Abstract
Chloroplasts are cellular organelles of plants and algae that are responsible for energy conversion and carbon fixation by the photosynthetic reaction. As a consequence of their endosymbiotic origin, they still contain their own genome and the machinery for protein biosynthesis. Here, we present the atomic structure of the chloroplast 70S ribosome prepared from spinach leaves and resolved by cryo‐EM at 3.4 Å resolution. The complete structure reveals the features of the 4.5S rRNA, which probably evolved by the fragmentation of the 23S rRNA, and all five plastid‐specific ribosomal proteins. These proteins, required for proper assembly and function of the chloroplast translation machinery, bind and stabilize rRNA including regions that only exist in the chloroplast ribosome. Furthermore, the structure reveals plastid‐specific extensions of ribosomal proteins that extensively remodel the mRNA entry and exit site on the small subunit as well as the polypeptide tunnel exit and the putative binding site of the signal recognition particle on the large subunit. The translation factor pY, involved in light‐ and temperature‐dependent control of protein synthesis, is bound to the mRNA channel of the small subunit and interacts with 16S rRNA nucleotides at the A‐site and P‐site, where it protects the decoding centre and inhibits translation by preventing tRNA binding. The small subunit is locked by pY in a non‐rotated state, in which the intersubunit bridges to the large subunit are stabilized.
Keywords: chloroplast, cryo‐EM, pY, ribosome, translation
Subject Categories: Plant Biology, Protein Biosynthesis & Quality Control, Structural Biology
Introduction
Chloroplasts are cellular organelles in algae and higher plants responsible for carbon fixation by the photosynthetic reaction (Eberhard et al, 2008). Consequently, these autotrophic organisms are the primary source of fixed carbon and chemical energy in most ecosystems on earth. The endosymbiotic theory states that plastids, including the chloroplast, evolved through the engulfment of a cyanobacterium by the eukaryotic progenitor cell (Sagan, 1967; Margulis, 1970). This primary endosymbiotic event occurred about 1 billion years ago and led subsequently to three evolutionary lines of plastid‐containing organisms: the glaucophytes, the rhodophytes (red algae) and the chlorophytes (green algae), from which the higher plants diverged approximatively 400–475 million years ago (Gould et al, 2008; Jensen & Leister, 2014). Although transfer of genes to the nuclear genome happened over time (Timmis et al, 2004; Bock & Timmis, 2008), plastids still contain their own genome (plastome). In plastids of higher plants, these approximately 100 genes encode proteins and RNA molecules of the transcription and translation machinery and components of the photosynthetic apparatus (Sugiura, 1989). To control the proper function of the chloroplasts under changing environmental conditions, algae and plant cells evolved to coordinate the expression of the plastid‐ and nuclear‐encoded genes by regulating the levels of transcription, mRNA stability and translation (Jarvis & Lopez‐Juez, 2013).
The protein biosynthesis in chloroplasts is catalysed by a bacterial‐type 70S ribosome (Tiller & Bock, 2014), called chloroplast ribosome, composed of a 50S large subunit and a 30S small subunit. Although the chloroplast and the bacterial 70S ribosomes share a common ancestor, they have diverged considerably from each other as evident from proteomic analysis (Yamaguchi & Subramanian, 2000, 2003; Yamaguchi et al, 2000) and structural characterization at low resolution (Manuell et al, 2007; Sharma et al, 2007). Very recently, a cryo‐EM reconstruction of the chloroplast large ribosomal subunit at 3.5 Å resolution was published (Ahmed et al, 2016) and their findings concerning the 50S subunit are in agreement with our study of the complete chloroplast 70S ribosome. The chloroplast ribosomal RNA (rRNA) (total 4,524 nucleotides in spinach) is about the same length as in the bacterial ribosome (total 4,566 nucleotides in Escherichia coli). However, it is fragmented to include a 4.5S rRNA molecule with sequence homology to the 3′ tail of the bacterial 23S rRNA. Although bL25 and uL30 are completely missing in the chloroplast 70S ribosome, the total protein mass is increased by more than 170,000 Da due to extension of ribosomal proteins sharing homology with bacteria and the acquisition of three plastid‐specific ribosomal proteins (PSRPs) to the small and two PSRPs to the large subunit (Yamaguchi & Subramanian, 2000, 2003; Yamaguchi et al, 2000). The new components, together with plastid‐specific translation factors, play an important role in the regulation of translation and stability of the ribosome (Yamaguchi & Subramanian, 2003; Manuell et al, 2007; Sharma et al, 2007).
To better understand the evolution and the function of ribosomes in plastids, we determined the atomic structure of the chloroplast 70S ribosome in complex with the plastid translation factor pY using cryo‐electron microscopy (cryo‐EM).
Results and Discussion
Structure of the chloroplast 70S ribosome
Chloroplast 70S ribosomes were purified from spinach (Spinacia oleracea) leaves and investigated by cryo‐EM single‐particle analysis (Appendix Fig S1). The three‐dimensional (3D) reconstruction of the complete chloroplast 70S ribosome was resolved to 3.4 Å (Figs 1A and EV1, and Appendix Fig S2). To improve the structural interpretation, the particle images were further classified using maximum‐likelihood‐based algorithms, masking, and signal subtraction approaches to yield cryo‐EM maps of the 50S large subunit at 3.2 Å resolution (Fig EV1 and Appendix Fig S2) and of the 30S small subunit at 3.6 Å resolution (Fig EV1 and Appendix Fig S3). The obtained maps were of sufficient quality to allow building and refinement of an almost complete model of the chloroplast 70S ribosome (Fig 1A, and Appendix Figs S4 and S5; Appendix Tables S1,S2,S3) revealing the chloroplast‐specific ribosomal features at molecular detail.
Figure 1. Architecture of the chloroplast 70S ribosome.
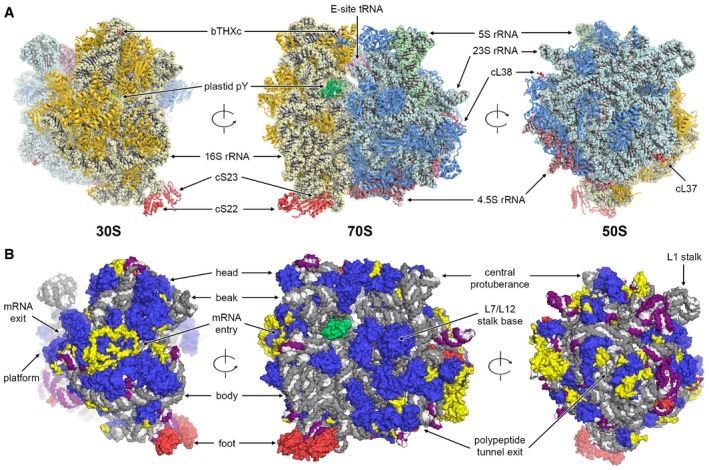
- Structure of the chloroplast 70S ribosome. 50S subunit proteins are in blue, 23S rRNA in cyan, 5S rRNA in green, 4.5S rRNA in red, 30S subunit proteins in gold, 16S rRNA in pale yellow, E‐site tRNA in pink and translation factor pY in green. Plastid‐specific ribosomal proteins cS22, cS23, bTHXc, cL37 and cL38 are shown in red.
- Protein and rRNA elements conserved between chloroplast and bacterial 70S ribosome are in blue and grey, respectively. Chloroplast‐specific rRNA elements are shown in purple. Plastid‐specific ribosomal proteins and additional protein extensions are in red and yellow, respectively. Translation factor pY is shown in green. Structural landmarks of the 70S ribosome are indicated.
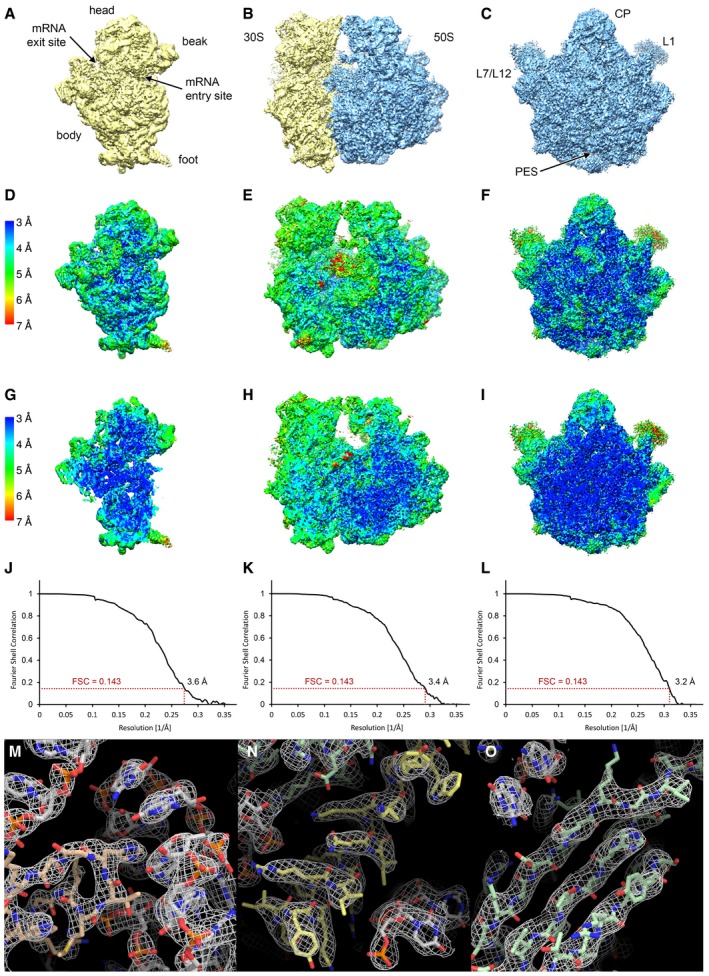
Figure EV1. Resolution and quality of the cryo‐EM reconstructions.
