Abstract
Glioblastoma is the most aggressive brain tumor and, even with the current multimodal therapy, is an invariably lethal cancer with a life expectancy that depends on the tumor subtype but, even in the most favorable cases, rarely exceeds 2 years. Epigenetic factors play an important role in gliomagenesis, are strong predictors of outcome, and are important determinants for the resistance to radio- and chemotherapy. The latest addition to the epigenetic machinery is the noncoding RNA (ncRNA), that is, RNA molecules that are not translated into a protein and that exert their function by base pairing with other nucleic acids in a reversible and nonmutational mode. MicroRNAs (miRNA) are a class of ncRNA of about 22 bp that regulate gene expression by binding to complementary sequences in the mRNA and silence its translation into proteins. MicroRNAs reversibly regulate transcription through nonmutational mechanisms; accordingly, they can be considered as epigenetic effectors. In this review, we will discuss the role of miRNA in glioma focusing on their role in drug resistance and on their potential applications in the therapy of this tumor.
1. Introduction
Epidemiological studies estimate that more than 250,000 new cases of central nervous system (CNS) tumors worldwide are expected every year with variable incidence rates ([1] and http://www.cancer.gov/types/brain/hp/adult-brain-treatment-pdq). Although glioblastoma is considered a rare tumor (Orphanet 360), it accounts for 4% of all cancer deaths making it as one of the deadliest human tumors. According to the current classification [2], approximately 38% of these tumors are at high grade (WHO III, anaplastic astrocytoma or AA and WHO IV, glioblastoma or GB) and hence are virtually lethal.
Given the extremely limited success of the standard treatment in prolonging survival in GB patients, considerable efforts were undertaken to develop targeted therapies that could significantly improve the outcome of these patients [3]. In this respect, epigenetics and epigenetic modulators have become a preferred field of investigation because of their influence in many aspects of cancer [4, 5].
Epigenetics, at large, is the mechanism utilized by living cells to decode and utilize properly the information contained in the raw DNA sequence. In practice, epigenetics consists in a “code” that lays on top of the genetic code and translates the simple information into function [6]. By definition, epigenetics does not change the “content of the information” (i.e., the sequence) and acts through reversible modifications like cytosine methylation at CpG doublets, postsynthetic modifications of the histones (acetylation, methylation, phosphorylation, etc.), and changes in the chromatin conformation. In the last years, a new class of effectors has been added to the epigenetic machinery: the microRNAs or in general, the noncoding RNAs that are capable of reversibly interfering with the transcription and translation of the genes without altering DNA sequence as expected for an epigenetic mechanism.
In this review, we will discuss some general aspects of miRNA in glioma focusing on the circuitry between miRNA and other epigenetic determinants like DNA methylation in this tumor, their role in drug resistance, and their potential therapeutic implications.
2. Epigenetics and Epigenetic Networks in Glioblastoma
Transcriptional profiling has delineated four major molecular subtypes of glioblastoma that could be better characterized by mutational, copy number variation, and methylation analyses [7–9]. In particular, this classification defines two clinical groups of GBs with distinct treatment response and outcome (Table 1). Overall, GB could be classified as “primary,” a group that includes three transcriptional subtypes (classical, mesenchymal, and neural) and “secondary” glioblastoma derived from the evolution of low-grade GB that include the transcriptional proneural subtype and that can be loosely subdivided in two subgroups according to the mutational and chromosomal status. The average survival is 31 months for secondary glioblastoma and only 15 months for the patients with primary GB. As can be seen in Table 1, the major features that distinguish primary from secondary GB can be considered, at large of epigenetic nature, namely, MGMT methylation status and the CpG island methylator phenotype (CIMP).
Table 1.
