1086–1123. [Google Scholar]
- [6].Holmes EC. Virus evolution In: Knipe DM, Howley PM, editors. Fields virology, 6 ed. Lippincott Williams & Wilkins, Philadelphia: 2013, vol. 1, p.286–313. [Google Scholar]
- [7].Melero JA, García-Barreno B, Martínez I, et al. . Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78(10):2411–2418. 10.1099/0022-1317-78-10-2411 [DOI] [PubMed] [Google Scholar]
- [8].Johnson PR, Spriggs MK, Olmsted RA, et al. . The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Nat Acad Sci. 1987;84(16):5625–5629. 10.1073/pnas.84.16.5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martinez I, Dopazo J, Melero JA. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol. 1997;78(10):2419–2429. 10.1099/0022-1317-78-10-2419 [DOI] [PubMed] [Google Scholar]
- [10].Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162(1–2):80–99. 10.1016/j.virusres.2011.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Palomo C, García-Barreno B, Peñas C, et al. . The G protein of human respiratory syncytial virus: significance of carbohydrate side-chains and the Cterminal end to its antigenicity. J Gen Virol. 1991;72(3):669–675. 10.1099/0022-1317-72-3-669 [DOI] [PubMed] [Google Scholar]
- [12].Melero JA, Moore ML. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity. Curr Top Microbiol Immunol. 2013;372:59–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mufson MA, Orvell C, Rafnar B, et al. . Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66(10):2111–2124. 10.1099/0022-1317-66-10-2111 [DOI] [PubMed] [Google Scholar]
- [14].Trento A, Viegas M, Galiano M, et al. . Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol. 2006;80(2):975–984. 10.1128/JVI.80.2.975-984.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hirano E, Kobayashi M, Tsukagoshi H, et al. . Molecular evolution of human respiratory syncytial virus attachment glycoprotein (G) gene of new genotype ON1 and ancestor NA1. Infect Genet Evol. 2014;28:183–191. 10.1016/j.meegid.2014.09.030 [DOI] [PubMed] [Google Scholar]
- [16].Duvvuri VR, Granados A, Rosenfeld P, et al. . Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics. Sci Rep. 2015;5:e14268 10.1038/srep14268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cui G, Zhu R, Deng J, et al. . Rapid replacement of prevailing genotype of human respiratory syncytial virus by genotype ON1 in Beijing, 2012–2014. Infect Genet Evol. 2015;33:163–168. 10.1016/j.meegid.2015.04.025 [DOI] [PubMed] [Google Scholar]
- [18].Zheng Y, Liu L, Wang S, et al. . Prevailing genotype distribution and characteristics of human respiratory syncytial virus in northeastern China. J Med Virol. 2017;89(2):222–233. 10.1002/jmv.v89.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].CDC protocol of real-time RTPCR for influenza A(H1N1). Geneva: World Health, Organization; 2009. Available at: https://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf?ua=1. [Google Scholar]
- [20].Kodani M, Yang G, Conklin LM, et al. . Application of Taqman low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. 2011;49(6):2175–2182. 10.1128/JCM.02270-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zlateva KT, Vijgen L, Dekeersmaeker N, et al. . Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J Clin Microbiol. 2007;45(9):3022–3030. 10.1128/JCM.00339-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reiche J, Schweiger B. Genetic variability of group a human respiratory syncytial virus strains circulating in Germany from 1998 to 2007. J Clin Microbiol. 2009;47(6):1800–1810. 10.1128/JCM.02286-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zlateva KT, Lemey P, Moes E, et al. . Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J Virol. 2005;79(14):9157–9167. 10.1128/JVI.79.14.9157-9167.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Venter M, Madhi SA, Tiemessen CT, Schoub BD et al. . Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol. 2001;82(9):2117–2124. 10.1099/0022-1317-82-9-2117 [DOI] [PubMed] [Google Scholar]
- [26].Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins. NetNGlycServer. 2004. Available from: https://www.cbs.dtu.dk/services/NetNGlyc/.Accessed:2014 Apr 6.