-
A–CSurface rendering of the final high‐resolution maps of the 30S subunit (A), the 70S ribosome (B) and the 50S subunit (C).
-
D–ILocal resolution plots showing the surface (D–F) and a cross section (G–I) of the cryo‐EM maps. Local resolution maps of the 30S (D, G), 70S (E, H) and 50S (F, I) are shown from the same view as in panels (A, B and C), respectively.
-
J–LFourier shell correlation (FSC) curves of the 30S (J), the 70S (K) and 50S (L) cryo‐EM reconstructions. The indicated resolutions are according to the FSC = 0.143 criterion (“gold‐standard”).
-
M–OExamples for the quality of the density: (M) ribosomal protein (salmon) interacting with rRNA (grey), (N) protein α‐helix (yellow) and (O) protein β‐sheet (green).
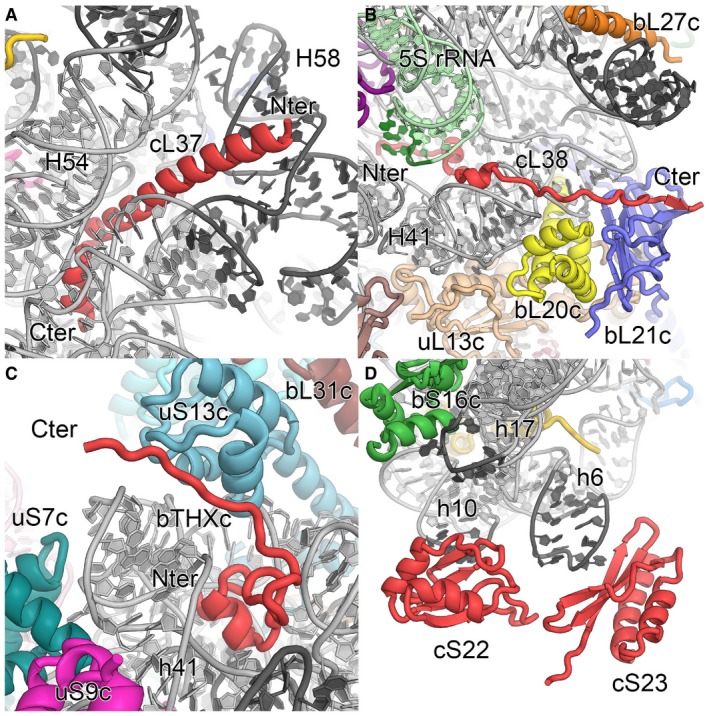
We could identify and position all plastid‐specific ribosomal proteins, which we also renamed to conform to the new convention of ribosomal protein nomenclature (Ban et al, 2014) (Fig 2). Several of these, bTHXc (PSRP4), cL37 (PSRP5) and cL38 (PSRP6), were de novo built into the electron density map (Figs 2A–C and EV2), whereas for the remaining two proteins, cS22 (PSRP2) and cS23 (PSRP3), which are bound to the more flexible foot of the small subunit, homology models were fitted as rigid bodies (Fig 2D and Appendix Fig S6). Compared to the bacterial 70S ribosome, the chloroplast 70S ribosome has different architectural features due to presence of additional proteins and N‐ and C‐terminal chloroplast‐specific extensions of ribosomal proteins with bacterial homologs (Fig 1B). The changes are particularly pronounced between the platform and shoulder of the 30S subunit and around the polypeptide exit site of the 50S subunit (Fig 1B). As a striking example, such protein extensions mediate the interactions between the ribosome and the plastid‐specific 4.5S rRNA (Whitfeld et al, 1978) (Fig EV3). In addition, we identified an intersubunit bridge unique for chloroplast ribosomes, called B7c, formed by a plastid‐specific extension of bS6c at the 30S platform and uL2c below the L1 stalk (Appendix Fig S7; Appendix Table S4).
Figure 2. Plastid‐specific ribosomal proteins.
-
A–CDe novo built and refined structures of the plastid‐specific ribosomal proteins cL37 (A), cL38 (B) and bTHXc (C) are shown in red, with N‐ and C‐termini indicated. 23S and 16S rRNAs in grey and 5S rRNA in green. Alterations in rRNA elements in comparison with bacteria are indicated with dark colour.
-
DRigid body fitted models of plastid‐specific ribosomal proteins cS22 and cS23 in red. Helices h6, h10 and h17 of 16S rRNA are indicated.
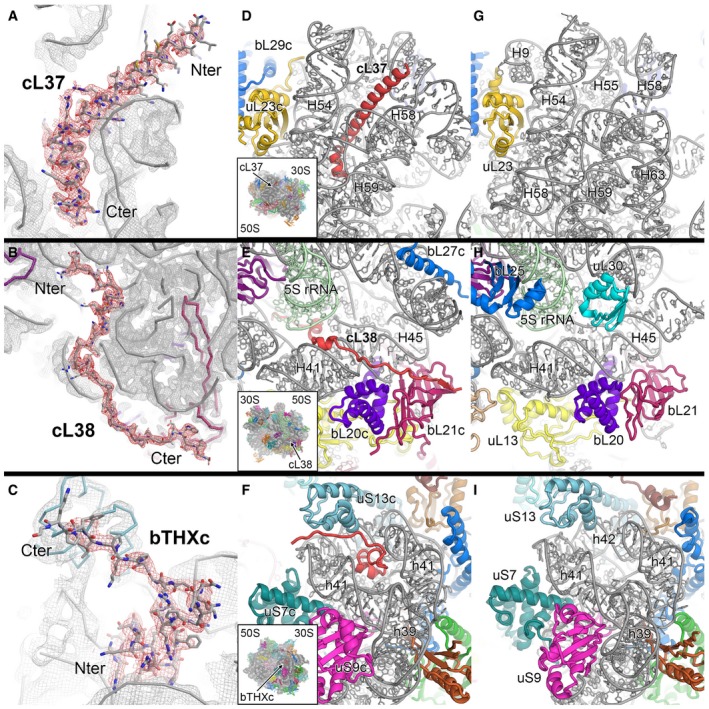
Figure EV2. De novo built plastid‐specific ribosomal proteins.
-
A–CThe density indicates clear side chain features and allows unambiguous tracing of cL37 (A), cL38 (B) and bTHXc (C).
-
D–FBinding sites of cL37 (D), cL38 (E) and bTHXc (F) in the chloroplast 70S ribosome.
-
G–ICorresponding sites to panels (D, E and F), respectively, in the bacterial 70S ribosome (PDB 4YBB; Noeske et al, 2015).
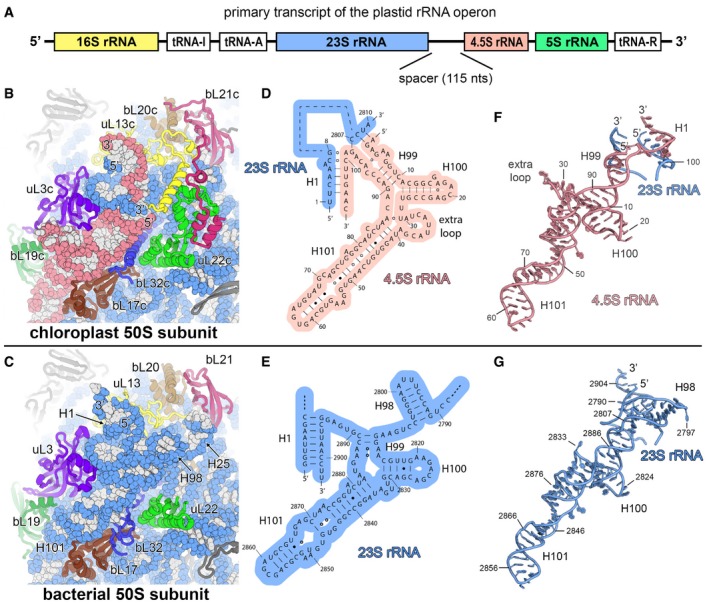
Figure EV3. The chloroplast 4.5S ribosomal RNA .