Molecular characteristics of glioblastoma subtypes according to methylation, expression, mutation, copy number variations patterns, and clinical outcome.
| IDH1/IDH2 status | Wild type | Mutated | |||
|---|---|---|---|---|---|
| MGMT | Mostly unmethylated | Methylated | |||
| Methylation status | CIMP− | CIMP+ | |||
| Mutations | TERT | ATRX-TP53 | CIC-FUBP1 | ||
| CNV | EGFR+ | NF1− | PDGFRA+ | PDGFRA+; 1p19q− | |
| Molecular subtype | Classical | Mesenchymal | Neural | Proneural | |
| Outcome | Very poor outcome | Significantly improved outcome | Significantly improved outcome. Better response to TMZ than 1p19q+ | ||
One of the first evidences of the primary role of epigenetic mechanisms in GB was the discovery of the effect of the inactivation by methylation of the MGMT gene on the sensitivity to the alkylating agent temozolomide (TMZ). In two seminal papers published in 2005 [10, 11], Stupp et al. and Hegi et al. established that the inactivation of the MGMT gene by DNA methylation in the tumor is associated with better survival in GB patients treated with TMZ and radiotherapy compared to the patients with unmethylated MGMT. Hypermethylation of MGMT occurs also in primary GB; however, it is a characteristic feature of secondary glioblastoma and is the “iceberg tip” of a more extensive alteration of the DNA methylation status known as “CpG island methylator phenotype” or CIMP.
The concept of CIMP was developed in 1999 by Toyota et al. that showed the concomitant presence of hypermethylation at many different CpG sites in a subset of colorectal cancer patients with distinct clinicopathological characteristics and favorable outcome [12]. Since then, CIMP was described in many other tumors (for a recent review, see [13]) although it is not clear if the CIMP phenotype is tissue-specific or if all CIMP+ tumors represent a class of tumors with similar characteristics. Moreover, the molecular parameters, including the methylation cut-off levels and the genes to be considered to positively assess the presence of CIMP in a given tumor are not well established. The clinical characteristics of CIMP+ tumors differ; indeed, it differs in GB [8], colon carcinoma [14], myeloid leukemia [15], and breast cancer [16]. The CIMP+ phenotype is a predictor of better outcome whereas in other tumors like neuroblastoma [17, 18] and melanoma [19], CIMP+ tumors are associated with poor prognosis. A possible explanation for the nonuniversal clinical significance of CIMP is the absence of accurate criteria that define CIMP so that the true phenotype of the tumor cannot be always assessed.
In GB, the CIMP phenotype clearly distinguishes the primary (CIMP−) from the secondary (CIMP+) tumors and is tightly associated with inactivating mutations of the IDH1 and IDH2 genes [8]. The mechanistic link between IDH mutations and CIMP was discovered in 2012 when it was demonstrated that IDH1 is an epigenetic controller that modulates the pattern of histone and DNA methylation. This occurs through the inhibition by D-2-hydroxyglutarate of the jumonji histone lysine demethylases (KDM) and of the TET-hydroxylases that convert 5-methylcytosine into 5-hydroxyl-methylcytosine thus leading to the accumulation of 5-methylcytosine.
Recently, histone modifications also have gained importance in GB and the possibility of pharmacological intervention on histone deacetylases (HDAC) has been exploited [20]. Moreover, the involvement of KDM genes in GB development and drug resistance has been demonstrated [21–23].
MicroRNAs, a class of noncoding RNAs, are considered epigenetic modifiers because they control the functionality of the genome by base pairing of nucleotides 2–8 of their sequence to the 3'UTR of mRNA forming the so-called “silencing complex” [24]. Since their inhibitory function is sequence-specific and does not involve the permanent alteration of the DNA sequence, miRNAs are considered an integral part of the epigenetic machinery.
In glioblastoma, as in many other tumors, the remodeling of the epigenome is an important aspect of the biology of the tumor [25, 26] and the interaction between epigenetic factors and the cell signaling cascade appears as a promising target for new therapeutic approaches [20, 23, 25, 27, 28].