- [27].Steentoft C, Vakhrushev SY, Joshi HJ, et al. . Precision mapping of the human O-GalNAc glycoproteome through simplecell technology. EMBO J. 2013;32(10):1478–1488. 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eshaghi A, Duvvuri VR, Lai R, et al. . Genetic variability of human respiratory syncytial virus a strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS ONE. 2012;7(3):e32807 10.1371/journal.pone.0032807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dapat IC, Shobugawa Y, Sano Y, et al. . New genotypes within respiratory syncytial virus group B genotype BA in Niigata. J Clin Microbiol. 2010;48(9):3423–3427. 10.1128/JCM.00646-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Houspie L, Lemey P, Keyaerts E, et al. . Circulation of HRSV in Belgium: from multiple genotype circulation to prolonged circulation of predominant genotypes. PLoS ONE. 2013;8(4):e60416 10.1371/journal.pone.0060416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Y, Yuan L, Zhang Y, et al. . Burden of respiratory syncytial virus infections in China: Systematic review and meta-analysis. J Glob Health. 2015;5(2):020417 10.7189/jogh.05.020417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cui G, Zhu R, Qian Y, et al. . Genetic variation in attachment glycoprotein genes of human respiratory syncytial virus subgroups A and B in children in recent five consecutive years. PLoS ONE. 2013;8(9):e75020 10.1371/journal.pone.0075020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Waris M. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J Infect Dis. 1991;163(3):464–469. 10.1093/infdis/163.3.464 [DOI] [PubMed] [Google Scholar]
- [34].Saso A, Kampmann B. Vaccination against respiratory syncytial virus in pregnancy: a suitable tool to combat global infant morbidity and mortality? Lancet Infect Dis. 2016;16(8):e153–e163. 10.1016/S1473-3099(16)00119-5 [DOI] [PubMed] [Google Scholar]
- [35].Gimferrer L, Campins M, Codina MG, et al. . Molecular epidemiology and molecular characterization of respiratory syncytial viruses at a tertiary care university hospital in Catalonia (Spain) during the 2013–2014 season. J Clin Virol. 2015;66:27–32. 10.1016/j.jcv.2015.02.018 [DOI] [PubMed] [Google Scholar]
- [36].Korsun N, Teodosieva A, Angelova S. Etiological role of ortho- and paramyxoviruses in acute respiratory tract infections among children aged <4 years in Bulgaria. Clin Lab. 2015;61(3–4):219–226. [DOI] [PubMed] [Google Scholar]
- [37].Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001;344(25):1917–1928. 10.1056/NEJM200106213442507 [DOI] [PubMed] [Google Scholar]
- [38].Agoti CN, Otieno JR, Gitahi CW, et al. . Rapid spread and diversification of respiratory syncytial virus genotype ON1. Emerg Infect Dis. 2014;20(6):950–959. 10.3201/eid2006.131438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Auksornkitti V, Kamprasert N, Thongkomplew S, et al. . Molecular characterization of human respiratory syncytial virus, 2010–2011: identification of genotype ON1 and a new subgroup B genotype in Thailand. Adv Virol. 2014;159(3):499–507. 10.1007/s00705-013-1773-9 [DOI] [PubMed] [Google Scholar]
- [40].Liu J, Mu Y, Dong W, et al. . Genetic variation of human respiratory syncytial virus among children with fever and respiratory symptoms in Shanghai, China, from 2009 to 2012. Infect Genet Evol. 2014;27:131–136. 10.1016/j.meegid.2014.07.011 [DOI] [PubMed] [Google Scholar]
- [41].Ren L, Xia Q, Xiao Q, et al. . The genetic variability of glycoproteins among respiratory syncytial virus subtype A in China between 2009 and 2013. Infect Genet Evol. 2014;27:339–347. 10.1016/j.meegid.2014.07.030 [DOI] [PubMed] [Google Scholar]
- [42].Tsukagoshi H, Yokoi H, Kobayashi M, et al. . Genetic analysis of attachment glycoprotein (G) gene in new genotype ON1 of human respiratory syncytial virus detected in Japan. Microbiol Immunol. 2013;57(9):655–659. [DOI] [PubMed] [Google Scholar]
- [43].Lee WJ, Kim YJ, Kim DW, et al. . Complete genome sequence of human respiratory syncytial virus genotype A with a 72-nucleotide duplication in the attachment protein G gene. J Virol. 2012;86(24):13810–13811. 10.1128/JVI.02571-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Prifert C, Streng A, Krempl CD, et al. . Novel respiratory syncytial virus a genotype, Germany, 2011–2012. Emerg Infect Dis. 2013;19(6):1029–1030. 10.3201/eid1906.121582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Valley-Omar Z, Muloiwa R, Hu NC, et al. . Novel respiratory syncytial virus subtype ON1 among children, Cape Town, South Africa, 2012. Emerg Infect Dis. 2013;19(4):668–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Khor CS, Sam IC, Hooi PS, et al. . Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989–2011. Infect Genet Evol. 2013;14:357–360. 10.1016/j.meegid.2012.12.017 [DOI] [PubMed] [Google Scholar]