-
APrimary transcript of the chloroplast rRNA operon. The canonical sequences of ribosomal RNAs are indicated by coloured boxes. The 4.5S rRNA is separated from the 23S rRNA by a 115‐nucleotide RNA spacer.
-
B, CComparison of the chloroplast 50S subunit (B) with the bacterial 50S subunit (PDB 4YBB; Noeske et al, 2015) (C) indicating the structural rearrangement of the ribosomal proteins that interact with the 23S rRNA (blue) and the 4.5S rRNA (red).
-
D, ESecondary structure diagram of the chloroplast 4.5S rRNA (D) and the 3′ end of the bacterial 23S rRNA (E). The interactions of the 5′ and 3′ ends of the 4.5S rRNA with the 23S rRNA are shown. Watson–Crick base pairs are indicated by lines (‐), G•U base pairs by dots (•) and non‐standard base pairs by rings (○).
-
F, GModel of the chloroplast 4.5S rRNA (F) and the 3′ end of the bacterial 23S rRNA (G). The same nucleotides are shown in the models as represented in the secondary structure diagrams (D, E).
In contrast to the solvent side, the rRNA core and the interface of the two subunits are structurally much more conserved relative to the bacterial ribosome (Fig 1B), implying also a conserved mechanism of mRNA decoding by the small subunit and the peptide bond synthesis by the large subunit.
Structural insights into the role of the plastid‐specific ribosomal proteins
Plastid translation activity is absolutely required for the regular development and function of plants, and mutations of plastid ribosomal proteins and defective assembly of the translation apparatus influence the plant anatomy and morphology (Ahlert et al, 2003; Rogalski et al, 2006; Tiller et al, 2012). Although biochemical studies have not yet been conducted for all plastid ribosomal proteins, it appears that more ribosomal proteins are essential for proper translation activity in the chloroplast than in the bacterial (E. coli) 70S ribosome (Tiller & Bock, 2014). Interestingly, of the five chloroplast‐specific ribosomal proteins, cS23 (PSRP3), bTHXc (PSRP4) and cL37 (PSRP5) are essential for plastid translation activity (Tiller et al, 2012), and furthermore, cS22 (PSRP2) plays a role in plant development under stress conditions and has RNA chaperone activity (Xu et al, 2013). Under the tested conditions, the knockdown of cL38 (PSRP6) had no measurable effect on plastid translation (Tiller et al, 2012). Our atomic structure reveals the role of these proteins in the context of the chloroplast 70S ribosome.
The two proteins associated with the large subunit, cL37 and cL38, are lacking a homologue in bacteria, indicating that they were probably acquired later in evolution. cL37 forms a long helical structure and is positively charged (10.72 pI) due to many lysine and arginine side chains that interact with the RNA backbone (Figs 2A and EV2). Contrary to previous studies (Sharma et al, 2007) in which cL37 was incorrectly positioned close to the L1 stalk, we identified cL37 bound to a deep groove formed by several rRNA elements of 23S rRNA domain III (Fig 1A). Binding of cL37 at this position is accompanied by changes in helix H58 and the expansion of the loop connecting H54 and H55 when compared to bacteria (Fig EV2). Furthermore, the complete helix H63, which interacts with helices H58, H59 and H60 of domain III in bacteria, is absent in chloroplasts (Fig EV2), and it appears that cL37 is required to stabilize this rRNA fold through extensive interactions with the RNA backbone. This is in agreement with the biochemical data by Tiller et al (2012), showing that knockdown of cL37 leads to reduced level of 50S subunits, probably due to incomplete folding and subsequent degradation of the 23S rRNA.
For plants with a knockdown of cL38, no obvious changes in the plant phenotype were observed under the experimental conditions, and only a slightly lower content of thylakoid complexes could be measured (Tiller et al, 2012). Our map revealed an elongated fold of cL38, which is bound to the L7/L12 stalk base via N‐terminal interactions to the 23S and the 5S rRNA and C‐terminal protein–protein contacts to bL20c and bL21c (Figs 1A, 2B, and EV2). The hairpin D loop of 5S rRNA domain γ interacting with cL38 is rearranged due to the insertion of an additional nucleotide in the plastid 5S rRNA. In bacteria, domain γ is further contacting ribosomal proteins bL25 and uL30, which are missing in chloroplast ribosomes. Although cL38 probably evolved to locally stabilize the stalk base and the attachment of the 5S rRNA, bL25 and uL30 are not structurally replaced by cL38 or by other plastid ribosomal proteins (Fig EV2).
Plastid ribosomal protein bTHXc is homologous to bacterial protein bTHX (Yamaguchi & Subramanian, 2003) found so far only in the Thermus bacterial genus (Leontiadou et al, 2001), indicating convergent evolution or gene capture at a later step in evolution. Bacterial bTHX is a small (26 amino acids) basic (12.1 pI) protein, and its structure has been visualized by X‐ray crystallography as part of the 30S subunit of Thermus thermophilus bound to a cavity formed by 16S rRNA elements of the head (Wimberly et al, 2000). In our cryo‐EM map, we identified bTHXc located at the same place (Figs 1A, 2C, and EV2). The overlay of bTHXc (46 amino acids) with bTHX (Wimberly et al, 2000) indicates a similar core fold interacting with the 16S rRNA helices h41 and h42 and a plastid‐specific C‐terminal extension forming hydrophobic interactions with uS13c (Fig EV2). Therefore, bTHXc stabilizes the 16S rRNA of the 30S head and the intersubunit bridge B1b between uS13c and the central protuberance (CP) of the 50S subunit. In plants with a bTHXc knockdown, reduced levels of plastid 30S subunits lead to a reduced plastid translation activity and severe growth defects (Tiller et al, 2012). Considering that bTHX is only found in thermophilic bacteria, plastid ribosomes possibly acquired this protein to stabilize the ribosomes as an adaptation to fluctuating temperatures that the plant cells are exposed to.
The foot of the chloroplast 30S subunit is highly reorganized due to the truncation of 16S rRNA helices h6, h10, h17 and the acquisition of new proteins and protein extensions (Fig 1). Despite the structural flexibility of the foot, the use of local 3D classification allowed us to obtain a map into which homology models of cS22 and cS23 could be fitted as rigid bodies (Fig 2D and Appendix Fig S6), at a position consistent with previous reports (Sharma et al, 2007). cS22 contains two RNA‐binding motifs (RBM) connected by a flexible linker of 17 amino acids and shows RNA chaperone activity (Xu et al, 2013). Because only one RBM domain could be fitted into the density close to loop h10 and no other unassigned density in close proximity is visible, we suggest that the second RBM domain is flexibly attached to the first and may be used to bind mRNA during translation initiation or for localizing the ribosome within the chloroplast through interactions with RNA or ssDNA. The positioning of cS23 in the context of the changed structure of the rRNA suggests that this protein may function as a replacement for the truncated helix h6 (Fig 2D and Appendix Fig S6). The knockdown of cS23 leads to defective chloroplast translation with severe alterations of leaf anatomy (Tiller et al, 2012), indicating that cS23 is an important ribosomal protein with possible additional roles in ribosome assembly or in translation.
New features of the plastid ribosomal RNA
In the chloroplast 70S ribosome, the loss of rRNA mass through truncations is almost balanced by the acquisition of plastid‐specific rRNA expansion segments, which mainly protrude from the solvent side of the 50S subunit (Fig 1B). The 16S rRNA of the 30S subunit (Appendix Fig S8) responsible for mRNA binding and stabilization of the codon–anticodon interactions shows relatively small structural adaptations compared to the bacterial 16S rRNA except for truncations of helices h6, h10 and h17, which are involved together with cS22 and cS23 in forming the remodelled foot of the small subunit (Fig 2D and Appendix Fig S6). In the 23S rRNA of the 50S subunit (Appendix Fig S9), rRNA helices H7, H9, H45, H63 and H98 are truncated or completely lost. New rRNA expansions segments, unique for chloroplast ribosomes, have evolved on the solvent side and are partially stabilized by ribosomal protein extensions (Fig 1B). The 5S rRNA forms large parts of the CP and is structurally rearranged in the area where it interacts with cL38 (Fig 2B). The 50S subunit also contains a third rRNA molecule, the 4.5S rRNA (Whitfeld et al, 1978). In the plastome, the sequence of the 4.5S rRNA shows homology to the 3′ tail of bacterial 23S rRNA (55% sequence identity); however, it is separated from the 23S rRNA by a 115 nt spacer sequence (Fig EV3). Our map revealed the complete fold and interactions of the 4.5S rRNA (Fig 3A), which occupies a similar position on the chloroplast ribosome as the 3′ tail of the bacterial 23S rRNA and forms interactions with uL3c, uL13c, bL17c, bL19c and bL32c. A loop formed by nine nucleotides (comprising residues 30–38) unique to the 4.5S rRNA interacts with the N‐terminal extension of protein bL19c and several loops of uL3c (Fig EV3). The interaction between the 5′ end of the 4.5S rRNA and 3′ end of the 23S rRNA is stabilized by a plastid‐specific α‐helical extension of uL13c that compensates the loss of helix H98 of the 23S rRNA and forms specific electrostatic interactions with the rRNA ends via two conserved basic residues, Arg48 and Lys49 (Fig 3B). This α‐helix of uL13c is positioned through interactions with two plastid‐specific domains of bL21c and uL22c, underscoring its importance for integration of the 4.5S rRNA into the subunit.