3. The Interplay between Tumor Suppressing and Oncogenic miRNA in Glioblastoma
From the current release, 21 of the miRNA database lists 2588 mature and 1881 precursor human miRNA sequences (http://www.mirbase.org/cgi-bin/browse.pl?org=hsa). Each of these miRNA can modulate the expression of several mRNAs, and each mRNA can be modulated by several miRNA generating an extraordinary complex regulatory network. In a literature survey of miRNA deregulated in GB, it was found that the majority of them (n = 253) were overexpressed compared to normal brain tissue, 95 were downmodulated, and conflicting results were reported for 17 of them [29].
The genes targeted by deregulated microRNAs in GB are involved in many pathways including cell proliferation, resistance to apoptosis, autophagy, invasion and metastasis, angiogenesis, and drug resistance. Since microRNAs have multiple targets in different tissues, they may have oncogenic (oncomiR) or antioncogenic effects depending upon the biological context.
Several miRNAs acting as tumor suppressor genes have been identified; some of them are unique of glioblastoma whereas others are involved also in other tumors. In principle, all miRNA interfering with the histone methyltransferase EZH2 (Table 2) can be considered as tumor suppressors, in particular let-7 which inhibits also oncogenes like MYC and K-RAS [30, 31] and is capable of inhibiting glioblastoma cell proliferation [32]. miR-128 and miR-34a are two examples of miRNA acting as tumor suppressor in glioblastoma. miR-128 is an antiproliferative miRNA that interferes with multiple pathways targeting genes involved in glioblastoma pathogenesis like EGFR and PDGFRA [33] and WEE1 [34] and E2F3a [35]. miR-34a interferes with cell proliferation through multiple targets (CDK6, CCND1, NOTCH, and others). When the functionality of miR-34a is restored, this miRNA acts as a tumor suppressor gene reducing cell proliferation and invasion [36]. MiR-124 and miR-137 are two microRNA significantly downregulated in high-grade gliomas and in vitro can induce phenotypic changes, growth arrest, and differentiation in glioma stem cells and thus can be considered oncosuppressive miRNA [37]. Most deregulated miRNAs in GB interfere with cell proliferation pathways, particularly those of EGFR and AKT. A prototype of this group of miRNA is miR-7 whose transfection in GBM cells leads to decreased invasiveness and increased apoptosis fulfilling the basic requirements of a tumor suppressor [38, 39].
Table 2.
miRNA involved in the regulation of EZH2.
| miRNA | Action | Reference |
|---|---|---|
| Let-7a | Direct targeting of EZH2 in nasopharyngeal carcinoma, inhibition of glioma growth by targeting K-RAS | [206, 207] |
| miR-26a | Inhibits growth of nasopharyngeal carcinoma targeting EZH2 | [208] |
| miR-101 | miR-101 downregulation in GB results in EZH2-induced proliferation regulating the methylation status of CPBE1 | [209, 210] |
| miR-124 | Modulates the proliferation of epatocarcinoma cells by direct targeting of EZH2 | [211] |
| miR-138 | Blocks GB tumorigenicity by EZH2-CDK4/6-pRb-E2F1 signaling cascade | [212] |
| miR-214 | Targeting of EZH2 in skeletal muscles | [213] |
| miR-708 | Inhibits GB cell proliferation targeting EZH2, AKT1, MMP2, CCND1, Parp-1, and Bcl-2 | [214] |
Many miRNAs are upregulated in glioblastoma and can be functionally classified as oncomiR. Historically, miR-21 was the first oncogenic miRNA to be identified [40] and can target a set of oncosuppressor genes including PTEN [41] and the metalloproteinase inhibitor TIMP3 which is involved in extracellular matrix remodeling, tumor infiltration, and angiogenesis [42, 43]. Thus, miR-21 targets an entire network of tumor suppressor genes and its inhibition by complementary oligonucleotides blocks GB cell growth in vitro and in vivo [44]. It is reasonable to hypothesize that the delivery of an inhibitor of miR-21 at the tumor site might be a useful addition to the standard therapy.
The targeting of multiple oncosuppressor genes easily explains the oncogenic mechanism of miR-21. On the contrary, the oncogenic properties of miR-221and miR-222, overexpressed in a variety of tumors including GB, have several oncogenic functions including the inactivation of the cell cycle suppressors p27 and p57 [45, 46].