- [47].Pierangeli A, Trotta D, Scagnolari C, et al. . Rapid spread of the novel respiratory syncytial virus A ON1 genotype, central Italy, 2011 to 2013. Euro Surveill. 2014;19(26):20843. [DOI] [PubMed] [Google Scholar]
- [48].Choudhary ML, Anand SP, Wadhwa BS, et al. . Genetic variability of human respiratory syncytial virus in Pune. Infect Genet Evol. 2013;20:369–377. 10.1016/j.meegid.2013.09.025 [DOI] [PubMed] [Google Scholar]
- [49].Balmaks R, Ribakova I, Gardovska D, et al. . Molecular epidemiology of human respiratory syncytial virus over three consecutive seasons in Latvia. J Med Virol. 2014;86(11):1971–1982. 10.1002/jmv.v86.11 [DOI] [PubMed] [Google Scholar]
- [50].Kim YJ, Kim DW, Lee WJ, et al. . Rapid replacement of human respiratory syncytial virus A with the ON1 genotype having 72 nucleotide duplication in G gene. Infect Genet Evol. 2014;26:103–112. 10.1016/j.meegid.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tabatabai J, Prifert C, Pfeil J, et al. . Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during Winter Season 2012–13. PLoS ONE. 2014;9(10):e109191 10.1371/journal.pone.0109191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Trento A, Galiano M, Videla C, et al. . Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84(11):3115–3120. 10.1099/vir.0.19357-0 [DOI] [PubMed] [Google Scholar]
- [53].Martinelli M, Frati ER, Zappa A, et al. . Phylogeny and population dynamics of respiratory syncytial virus (Rsv) A and B. Virus Res. 2014;189:293–302. 10.1016/j.virusres.2014.06.006 [DOI] [PubMed] [Google Scholar]
- [54].Tran DN, Pham TMH, Ha MT, et al. . Molecular epidemiology and disease severity of human respiratory syncytial virus in Vietnam. PLoS ONE. 2013;8(1):e45436 10.1371/journal.pone.0045436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Trento A, Casas I, Calderon A, et al. . Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84(15):7500–7512. 10.1128/JVI.00345-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Baek YH, Choi EH, Song MS, et al. . Prevalence and genetic characterization of respiratory syncytial virus (RSV) in hospitalized children in Korea. Adv Virol. 2012;157(6):1039–1050. 10.1007/s00705-012-1267-1 [DOI] [PubMed] [Google Scholar]
- [57].Ohno A, Suzuki A, Lupisan S, et al. . Genetic characterization of human respiratory syncytial virus detected in hospitalized children in the Philippines from 2008 to 2012. J Clin Virol. 2013;57(1):59–65. 10.1016/j.jcv.2013.01.001 [DOI] [PubMed] [Google Scholar]
- [58].Malasao R, Okamoto M, Chaimongkol N, et al. . Molecular characterization of human respiratory syncytial virus in the Philippines, 2012–2013. PLoS ONE. 2015;10(11):e0142192 10.1371/journal.pone.0142192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Esposito S, Piralla A, Zampiero A, et al. . Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in Northern Italy in five consecutive winter seasons. PLoS ONE. 2015;10(6):e0129369 10.1371/journal.pone.0129369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nagasawa K, Hirano E, Kobayashi M, et al. . Molecular evolution of the hypervariable region of the attachment glycoprotein gene in human respiratory syncytial virus subgroup B genotypes BA9 and BA10. Infect Genet Evol. 2015;36:217–223. 10.1016/j.meegid.2015.09.020 [DOI] [PubMed] [Google Scholar]
- [61].Fall A, Dia N, Cisse EHAK, et al. . Epidemiology and molecular characterization of human respiratory syncytial virus in Senegal after four consecutive years of surveillance, 2012–2015. PLoS ONE. 2016;11(6):e0157163 10.1371/journal.pone.0157163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Slovic A, Ivancic-Jelecki J, Ljubin-Sternak S, et al. . A molecular epidemiological study of human respiratory syncytial virus in Croatia, 2011–2014. Infect Genet Evol. 2016;44:76–84. 10.1016/j.meegid.2016.06.036 [DOI] [PubMed] [Google Scholar]
- [63].Hotard AL, Laikhter E, Brooks K, et al. . Functional analysis of the 60-nucleotide duplication in the respiratory syncytial virus buenos aires strain attachment glycoprotein. J Virol. 2015;89(16):8258–8266. 10.1128/JVI.01045-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.