Figure 3. The 4.5S ribosomal RNA and its interactions with ribosomal proteins.
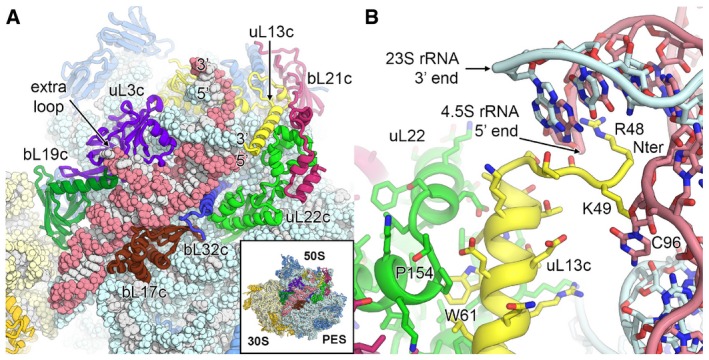
- Position of the 4.5S rRNA (red) at the surface of the 50S large subunit. The 3′ and 5′ ends of the 4.5S rRNA and the 23S rRNA (blue) are labelled. Ribosomal proteins interacting with or in close proximity to the 4.5S rRNA are shown in different colours.
- Stabilization of the 5′ end of the 4.5S rRNA and the 3′ end of the 23S rRNA by the plastid‐specific N‐terminal tail of uL13c (yellow). Specific residues of uL13c, uL22c and 4.5S rRNA are labelled.
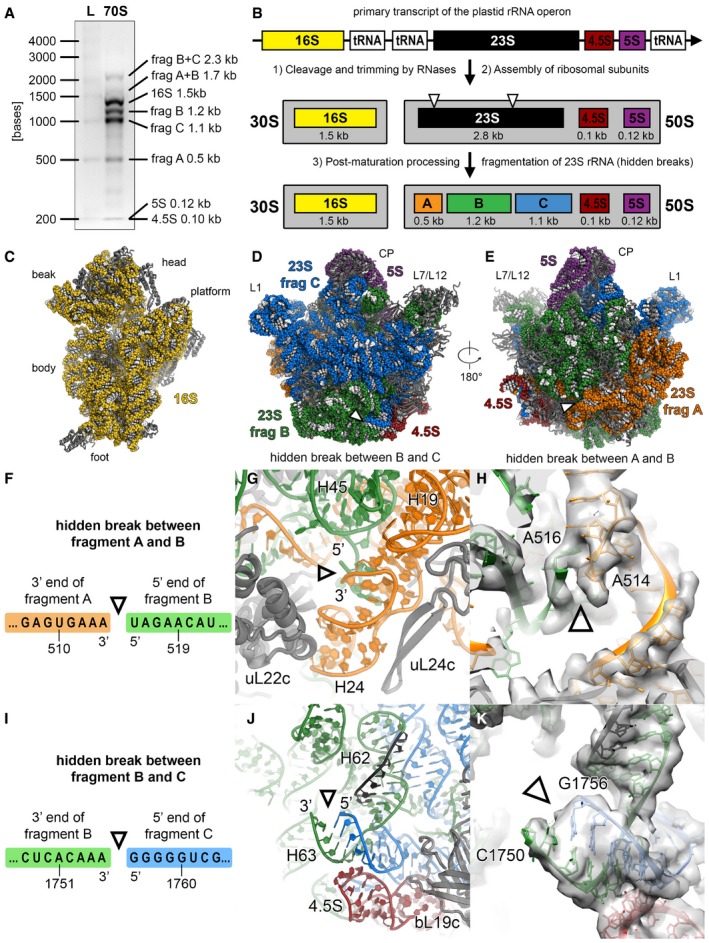
It was previously shown that the plastid 23S rRNA contains strand breaks (Leaver, 1973), termed “hidden breaks”, that are introduced after assembly of the 50S subunit (Nishimura et al, 2010; Liu et al, 2015) and that segment the 23S rRNA into three fragments of 0.5, 1.2 and 1.1 kb (from 5′ to 3′‐end) (Fig EV4). These hidden breaks are necessary for efficient chloroplast translation and are generated through specific action of enzymes such as RNA helicase RH39 (Nishimura et al, 2010) that binds to H62 to introduce a hidden break in H63. Indeed, we can visualize this hidden break in our cryo‐EM map (Fig EV4) as a gap in the rRNA backbone density at the putative cleavage site. Furthermore, our structure shows that the RNA‐binding sequence of RH39 is accessible to the solvent and well‐positioned for processing of the hidden break.
Figure EV4. The hidden breaks of the 23S rRNA .
-
AAnalysis of ribosomal RNA by agarose gel electrophoresis. RNA was extracted from chloroplast 70S ribosome sample and separated on a 2% (w/v) agarose gel (L: high range RNA ladder; 70S: RNA of chloroplast 70S sample).
-
BSchematic of rRNA processing and assembly in the chloroplast. Because of two specific cleavage sites on the 23S rRNA, called “hidden breaks”, the 23S rRNA gets separated into three fragments: A (0.5 kb), B (1.2 kb) and C (1.1 kb).
-
C–EViews of the 30S (C) and the 50S subunits (D) from the subunit interface and of the 50S subunit (E) from the solvent accessible side. The rRNA is shown as spheres and coloured according to the elements indicated in panel (B). The positions of the hidden breaks on the 23S rRNA are marked with triangles.
-
F–HThe hidden break indicated with a triangle between fragments A and B is introduced in the connection between helices H2 and H24.
-
I–KThe hidden break indicated with a triangle between fragments B and C is positioned at the stem loop of helix H63. The binding site of the helicase RH39 on helix H62 is coloured black. The electron density map shown in panels (H and K) is low‐pass filtered to 4 Å, and the nucleotides at the hidden break sites are labelled. The exact positions of the hidden breaks on the 23S rRNA sequence shown in panels (F and I) were stated in a previous publication (Liu et al, 2015).
Adaptions of the mRNA entry and exit sites to chloroplast‐specific translation initiation
Our cryo‐EM map of the chloroplast 30S subunit reveals new protein features around uS2c, which modify the mRNA entry and exit sites and represent parts of bS1c and uS5c (Fig 4A). In bacteria, ribosomal protein bS1 is essential for translation initiation as it binds (Boni et al, 1991) and unfolds the 5′‐UTR of mRNA (Qu et al, 2012; Duval et al, 2013) to recruit it to the mRNA channel of the small subunit. Chloroplast bS1c contains three OB‐folds and long C‐ and N‐terminal extensions. We observe that in the chloroplast ribosome interactions between bS1c and uS2c are much more extensive compared to the bacterial system and involve not only one of the OB‐folds of bS1c, but also chloroplast‐specific C‐ and N‐terminal extensions that wrap around uS2c (Fig 4B). The remaining two OB‐folds of bS1c are not visible in the maps; however, their position is optimal for interactions with plastid mRNAs as predicted biochemically (Shteiman‐Kotler & Schuster, 2000; Merendino et al, 2003). The disordered regions of bS1c could also play a role in the delivery and the correct positioning of mRNA through interplay with proteins that regulate translation by binding to RNA elements located in the 5′‐UTR (Marin‐Navarro et al, 2007).
Figure 4. Architecture of the mRNA entry and exit site.