Apoptosis and autophagy are two mechanisms utilized to eliminate dysfunctional or otherwise stressed cells, and resistance to apoptosis is one of the hallmarks of cancer cells. Unsurprisingly miRNA can target several genes at the center of both mechanisms. Oncogenic antiapoptotic miRNAs like mir-21 [42], miR-221/mir-222 [47], and miR-335 [48] are overexpressed in glioblastoma and interfere with the p53/Bcl-2/PUMA and TGF-β signaling (miR-21/miR-221/miR-222) or with the potential tumor suppressor DAAM1 (miR-335). Conversely, proapoptotic oncosuppressive miRNAs like miR-218 and miR-451 are downmodulated in GB [49, 50]. Interestingly, downmodulation of miR-221/miR-222 restores the p53 pathway, activates apoptosis, and sensitizes GB cells to TMZ [51]. In addition to its antiapoptotic effect, miR-21, along with miR-17, inhibits autophagy. Downregulation of these miRNA increases the sensitivity of GB cells to TMZ and radiation expanding the cell population undergoing apoptosis [52, 53].
Experimental models indicate that in GB exists a subpopulation of cells possessing the characteristics of neural stem cells that are responsible for continuous proliferation and drug resistance [54, 55]. miRNA profiling revealed that glioma cells have an expression profile remarkably similar to that of embryonic and neural precursor cells and distinct from that of a normal adult brain [56]. Interestingly, a set of 71 miRNA deregulated in human spontaneous GB is remarkably similar to that of chemically induced mouse glioma suggesting a common pattern of cancer development [56]. The miRNAs deregulated in GB and in neural precursor cells are clustered in seven genomic regions and have been associated with many other cancers like the mir-17 family cluster [57–59], miR-182-183 cluster [60], miR-302-367 and miR-372 [59, 61, 62], and the Dlk1 domain [63].
In GB, recurrent chromosomal aberrations are lacking; nevertheless, chromosomal instability (CIN) is considered an important mechanism for the establishment and maintenance of tumor heterogeneity [64]. CIN has the dual function of responding to various stressing conditions while being, at the same time, the origin of further genome destabilization. The comparison by genome-wide analysis between long-survival GB outlier patients (OS > 33 months) and short-term survivors (OS < 7 months) showed significantly lower genetic alterations in the short-term survivors than in long-term survivors [65]. The increased genomic instability of long-surviving patients might be responsible for the increased vulnerability of the cells to the standard radio- and chemotherapy. Along with this line, it was shown that glioma stem cells have high CIN that accounts for maintaining tumor heterogeneity and that increasing CIN further abolishes tumorigenicity as if an upper limit of genetic instability exists to initiate tumor formation [66].
Unsurprisingly, miRNAs are implicated in the molecular mechanisms of CIN and the intervention of these molecules into chromosomal instability has been studied in several tumors but, to the best of our knowledge, not yet in glioblastoma. Several miRNAs, like miR-26a and miR-28-5p, interfere with genes involved in cell replication and cell cycle checkpoints [67, 68] while others like miR-1255b, miR-148b∗, and miR-193b∗ reduce homologous recombination at G1 thus maintaining genomic stability [69]. Other miRNAs like miR-214 [70], miR-137 [71], miR-1255b, miR-148b∗, and miR-193b∗ [69] regulate at a different level the DNA repair mechanisms.
