Abstract
Occasional auto‐modification of ubiquitin ligases typically leads to their proteasomal destruction, but new findings published in The EMBO Journal now show that in the case of Rsp5/Nedd4, auto‐ubiquitylation instead triggers oligomerization and concomitant reduction of ligase activity. This novel mechanism therefore creates silenced ligases that remain poised for reactivation.
Subject Categories: Post-translational Modifications, Proteolysis & Proteomics
It is well known that protein kinases and phosphatases may dynamically regulate enzyme activity through cycles of protein phosphorylation and de‐phosphorylation. There are approximately 600 protein kinases identified in the human proteome, which is interestingly roughly equivalent to the known number of human ubiquitin ligases (aka E3 enzymes), the enzymes responsible for transfer of the highly conserved, 76 amino acid protein ubiquitin onto lysine residues of protein substrates in eukaryotic cells (Kleiger & Mayor, 2014). It therefore appears surprising that protein ubiquitylation, at least so far, has not yet been found to play similar roles in reversible control of enzyme activity. However, thanks to the hard work of Attali and colleagues (Attali et al, 2017), this belief may be about to change.
Attali et al (2017) provide major new insights into the mechanism regulating the Nedd4 family of ubiquitin ligases. These important enzymes control a myriad of cellular functions including intracellular trafficking and endocytosis, and their amino acid sequences are highly conserved from yeast to humans. In fact, the activity of the yeast Nedd4 ortholog Rsp5 is essential for viability of this organism. For understanding the details of the authors' discovery, a little background information is necessary: There are two major classes of E3s catalyzing protein ubiquitylation, the HECT (homologous to E6AP C‐terminus; Kee & Huibregtse, 2007) and RING (really interesting new gene; Deshaies & Joazeiro, 2009) E3 families. The function of RING E3s is to recruit both ubiquitin‐conjugating (E2) enzymes and protein substrates, and to promote ubiquitin transfer directly from an E2 onto the substrate. HECT E3s, including the Nedd4 family, differ from RING E3s in that ubiquitin is first transferred from the E2 onto an active‐site cysteine residue of E3, and from there finally onto E3‐bound substrates. While the fate of ubiquitylated proteins is typically degradation by the 26S proteasome, ubiquitylation may also direct cellular trafficking or regulate protein–protein interactions. Finally, note that ubiquitylation is not necessarily an irreversible event, since de‐ubiquitylating enzymes (DUBs) that can trim off ubiquitin moieties from covalently modified proteins are present in all eukaryotic cells.
Interestingly, many characterized E3s have the propensity to ubiquitylate themselves (referred to as auto‐ubiquitylation) in addition to their substrates. In cells, such auto‐ubiquitylation often leads to proteasomal degradation of the ligase; however, this is not always the case. For instance, Rsp5 is found auto‐ubiquitylated in yeast without affecting its stability (Wang et al, 1999). Rsp5 has also been found to assemble into oligomers (Dunn & Hicke, 2001), although the functional consequences of auto‐ubiquitylation and self‐assembly were up until now shrouded in mystery. Through elegant biochemical and cell biological assays, Attali et al (2017) demonstrate that ubiquitin conjugation to lysine side chains on an α‐helix adjacent to the Rsp5 catalytic domain triggers conformational changes, which in turn enable trimerization of ubiquitylated Rsp5 subunits and subsequent inhibition of Rsp5 activity (Fig 1).
Figure 1. Schematic describing how NEDD4 family ubiquitin ligases self‐regulate through auto‐ubiquitylation.
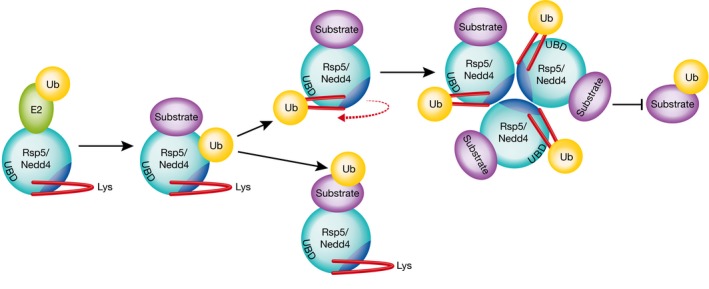
Ubiquitin (UB) is transferred from a ubiquitin‐conjugating enzyme (E2) to either the Rsp5/Nedd4 active‐site cysteine and then to E3‐bound substrate, or onto a flexible α‐helix on the E3. Upon auto‐ubiquitylation of the α‐helix, the position of the helix rotates to interact with a conserved ubiquitin‐binding domain (UBD) opening access to an oligomerization domain and trimerization of Nedd4. Self‐association renders the E3 inactive.