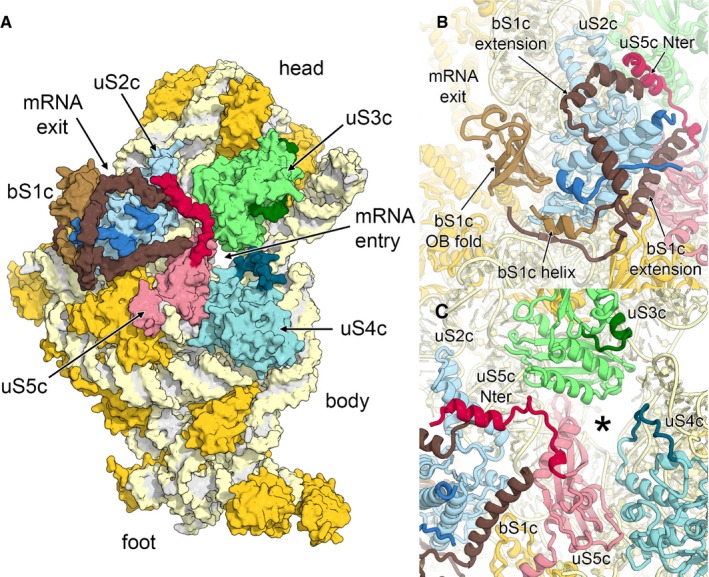
- Surface representation of the solvent accessible side of the 30S small subunit. Ribosomal proteins bS1c (brown), uS2c (blue), uS3c (green), uS4c (cyan) and uS5c (red) are labelled, and plastid‐specific elements are indicated by darker colour shades.
- Helical extensions of bS1c cluster around uS2c.
- Chloroplast‐specific extensions of uS4c and uS5c remodel the mRNA entry site (marked with an asterisk).
In the chloroplast ribosome, the mRNA entry site is encircled by the ribosomal proteins uS3c, uS4c and uS5c. Ribosomal protein uS5c contains a plastid‐specific N‐terminal tail that extends to uS3c on the head and further interacts with uS2c and an extension of bS1c (Fig 4C). Therefore, the mRNA entry site is narrowed in comparison with bacterial 30S (Appendix Fig S10). An insertion within uS4c further reshapes the architecture of the mRNA entry channel. Considering that translation initiation in chloroplasts does not rely on Shine‐Dalgarno (SD)‐like interactions between the mRNA and the anti‐SD sequence of the plastid 16S rRNA and considering that two‐thirds of all transcripts lack a SD‐like sequence (Ruf & Kossel, 1988; Hirose et al, 1998; Drechsel & Bock, 2011), alternative mechanisms for plastid translation initiation have been proposed (Zerges, 2000; Sugiura, 2014), in which cis‐elements in the 5′‐UTRs of plastid mRNAs are proposed to be the major determinants of correct translation initiation in plastids. Nuclear‐encoded trans‐acting factors, which are partially regulated by abiotic factors as light or temperature, specifically bind to these cis‐elements and enable efficient translation initiation either by rearranging the structure of the mRNA 5′‐UTR or by mediating the interaction between the mRNA and structural elements of the small ribosomal subunit. Such a structural element of the small subunit that could play a role in recruitment of mRNAs in chloroplasts, in addition to the above‐mentioned bS1c at the mRNA channel exit, is the constriction of the mRNA entry site formed by the extension of uS5c. Interestingly, a similar structural feature was observed in the mammalian mitochondrial ribosome where uS5m forms a latch at the mRNA entry site involved in recruitment of leaderless mitochondrial mRNAs (Appendix Fig S10) (Greber et al, 2015).
Modifications of the SRP‐binding site at the polypeptide tunnel exit
In chloroplasts, the polypeptide exit tunnel region is considerably different compared to the bacterial ribosome because of the truncation of 23S rRNA helix H7 and extensions of uL24c and uL29c (Fig 5A and Appendix Fig S11). Furthermore, in the course of evolution, the bacterial‐ancestral uL23 was substituted by a variant of the eukaryotic‐cytoplasmic uL23 (Bubunenko et al, 1994), which has a truncated hairpin loop pointing towards the polypeptide tunnel and an elongated α‐helical C‐terminus (Fig 5B). These architectural modifications at the tunnel exit region likely coevolved with the protein targeting mechanism that is specific for chloroplasts. In contrast to the bacterial system, the chloroplast lacks the RNA component of the signal recognition particle (SRP) (Richter et al, 2010) and consists only of protein cpSRP54. Consequently, two known docking sites of the bacterial SRP RNA (Halic et al, 2006; Jomaa et al, 2016), the C‐terminal domain of bL32 and stem‐loop H100 of 23S rRNA, have changed in chloroplasts (Fig 5A and Appendix Fig S11). In particular, bL32c has an elongated α‐helix interacting with the surrounding rRNA backbone via many positively charged residues, and bacterial stem‐loop H100 of 23S rRNA is structurally replaced by the extra loop of the plastid‐specific 4.5S rRNA.
Figure 5. Architecture of the polypeptide tunnel exit.
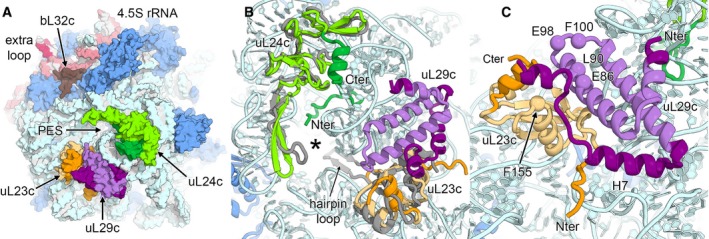
- Surface representation of the polypeptide exit site (PES) of the 50S large subunit. Ribosomal proteins located around the PES are uL23c (orange), uL24c (green) and uL29c (violet). Plastid‐specific features are indicated by darker colour shades.
- Structural differences of the PES (marked with an asterisk) of the chloroplast 50S in comparison with the bacterial 50S subunit. The bacterial ribosomal proteins uL23 and uL24 (both in grey) are overlaid.
- Adaptations of putative binding sites of cpSRP54. Corresponding residues involved in NG‐domain binding in the bacterial ribosome are shown as spheres and are labelled.
The NG‐domain of bacterial SRP Ffh, which is homologous to cpSRP54, binds to conserved residues of uL29 and a binding pocket formed by uL23 and uL29 (Kramer et al, 2002; Jomaa et al, 2016). In chloroplasts, the residues on uL29c are partially conserved, but the putative binding pocket of uL23c is adapted by a plastid‐specific extension of uL29c (Fig 5C). The C‐terminal helix of uL29c interacts with the C‐terminus of uL23c, thereby shielding the residues that mediate the interactions with the NG‐domain.
Plastid translation factor pY bound to the mRNA channel
Protein synthesis in chloroplasts responds to changes in light and temperature and is mainly regulated at the translational level, while the mRNA content in the organelle is maintained constant (Fromm et al, 1985; Kim & Mayfield, 1997; Marin‐Navarro et al, 2007). We purified chloroplast ribosomes from plant tissue incubated in the cold and in darkness, conditions under which the protein synthesis is reduced (Fromm et al, 1985). As observed previously (Sharma et al, 2007), under these conditions ribosomes are associated with plastid translation factor pY (previously called PSRP1) (Fig 6A and Appendix Fig S12), which is a homologue of bacterial cold shock protein Y (pY or YifA) that stabilizes 70S ribosomes under stress conditions by binding to the subunit interface (Agafonov et al, 2001; Vila‐Sanjurjo et al, 2004; Sharma et al, 2010; Polikanov et al, 2012).
Figure 6. Plastid translation factor pY.
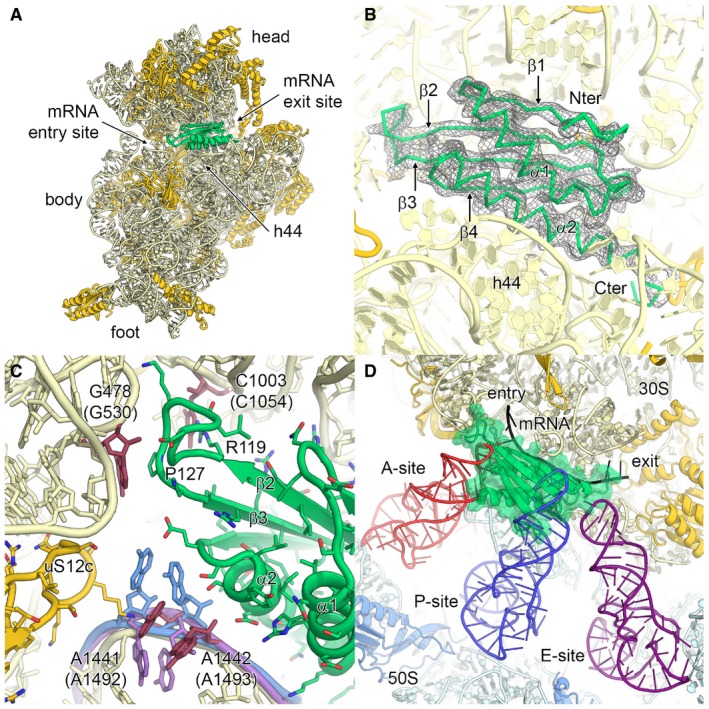
- Binding of plastid translation factor pY, shown in lime green, to the mRNA channel of the small subunit. The small subunit is shown from the intersubunit side. The 16S rRNA is coloured in pale yellow, and ribosomal proteins are in gold.