4. miRNA Targeting Immune Checkpoints and Inflammatory Molecules in Glioblastoma
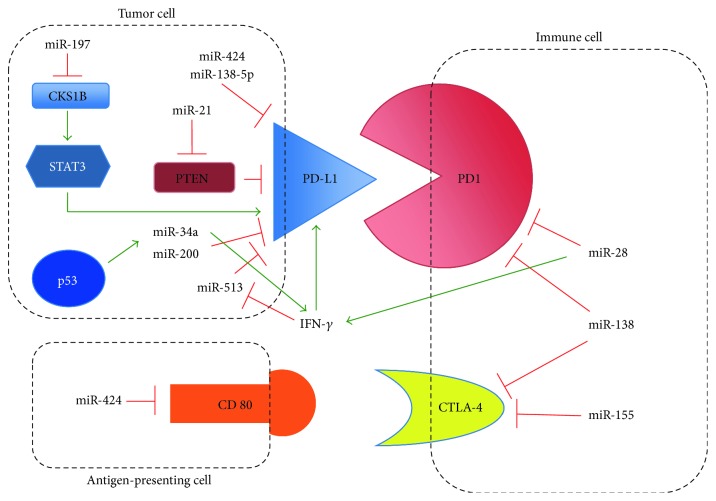
Targeting the immune checkpoint gene PD1, its ligand PDL1, and other genes like CTLA-4 has raised considerable attention in the recent years because of the dramatic antitumor effect exerted by antibodies against these molecules particularly in tumors with limited therapeutic options like lung cancer and melanoma (reviewed in [72, 73]). In this respect, glioblastoma is not an exception and GB escapes T-cell killing by activating immune checkpoints [74]. In an experimental model of glioma, the blockade of three immune checkpoints (IDO, CTLA-4, and PD1) significantly increased the survival of tumor-bearing mice [75]. However, these findings might not be generally applicable because of the extreme heterogeneity of this tumor and the absence of solid predictive biomarkers of sensitivity to immune checkpoints inhibitors [76]. Immune checkpoints can be modulated by microRNAs [77], and Figure 1 summarizes some of the interaction of the complex network between miRNA, genes, and immune checkpoints.
Figure 1.
Interactions between miRNA and immune checkpoints. This nonexaustive scheme shows the major interactions between miRNA and immune checkpoint molecules. Red and green arrows indicate the suppressive or activating interactions, respectively.
Intuitively, this complex network requires an extremely precise tuning since immune checkpoint molecules can be blocked by a given miRNA (e.g., miR-34a and miR-138) that, at the same time, can indirectly promote the expression of cytokines that in turn induce the expression of the same checkpoint molecules that are targeted by the miRNA.
Microglia and astrocytes, along with macrophages, are part of the glioma microenvironment, astrocytes are part of the brain immune system as they express cytokines and chemokines, and glioma cells produce and are targets of inflammatory molecules [78, 79]. Glioma cells produce IL-1 which is a potent inducer of angiogenesis and invasion and in glial cells, strongly upregulates miR-155 implicated in inflammation-mediated cancer development [80]. Besides miR-155, other IL-1-induced miRNAs involved in inflammation, miR-21, and miR-146 are upregulated in gliomas [81]. Interestingly, miR-146 is a negative regulator of astrocyte-mediated inflammation [82], and upregulation of this miRNA decreases the expression of its target TRAF6 that is linked to seizure frequency in glioma patients suggesting that miR-146 could be involved in the epileptogenic focus surrounding the tumor [81].
5. Invasiveness and the Blood Brain Barrier as Escape Mechanisms from Therapy
An important mechanism contributing to the failure of treatment in GB is the invasiveness of the tumor. Brain is a particular environment that is made impermeable to external molecules by the blood brain barrier (BBB); this prevents the efficient targeting of glioma cells with antineoplastic drugs unless the BBB is severely damaged as in glioma above 2–4 mm [83]. Nevertheless, GB can escape treatment also because of its capacity to infiltrate the brain forming niches in regions where the BBB is intact. Invasiveness is part of the epithelial to mesenchymal transition (EMT), a mechanism through which cells lose the epithelial phenotype and acquire that of mesenchymal cells. Several miRNAs are involved in EMT or in general into the mechanisms of invasion; they include miR-21, miR-146, miR-10b, and miR-7 that target directly or indirectly metalloproteinase inhibitors [84, 85], adhesion molecules, and other genes involved in metastasis and cell invasion [86–88].