The molecular mechanism behind this rearrangement involves the known ubiquitin‐binding domain (UBD) of Rsp5. In the absence of auto‐ubiquitylation, the α‐helix in the Rsp5 HECT domain constrains the cryptic intermolecular interface such that steric clashes would occur upon oligomerization. Ubiquitylation on the N‐terminal side of this α‐helix results in the formation of a complex between ubiquitin and the UBD, alleviating the previous steric clashes and permitting oligomerization (Fig 1). Importantly, the authors provide evidence that this model applies to both Rsp5 and its mammalian homolog Nedd4, and furthermore show that mutation of the auto‐ubiquitylated lysine residues on Nedd4 has physiological consequences consistent with unregulated Nedd4 activity in human cells.
Similar to how protein phosphorylation controls enzyme activity, Rsp5/Nedd4 auto‐ubiquitylation not only complements previously described modes of regulation, which unsurprisingly also include ligase phosphorylation (Kee & Huibregtse, 2007), but it may also provide the means to control E3 activity reversibly instead via irreversible degradation of the ligase. Such a mechanism would be highly attractive for signal transduction pathways that require quick responses to stimuli: If E3s were rapidly degraded upon auto‐ubiquitylation, a significant amount of time would be required for fresh rounds of transcription and protein synthesis to occur, whereas DUB‐catalyzed ubiquitin removal from a protein can occur in a matter of seconds. In fact, Rsp5 is known to associate with the DUB enzyme Ubp2 (Kee et al, 2005), and it will be interesting to test whether Ubp2 is involved in reactivation of inhibited Rsp5, and whether similar DUB interactions occur for Nedd4 as well. On the other hand, perhaps auto‐ubiquitylated Rsp5/Nedd4 is akin to a mouse trap, where binding of at least some substrates is sufficient to release the trap and trigger reactivation of the enzyme.
Regardless of the precise mechanism, reversible regulation of E3 activity through self‐modification is tantalizing since the phenomenon of auto‐ubiquitylation is rather common. In fact, a somewhat similar mechanism exists to regulate the activities of Cullin–RING ligases (CRLs), the largest class of RING E3s (Deshaies & Joazeiro, 2009). Here, inactive CRLs are covalently modified by the ubiquitin‐like protein Rub1/Nedd8, which serves to activate them (Saha & Deshaies, 2008). This mechanism is, of course, distinct in comparison with E3 auto‐ubiquitylation, since a dedicated separate E3, Dcn1, is responsible for catalyzing Nedd8 conjugation to CRLs (Scott et al, 2011). Nevertheless, it is interesting to compare these mechanisms, as in the case of CRLs, a protease called the COP9 signalosome is responsible for removing Nedd8 from CRLs, producing a rapid molecular switch to regulate CRL activity (Lyapina et al, 2001).
As with any groundbreaking paper, many questions remain regarding how Rsp5/Nedd4 auto‐ubiquitylation results in inhibition of E3 activity. Specifically, how does trimerization inhibit the enzyme? Structural studies on Rsp5 suggest that the mechanism of ubiquitylation involves large conformational changes, and trimerization may inhibit these from occurring (Kamadurai et al, 2013). Furthermore, HECT E3s are highly complex enzymes with functions including (but not limited to) recruitment of both substrate and the E2–ubiquitin complex, promoting ubiquitin transthiolation from E2 to E3, and transfer of ubiquitin from E3 to a lysine residue on the substrate. Additional biochemical and structural work will be necessary to uncover how trimerization affects these critical steps during catalysis. Finally, it was recently determined that both Rsp5 and Nedd4 play important roles in ridding the cytoplasm of misfolded proteins (Fang et al, 2014), and determining how E3 regulation fits into this physiological context will be highly important as well.
Acknowledgements
This work, including the efforts of Spencer Hill and Gary Kleiger, was funded by HHS | National Institutes of Health (NIH) (R15 GM117555‐ 01).
See also: I Attali et al (February 2017)
References
- Attali I, Tobelaim WS, Persaud A, Motamedchaboki K, Simpson‐Lavy KJ, Mashahreh B, Levin‐Kravets O, Keren‐Kaplan T, Pilzer I, Kupiec M, Wiener R, Wolf DA, Rotin D, Prag G (2017) Ubiquitylation‐dependent oligomerization regulates activity of Nedd4 ligases. EMBO J 36: 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 1 [DOI] [PubMed] [Google Scholar]
- Dunn R, Hicke L (2001) Domains of the Rsp5 ubiquitin‐protein ligase required for receptor‐mediated and fluid‐phase endocytosis. Mol Biol Cell 12: 421–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Chan GT, Zhu M, Comyn SA, Persaud A, Deshaies RJ, Rotin D, Gsponer J, Mayor T (2014) Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat Cell Biol 16: 1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Qiu Y, Deng A, Harrison JS, Macdonald C, Actis M, Rodrigues P, Miller DJ, Souphron J, Lewis SM, Kurinov I, Fujii N, Hammel M, Piper R, Kuhlman B, Schulman BA (2013) Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife 2: e00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J 24: 2414–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Huibregtse JM (2007) Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun 354: 329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Mayor T (2014) Perilous journey: a tour of the ubiquitin‐proteasome system. Trends Cell Biol 24: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD‐CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA (2011) N‐terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM (1999) Functional domains of the Rsp5 ubiquitizcn‐protein ligase. Mol Cell Biol 19: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]