- EM density for plastid pY. Secondary structure elements and N‐ and C‐termini are indicated.
- Molecular interaction of plastid pY with 16S rRNA. The conserved nucleotides involved in A‐site decoding are coloured in red and labelled (bacterial numbering in brackets). The bacterial nucleotides A1492 and A1493 of the empty 30S subunit (PDB 1J5E; Wimberly et al, 2000) and of the 70S ribosome in complex with mRNA and tRNAs (PDB 4V51; Selmer et al, 2006) are overlaid and coloured in purple and blue, respectively. Pro127 and Arg119 of plastid pY are indicated.
- Superposition of the chloroplast 70S:pY complex with bacterial A‐, P‐ and E‐site tRNAs and mRNA from the crystal structure of the Thermus thermophilus 70S ribosome (PDB 4V51; Selmer et al, 2006).
The high‐resolution maps revealed the binding site of factor pY bound to the mRNA channel of the small subunit and allowed us to build an almost complete structure (Fig 6B). The identity of plastid pY was established by direct inspection of the density and confirmed by mass spectrometry analysis of the ribosome sample (Appendix Fig S12). The structure reveals the molecular interactions of pY with the 16S rRNA residues that form the decoding centre. Helix α1 binds to the rRNA backbone of helix h44 via positively charged residues, helix α2 extends above the mRNA channel, thereby preventing mRNA binding, and the four stranded β‐sheet (β1‐4) points towards the head (Fig 6A and B). In the A‐site of the ribosome, negatively charged residues of helix α2 and the loop between helix α2 and sheet β4 (Fig 6C) stabilize the universally conserved bases A1441 (A1492 in E. coli) and A1442 (A1493), which are involved in decoding in a partially flipped out conformation that is between the empty and mRNA‐tRNA bound states observed for bacterial ribosomes (Wimberly et al, 2000; Selmer et al, 2006). Furthermore, bases G478 (G530) and C1003 (C1054), which are also involved in decoding, are stabilized through respective contacts with Pro127 and Arg119 of pY. The loop between β2 and β3 located at the A‐site is larger than in the eubacterial pY homologues and forms contacts to helices h18 and h34 (Fig 6C) at the mRNA entry site. Reaching towards the mRNA exit site, the C‐terminal extension of helix α2 contains two histidine residues, His178 and His181, that mimic the mRNA bases at the E‐site. All these specific interactions enable plastid pY to fulfil two of its functions: first, functionally most important areas of the 70S, including the key nucleotides of the decoding centre, are protected by plastid pY from being degraded during the translational arrest, in a similar way as suggested for bacterial pY (Vila‐Sanjurjo et al, 2004), and second, the binding of pY to the mRNA channel prevents binding of the A‐site and P‐site tRNAs (Fig 6D) and inhibits translation. The C‐terminal domain of plastid pY is disordered in our structure, in agreement with its possible role in pY activation that likely involves interactions with other factors (Bubunenko & Subramanian, 1994; Sharma et al, 2010).
Comparing the maps of the chloroplast ribosome in the pY‐bound state with a reconstruction of an empty state chloroplast ribosome at lower resolution reveals that the small subunit is in a rotated state relative to the large subunit in the absence of pY (Fig EV5A–D). In this conformation, the body is rotated by 5.8° (ratcheting) and the head by 7.0° (swivelling) in comparison with the non‐rotated state with bound pY, which reduces the number of intersubunit contacts (Fig EV5E–H and Appendix Table S4). Especially, the bridges B1a and B1b between the small subunit head and the large subunit A‐site finger and the CP, respectively, as well as bridges B7a, B7b and B7c are weakened by the small subunit rotation. Coupled with body rotation and head swivelling, the tRNA moves from an E/E‐state in the non‐rotated to a P/E‐state in the rotated conformation (Fig EV5I and J). Because we do not see empty 70S ribosome in a non‐rotated state, it is likely that plastid pY stabilizes the chloroplast ribosomes from dissociation in the non‐rotated state with increased intersubunit contacts under environmental conditions that do not require active protein synthesis.
Figure EV5. Plastid translation factor pY .
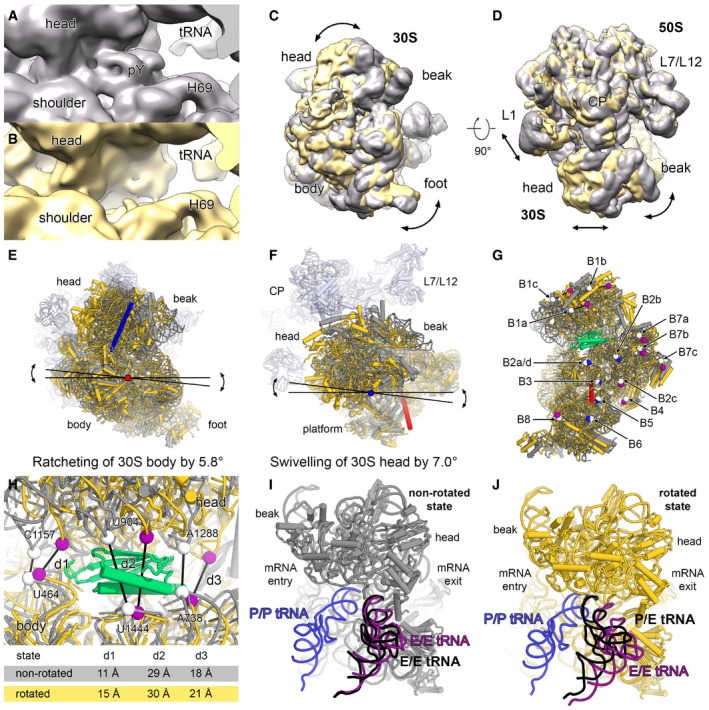
-
A–DThe 70S map with density for pY in the mRNA channel is shown in grey (A) and the empty 70S map is shown in yellow (B). The 70 maps were overlaid using only the density of the 50S. Comparison of the overlaid maps, shown in 30S subunit view (C) and top view (D). The rotation of the 30S subunit is indicated with arrows.
-
E, FThe body and the head domains of the 30S model were independently fitted into the cryo‐EM map of the rotated state. The rotation angle of the 30S body rotation (ratcheting) (E) and the 30S head rotation (swivelling) (F) were measured using PyMOL, and the rotation axes are shown in red and blue, respectively.
-
GIntersubunit bridges are affected by the 30S rotation. A selected residue of each intersubunit bridge is represented as white sphere in the non‐rotated state. The same residues are coloured in the rotated state either in blue, if the contact with the large subunit is maintained, or in purple, if the contact is lost. The intersubunit bridges and the differences between the rotated and the non‐rotated state are described in Appendix Table S4.
-
HDistances (d1, d2 and d3) between backbone phosphates of selected rRNA residues (spheres) in the non‐rotated (white) and the rotated state (purple) indicating a slight opening of the mRNA channel.
-
I, JtRNAs in the P/P‐ and E/E‐state from a crystal structure of the bacterial 70S ribosome in complex with mRNA and tRNAs (PDB 4V51; Selmer et al, 2006) are overlaid and shown in blue and purple, respectively. A tRNA (black) was fitted as rigid body into the 70S maps of the non‐rotated (I) and of the rotated state (J).
Relationship to apicoplast ribosomes
The structure of the chloroplast ribosome also allows for better understanding of ribosomes found in a relict plastid, called apicoplast, in protozoan parasites (Wilson, 1993; McFadden et al, 1996) responsible for severe diseases like malaria (Plasmodium falciparum) and toxoplasmosis (Toxoplasma gondii). The “plantlike” apicoplast critical for proliferation of these organisms probably originated from red algae by secondary endosymbiosis (Waller et al, 2003) and contains its own active transcription and translation machinery. The chloroplast ribosome described here now provides the best starting point for understanding the structure of apicoplast ribosomes that have an rRNA reduced in length and are lacking several proteins (12 ribosomal proteins with bacterial homolog for P. falciparum and 14 for T. gondii) that are present in chloroplast ribosomes (Habib et al, 2016). Considering that several compounds targeting the bacterial ribosome also show activity against the malaria parasite (Goodman et al, 2007; Kalanon & McFadden, 2010; Wilson et al, 2015), the structure presented here can also be used as a starting model for designing better drugs capable of targeting the translation apparatus of plastids.
Conclusions
The structure presented here reveals the architecture of the chloroplast 70S ribosome with important implications for understanding its relationship to ancestral bacterial ribosomes. We also observe important differences in the structure related to plastid‐specific mechanism of translation initiation and membrane protein targeting. Furthermore, we reveal the molecular mechanism of how translation factor pY inhibits translation by binding to the mRNA channel region of the small subunit to protect the decoding site and stabilize the ribosome in an inactive form during the dark phase of the chloroplast day cycle. These results contribute to the mechanistic understanding of translation in chloroplast and its regulation and pave the way for future structure‐based biochemical and genetic studies.