GB is a highly vascularized tumor, and this feature likely contributes to the invasive and proliferative capacity of the tumor and to the shielding of GB cells behind the BBB. A set of miRNA (miR-296, miR-125b, and others) can directly or indirectly fine-tune angiogenic factors and promote neoangiogenesis in GB [89, 90]. In GB, angiogenesis can be stimulated also by other mechanisms like hypoxia through the targeting of HIF3A by miR-210-3p that results in the overexpression of VEGF [91]. Interference with VEGF is not the only angiogenic mechanisms exerted by miRNA in glioma; indeed, neovascularization can be promoted by miR-93 that targets integrin-β8 involved in cell-cell and cell-matrix interactions [92]. The transport mechanisms of miRNA across the BBB is still debated and could involve extracellular vesicles (EV) like exosomes that could serve as a communication tool in nonpathologic situations [93] or between tumor cells and their environment and trigger cell proliferation [94].
MicroRNAs have also an important role in drug and radioresistance as will be described in another part of this review.
6. Circulating miRNA as Glioma Biomarkers
Circulating nucleic acids and circulating tumor cells [95, 96] are extensively exploited as tumor markers to predict outcome and to monitor the response to therapy. Importantly, in GB, response to therapy often results in enhancement of the captation of the contrast medium that can be disguised as progression (“pseudoprogression”) challenging the imaging assessment of the disease [97]. The distinction between true progression and pseudoprogression is a diagnostic need required for optimizing patient's care.
Overall, both blood and cerebrospinal fluid are a source of circulating biomarkers and relevant glioma mutations like those of IDH1/IDH2 can be detected in circulating DNA [98]. On the contrary, circulating tumor cells are so scarce in glioma that, with the available technologies, their clinical potentials remain an open issue. Similarly, serum proteomics has not yet provided clinically useful results [99].
Extracellular vesicles (EV) are an attractive source of circulating biomarkers because they act as a cargo for many types of molecules that are protected from degradation [100]. EV are released by all cells to enable the communication between nonadjacent cells, and EV secretion is an early response of cancer cells to a variety of stress conditions including treatment [101]. Interestingly, EV are not randomly loaded and their content seems to reflect the biology of the donor cells making EV an ideal source of circulating biomarkers [102]. Although the utilization of EV in the clinical context is still in its infancy, promising results were obtained in two small GB trials. One of these was aimed at comparing the level of the DNA repair enzymes MGMT and APNG in the parental tissue and in EV before and after TMZ treatment [103]. In the second study, exosome mRNA was examined to study the changes of expression of immune markers and cytokines after inoculation of a tumor vaccine in glioblastoma patients [104]. Both studies demonstrate that in principle, molecules carried by EV can be utilized to develop robust assays to monitor disease progression in glioblastoma.
Different studies reported the miRNA profiling in the plasma of glioma patients or evaluated the level of defined circulating miRNA known to be involved in this tumor. A restricted signature of eleven miRNAs was selected through a systematic review of the literature and utilized to screen a small cohort of GB patients, and the results showed that the level of expression of miR-497 and miR-125b could distinguish between low- and high-grade glioma [105]. In principle, these types of markers could be very useful to monitor the evolution of primary low-grade glioma into secondary GB for better timing the beginning of therapeutic procedures. The expression of miRNA deregulated in GB was included in several studies on circulating biomarkers. Indeed, miR-21 was found overexpressed in plasma of glioblastoma patients compared to normal controls while miR-128 and miR-342-3p were downmodulated in the same set of patients [106]. Expression of these miRNA returned to baseline levels after treatment suggesting that circulating miRNA can be utilized to monitor disease response to treatment and disease relapse [106]. Interestingly, a recent whole miRNA profiling from the plasma of a relatively large set of glioblastoma patients identified a signature predicting disease-free and overall survival independently from other clinicopathological factors [107].
In conclusion, there are strong indications that circulating biomarkers have the potential to recapitulate the molecular complexity of GB and that they could gain clinical relevance. Nevertheless, more informative biomarkers are needed to develop robust and reproducible assay before a liquid biopsy could become a standard clinical practice.