Materials and Methods
Preparation of chloroplast 70S ribosomes
Fresh leaves of spinach (S. oleracea) were obtained from the local supermarket and stored for a few hours in the dark at 4°C. The purification of chloroplasts from leaf tissue was done according to a previously described protocol (Bartsch et al, 1982). The enriched chloroplasts were lysed by gentle stirring (180 rpm, 4°C, 90 min) in lysis buffer (10 mM Tris–HCl pH 7.6, 25 mM KCl, 25 mM MgCl2, 2 mM DTT, 0.1 mM PMSF, 2 mM spermidine, 0.05 mM spermine, 2% (w/v) Triton X‐100). The suspension was cleared by centrifugation (25,350 g, 30 min, 4°C) using a Beckman Type 45Ti rotor (Beckman‐Coulter), and the supernatant was loaded onto 50% (w/v) sucrose cushions and centrifuged (101,390 g, 15 h, 4°C) using a Beckman Type 45Ti rotor (Beckman‐Coulter). The ribosome pellets were dissolved in monosome buffer (25 mM Tris–HCl pH 7.6, 25 mM KCl, 25 mM MgOAc2, 2 mM DTT, 2 mM spermidine, 0.05 mM spermine). The sample was layered onto 10–40% (w/v) sucrose gradients and centrifuged (51,610 g, 15 h, 4°C) using a Beckman Type SW‐32Ti rotor (Beckman‐Coulter). The fractions containing most chloroplast 70S ribosomes were pooled (Appendix Fig S1), and the buffer was exchanged to sucrose‐free monosome buffer using Amicon Ultra‐4 spinning centrifugal filter units with 100,000 molecular weight cutoff (Merck Millipore).
Cryo‐EM data acquisition and processing
The sample was diluted with monosome buffer to a final 70S ribosome concentration of 50 nM, and 5 μl was applied to Quantifoil R2/2 holey carbon grids (Quantifoil Micro Tools), which had been previously coated with a thin home‐made carbon film and glow‐discharged (negative, 25 mA, 30 s) using an Emitech K100X (Quorum Technologies). The grids were automatically blotted and flash‐frozen by plunging into a 2:1 mixture of liquid ethane and propane using a Vitrobot (FEI Company). Cryo‐EM data were collected on a Titan Krios cryo‐transmission electron microscope (FEI Company) equipped with a Falcon II direct electron detector and operated at 300 keV with a magnification of 100,720× and a defocus range between −0.8 and −3.5 μm. The EPU software (FEI Company) was used for automated data acquisition by collecting seven movie frames with a combined dose of 20 electrons per Å2 per exposure (770 ms combined exposure time) after discarding the first frame (55 ms).
We used the software DOSEFGPU DRIFTCORR (Li et al, 2013) to correct for beam‐induced specimen motion and CTFFIND (Mindell & Grigorieff, 2003) to estimate the CTF parameters from the drift‐corrected micrographs. Micrographs were selected by evaluating the quality of the power spectra. From finally 2,796 selected micrographs, 326,094 particles were automatically selected using Batchboxer from the EMAN software package (Ludtke et al, 1999). Further steps of image processing were performed in RELION 1.4 (Scheres, 2012). In an initial 2D classification, binned particle images (5.56 Å/px on the object scale, 80 px frame size) were classified into 200 classes. Particles assigned to classes showing separated 30S and 50S subunits, 80S ribosomes and non‐ribosomal particles were removed from the dataset. Multiple 3D classification steps in combination with masking and signal subtraction approaches (Appendix Figs S2 and S3) were applied to obtain homogenous particle image subsets for the 3D reconstructions of the complete 70S ribosome, the 50S subunit and 30S subunit. The final high‐resolution refinements of these particle subsets at full‐pixel size (1.39 Å/px on the object scale, 320 px frame size) resulted in 3D reconstructions of the chloroplast 70S ribosome at 3.4 Å resolution from 140,583 particle images, of the 50S subunit at 3.2 Å resolution from 154,332 particle images and of the 30S subunit at 3.6 Å resolution from 127,031 particle images according to the Fourier Shell Correlation (FSC) = 0.143 criterion (Fig EV1). Subsequently, the maps were sharpened and used for manual atomic model building, refinement and validation.
Structure building and refinement
To obtain a full atomic model of the 70S ribosome, the structures of the 30S and 50S subunits were initially built into the EM maps of the individual subunits using O (Jones et al, 1991; Jones, 2004) and COOT (Emsley et al, 2010) and the coordinates of an E. coli ribosome as a guide (PDB 4YBB; Noeske et al, 2015). The maps were of excellent quality and allowed building of almost all RNA and protein residues. In peripheral regions of the subunits with lower local resolution, a few protein extensions were built as unassigned UNK residues, and in the case of proteins cS22, cS23 and bS1c, PHYRE models were docked as rigid bodies (Kelley et al, 2015) (Appendix Tables S2 and S3). The unambiguous density for factor pY (PSRP1) allowed docking of a PHYRE model followed by rebuilding at atomic level (Appendix Fig S12). The atomic models were subsequently refined and validated using PHENIX (Adams et al, 2010) as described previously (Greber et al, 2014). In brief, the coordinates were refined in reciprocal space against structure factors back‐calculated from the EM maps using the mlhl target to restrain the phases. The phases were weighted according to the FSC dropoff as described (Greber et al, 2014). During coordinate refinement of the subunits, an optimal geometry weighting value of wxc = 1.25 was established, which resulted in model geometry and R‐factor values typical for the chosen resolution ranges (Urzhumtseva et al, 2009) (Appendix Fig S4; Appendix Table S1). Using high geometry weighting values results in low R‐factors but worse model geometry and possible overrefinement, while at low geometry weighting values the R‐factors are increased and the model geometry is overtightened. The refinement of the complete 70S was performed using the coordinates of the individually refined 50S and 30S subunits, which were rigid body fitted into the 3.4 Å cryo‐EM reconstruction of the chloroplast 70S ribosome (Fig EV1). At the E‐site of the 70S intersubunit space, a density representing a mixture of tRNAs was observed. To account for this density, an optimized canonical E. coli tRNA‐Phe derived from PDB 2J00 was docked. Protein contacts between both subunits were adjusted, and the linker of bL31c, which bridges both subunits, was added. The complete 70S model was then fully refined against the 3.4 Å cryo‐EM map using PHENIX (Appendix Fig S4; Appendix Table S1) in a similar procedure as described above for the subunits, using an optimal geometry weighting value of wxc = 1.4.
Creation of figures
Figures showing cryo‐EM reconstructions and molecular models were created using UCSF Chimera (Pettersen et al, 2004) and PyMOL (The PyMOL Molecular Graphics System, Version 1.7 Schrödinger, LLC). Local resolution plots were generated in ResMap (Kucukelbir et al, 2014).
Mass spectrometry analysis
Purified chloroplast 70S ribosomes (~50 μg) were mixed with SDS gel‐loading buffer (final concentration: 50 mM Tris–HCl pH 6.8, 2% (w/v) sodium dodecyl sulphate, 0.1% (w/v) bromophenol blue, 10% (v/v) glycerol, 100 mM β‐mercaptoethanol) and heated for 10 min at 70°C before loading the sample on a 12% polyacrylamide gel (GenScript). The gel was stained with Coomassie brilliant blue G‐250 (Sigma‐Aldrich) and protein bands in the molecular weight range between 20 and 40 kDa and one band at 50 kDa have been cut out. The sliced protein bands were sent for protein identification by mass spectrometry (liquid chromatography MS/MS) performed at the Functional Genomics Center Zurich (FGCZ). The Mascot software packages (Perkins et al, 1999) was used for the database searches in SwissProt and Trembl, and the results were analysed applying stringent settings [1% protein false discovery rate (FDR), a minimum of two peptides per protein, 0.1% peptide FDR].
Accession numbers
The 3.4 Å cryo‐EM map of the chloroplast 70S ribosome, the 3.2 Å cryo‐EM map of the 50S subunit and the 3.6 Å cryo‐EM map of the 30S subunit have been deposited in the Electron Microscopy Databank with accession codes EMD‐3533, EMD‐3531 and EMD‐3532, respectively. The refined coordinates of the atomic structure of the 50S subunit and the 30S subunit have been deposited in the Protein Databank as PDBs 5MMI and 5MMJ, respectively. The coordinates of the atomic model of the complete chloroplast 70S ribosome have been deposited as PDB 5MMM. A PyMOL script for display of the chloroplast 70S ribosome is available from the Ban Lab website (www.bangroup.ethz.ch).