7. MicroRNA and DNA Methylation: Interplay between Epigenetic Factors
The effects of miRNA dysregulation have been extensively studied initially at the level of single interaction between miRNA and its target gene or genes in a countless number of pathologic and physiologic conditions and more recently as components of signatures or within functional pathways. Intriguingly, miRNA can also be subjected to epigenetic control through DNA methylation and histone modifications [108] thus establishing a complex interplay capable of interfering, directly or indirectly, with multiple pathways in extraordinary complex networks that have been partially explored in several tumors including glioblastoma [109–115].
The effect of miRNA on epigenetic modifier genes and the influence of DNA methylation on miRNA expression in glioblastoma have been studied in some detail. In particular, targeting of DNMT3a and DNMT3b by miRNA-29, miRNA-29a, and miRNA-148 was observed, and it is generally believed that this interaction may contribute to the general hypomethylation seen in cancer [116–118]. However, the direct link between the expression of the miRNA-29 family and of miRNA-148 and the methylation status of glioblastoma cells has not yet been studied.
EZH2 is a histone methyltransferase that catalyzes the trimethylation of H3 at lysine 27 (H3K27), a postsynthetic modification of H3 leading to transcriptional inactivation [119]. Furthermore, EZH2 promotes the de novo DNA methylation interacting with DNMT3A and DNMT3B [120]. In glioblastoma, and other tumors, EZH2 is overexpressed and acts as an oncogene with multiple mode of actions including cell invasion utilizing largely tumor-specific mechanisms [121, 122], cell cycle progression, maintenance of cell stemness [123], and, last but not least, the development of drug resistance [123, 124] and inhibition of apoptosis [119]. It thus appears that EZH2 is at the center of many cancer-related pathways and that it must be kept under stringent transcriptional control. Several miRNAs, reported in Table 2, and lncRNAs are integral components of mechanisms that regulate EZH2 expression; however, the role of some of them in GB has not yet been investigated or experimentally proven.
Although manipulating EZH2 expression may seem a promising and logical strategy for the therapy of GB and other tumors, it must be reminded that knocking down a gene that masters DNA and histone methylation will epigenetically influence a vast number of genes with unpredictable effects. Indeed, it was shown that prolonged inhibition of EZH2 results in GB tumor progression whereas short-term inhibition improves survival in animal models [125]. However, it is likely that the major benefits from EZH2 inhibition will derive from appropriate scheduling of cytotoxic and epigenetic drugs as recently proposed [27].
Acetylation and deacetylation of histones H3 and H4 are postsynthetic modifications that contribute to the switching between permissive (acetylated) and repressed (deacetylated) conformation of the chromatin [126]. Acetylation and deacetylation are driven by two sets of enzymes: histone acetyltransferase (HAT) and histone deacetylases (HDAC) that include several variants. In glioblastoma, the expression of HDAC1 and HDAC3 is inversely correlated with survival of GB patients, whereas that of HDAC4, HDAC5, HDAC6, and HDAC11 is positively correlated with survival of glioma patients [127]. HDAC1 is a known target of miR-449 and miR-874 [128, 129] but the clinical relevance of the expression pattern of these miRNA in GB is not known. HDAC4 is targeted by miR-1 and miR-155 [130, 131]; in contrast with the HDAC expression data, exogenously expressed miR-1 that putatively should interfere with HDAC4 acts as a tumor suppressor gene prolonging survival in an animal model [132]. On the contrary, the expression of miR-155 in glioma is prevalent in high-grade tumors with a worse prognosis [133].
Several other genes belonging to the epigenetic machinery are targeted by miRNA; their involvement in glioblastoma is not yet well established, and they will not be discussed here.