Author contributions
PB and ML established the purification procedures. PB, ML and MS performed the preparation and the biochemical analysis of the chloroplast ribosome. PB and MS prepared cryo‐EM samples. PB and DB acquired the cryo‐EM data. PB and DB calculated the cryo‐EM reconstructions. ML, PB, MS and NB interpreted the structures. PB wrote the manuscript. All authors contributed to the final version of the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Ahmad Jomaa, David Ramrath and Basil Greber for discussions. Cryo‐EM data were collected at the electron microscopy facility of ETH Zurich (ScopeM), and we thank Peter Tittmann (ScopeM) for support. The cryo‐EM data was processed on Euler computing cluster of the Swiss National Supercomputing Centre (CSCS). The mass spectrometry analysis was done at the FGCZ, and we thank Peter Hunziker for support. This work was supported by the Swiss National Science Foundation (SNSF) and the National Center of Excellence in Research (NCCR) RNA and disease programme of the Swiss National Science Foundation (SNSF).
The EMBO Journal (2017) 36: 475–486
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agafonov DE, Kolb VA, Spirin AS (2001) Ribosome‐associated protein that inhibits translation at the aminoacyl‐tRNA binding stage. EMBO Rep 2: 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlert D, Ruf S, Bock R (2003) Plastid protein synthesis is required for plant development in tobacco. Proc Natl Acad Sci USA 100: 15730–15735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Yin Z, Bhushan S (2016) Cryo‐EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep 6: 35793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D et al (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Kimura M, Subramanian AR (1982) Purification, primary structure, and homology relationships of a chloroplast ribosomal protein. Proc Natl Acad Sci USA 79: 6871–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Timmis JN (2008) Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Boni IV, Isaeva DM, Musychenko ML, Tzareva NV (1991) Ribosome‐messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res 19: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko MG, Schmidt J, Subramanian AR (1994) Protein substitution in chloroplast ribosome evolution—a eukaryotic cytosolic protein has replaced its organelle homolog (L23) in spinach. J Mol Biol 240: 28–41 [DOI] [PubMed] [Google Scholar]
- Bubunenko MG, Subramanian AR (1994) Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J Biol Chem 269: 18223–18231 [PubMed] [Google Scholar]
- Drechsel O, Bock R (2011) Selection of Shine‐Dalgarno sequences in plastids. Nucleic Acids Res 39: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, Springer M, Marzi S (2013) Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11: e1001731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm H, Devic M, Fluhr R, Edelman M (1985) Control of psbA gene expression: in mature Spirodela chloroplasts light regulation of 32‐kd protein synthesis is independent of transcript level. EMBO J 4: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, Su V, McFadden GI (2007) The effects of anti‐bacterials on the malaria parasite Plasmodium falciparum . Mol Biochem Parasitol 152: 181–191 [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI (2008) Plastid evolution. Annu Rev Plant Biol 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515: 283–286 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N (2015) Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348: 303–308 [DOI] [PubMed] [Google Scholar]
- Habib S, Vaishya S, Gupta K (2016) Translation in organelles of apicomplexan parasites. Trends Parasitol 32: 939–952 [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R (2006) Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444: 507–511 [DOI] [PubMed] [Google Scholar]
- Hirose T, Kusumegi T, Sugiura M (1998) Translation of tobacco chloroplast rps14 mRNA depends on a Shine‐Dalgarno‐like sequence in the 5′‐untranslated region but not on internal RNA editing in the coding region. FEBS Lett 430: 257–260 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Lopez‐Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jensen PE, Leister D (2014) Chloroplast evolution, structure and functions. F1000Prime Rep 6: 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa A, Boehringer D, Leibundgut M, Ban N (2016) Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat Commun 7: 10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47(Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Jones TA (2004) Interactive electron‐density map interpretation: from INTER to O. Acta Crystallogr D Biol Crystallogr 60: 2115–2125 [DOI] [PubMed] [Google Scholar]
- Kalanon M, McFadden GI (2010) Malaria, Plasmodium falciparum and its apicoplast. Biochem Soc Trans 38: 775–782 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mayfield SP (1997) Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278: 1954–1957 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze‐Specking A, Ban N, Deuerling E, Bukau B (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174 [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD (2014) Quantifying the local resolution of cryo‐EM density maps. Nat Methods 11: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver CJ (1973) Molecular integrity of chloroplast ribosomal ribonucleic acid. Biochem J 135: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontiadou F, Triantafillidou D, Choli‐Papadopoulou T (2001) On the characterization of the putative S20‐thx operon of Thermus thermophilus . Biol Chem 382: 1001–1006 [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y (2013) Electron counting and beam‐induced motion correction enable near‐atomic‐resolution single‐particle cryo‐EM. Nat Methods 10: 584–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, Bock R, Huang J, Cheng Y (2015) The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis . Plant Physiol 168: 205–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high‐resolution single‐particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Manuell AL, Quispe J, Mayfield SP (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol 5: e209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L (1970) Origin of eukaryotic cells evidence and research implications for a theory of the origin and evolution of microbial, plant, and animal cells on the Precambrian earth. New Haven, CT: Yale University Press; [Google Scholar]
- Marin‐Navarro J, Manuell AL, Wu J, Mayfield SP (2007) Chloroplast translation regulation. Photosynth Res 94: 359–374 [DOI] [PubMed] [Google Scholar]
- McFadden GI, Reith ME, Munholland J, Lang‐Unnasch N (1996) Plastid in human parasites. Nature 381: 482 [DOI] [PubMed] [Google Scholar]
- Merendino L, Falciatore A, Rochaix JD (2003) Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii . Plant Mol Biol 53: 371–382 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Ashida H, Ogawa T, Yokota A (2010) A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JH (2015) High‐resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability‐based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Blaha GM, Steitz TA (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336: 915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr (2012) Ribosomal protein S1 unwinds double‐stranded RNA in multiple steps. Proc Natl Acad Sci USA 109: 14458–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CV, Bals T, Schunemann D (2010) Component interactions, regulation and mechanisms of chloroplast signal recognition particle‐dependent protein transport. Eur J Cell Biol 89: 965–973 [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf M, Kossel H (1988) Structure and expression of the gene coding for the alpha‐subunit of DNA‐dependent RNA polymerase from the chloroplast genome of Zea mays. Nucleic Acids Res 16: 5741–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L (1967) On the origin of mitosing cells. J Theor Biol 14: 255–274 [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012) RELION: implementation of a Bayesian approach to cryo‐EM structure determination. J Struct Biol 180: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK (2007) Cryo‐EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid‐specific ribosomal proteins. Proc Natl Acad Sci USA 104: 19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Donhofer A, Barat C, Marquez V, Datta PP, Fucini P, Wilson DN, Agrawal RK (2010) PSRP1 is not a ribosomal protein, but a ribosome‐binding factor that is recycled by the ribosome‐recycling factor (RRF) and elongation factor G (EF‐G). J Biol Chem 285: 4006–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteiman‐Kotler A, Schuster G (2000) RNA‐binding characteristics of the chloroplast S1‐like ribosomal protein CS1. Nucleic Acids Res 28: 3310–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M (1989) The chloroplast chromosomes in land plants. Annu Rev Cell Biol 5: 51–70 [DOI] [PubMed] [Google Scholar]
- Sugiura M (2014) Plastid mRNA translation. Methods Mol Biol 1132: 73–91 [DOI] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schottler MA, Bock R (2012) The plastid‐specific ribosomal proteins of Arabidopsis thaliana can be divided into non‐essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Urzhumtseva L, Afonine PV, Adams PD, Urzhumtsev A (2009) Crystallographic model quality at a glance. Acta Crystallogr D Biol Crystallogr 65: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila‐Sanjurjo A, Schuwirth BS, Hau CW, Cate JH (2004) Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, van Dooren GG, McFadden GI (2003) Comment on “A green algal apicoplast ancestor”. Science 301: 49 [DOI] [PubMed] [Google Scholar]
- Whitfeld PR, Leaver CJ, Bottomley W, Atchison B (1978) Low‐molecular‐weight (4.5S) ribonucleic acid in higher‐plant chloroplast ribosomes. Biochem J 175: 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I (1993) Plastids better red than dead. Nature 366: 638 [DOI] [PubMed] [Google Scholar]
- Wilson DW, Goodman CD, Sleebs BE, Weiss GE, de Jong NW, Angrisano F, Langer C, Baum J, Crabb BS, Gilson PR, McFadden GI, Beeson JG (2015) Macrolides rapidly inhibit red blood cell invasion by the human malaria parasite, Plasmodium falciparum . BMC Biol 13: 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan‐Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Xu T, Lee K, Gu L, Kim JI, Kang H (2013) Functional characterization of a plastid‐specific ribosomal protein PSRP2 in Arabidopsis thaliana under abiotic stress conditions. Plant Physiol Biochem 73: 405–411 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28455–28465 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2003) Proteomic identification of all plastid‐specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur J Biochem 270: 190–205 [DOI] [PubMed] [Google Scholar]
- Zerges W (2000) Translation in chloroplasts. Biochimie 82: 583–601 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