Besides controlling epigenetic modifier genes, miRNA can be subjected to epigenetic control. This control can be exerted at three levels: DNA methylation, histone modification, and combined DNA methylation and histone modification. Approximately half of the known miRNAs are hosted in CpG -rich regions and are thus potential targets of DNA methylation; indeed, the effect of DNMT inhibitors has been tested on several cancer cells showing the activation of the miRNA-target gene axis [114, 115, 134]. To the best of our knowledge, the systematic analysis of miRNA silenced by DNA methylation in glioma has not yet been performed; nevertheless, several examples of miRNA silenced by DNA methylation in GB have been described along with the functional effects of their reexpression [135].
miR-211 targets MMP9, activates the caspase-9/caspase-3 apoptotic cascade, and was found to be hypermethylated in GB [136]. miR-204, methylated and downregulated in glioma, when activated suppresses the expression of stem transcription factor SOX4, reduces cell invasion, and prolongs survival in animal models [137]. miR-23 is hypermethylated in GB and is reactivated by 5-Azacytidine treatment leading to cell cycle arrest [138]. miR-145 is underexpressed in astrocytoma compared to normal brain, functionally acts as a tumor suppressor gene targeting SOX2, a stem-maintaining gene, and reduces proliferation and migration of GB cells targeting CTGF and NEDD9 [135, 139, 140]. miR-137 is epigenetically inactivated in many cancers, and its expression is diminished in GB and in glioma stem cells. Reexpression of miR-137, hypermethylated in GB tumor samples, promotes neural differentiation and decreases the expression of stem cell markers (Oct4, SOX2, and Nanog) [141]. Furthermore, miR-137 is also an inhibitor of EZH2 [142]. In one of the most comprehensive methylation analysis of miRNA promoter regions to date [143], 29 miRNAs differentially methylated in high-grade glioma were identified. The hypermethylation (and low expression) of three of them, miR-155, miR-210, and miR-355, was a strong predictor of better outcome and longer PFS. However, upon validation in different patient series and in multivariate models, only miR-155 remained of prognostic value independently from other indicators like histology, MGMT methylation, and IDH1/IDH2 mutation. Therefore, miR-155 can be considered both as an oncomiR in GBM with multiple biological roles including the activation of the NFkB pathway [143]. On the other hand, miR-155 could act also on the immune cell compartment by downmodulating the immune checkpoint molecule CTLA-4 exerting the function of a tumor suppressor miRNA (Figure 1 and [144]). miR-181c, another example of miRNA downregulated by epigenetic mechanisms in glioblastoma, targets the NOTCH2 pathway and is important in self-renewal, proliferation, and invasion of GB cells [145]. This miRNA was sorted out by chromatin immunoprecipitation/sequencing screening as a region containing H3K4me3 and H3K27ac marks partially overlapping with a CpG-rich region close to miR-181c that is hypermethylated in GB [146].
8. miRNA and Chemo- and Radioresistance in Glioblastoma
The response to treatment in GB patients is variable and probably depends from tumor heterogeneity that originates from genetic and epigenetic alterations which can influence the behavior of the disease. In this respect, the relation between miRNA and chemo- and radiotherapy has been extensively exploited to search for new possible therapeutic targets or to predict and improve the response to treatment.
Earlier preclinical studies showed that cisplatin could increase the efficacy of TMZ by decreasing the activity of MGMT [147] but several clinical trials have tested the activity of cisplatin in GB patients with limited success [148, 149]. Indeed, GB cells after an initial and positive response to cisplatin develop chemoresistance. Many biological pathways underlie the resistance to cisplatin and platinum derivatives [150], and several miRNAs contribute to the reduction of platinum sensitivity. Let-7b seems to be involved in cisplatin resistance affecting the cyclin D1 pathway [151], and miR-873, which targets Bcl-2, is downregulated in a time-dependent manner by cisplatin and, if overexpressed, increases apoptosis in cisplatin-resistant GB cells [152].
Temozolomide is, at the moment, the first-line drug for high-grade glioma treatment independently from the methylation status of MGMT (https://www.cancer.gov/types/brain/hp/adult-brain-treatment-pdq#link/_1089_toc). Several mechanisms of resistance to TMZ have been identified (reviewed in [153]), and epigenetic mechanisms, besides MGMT methylation, are explored as possible effectors of constitutive or acquired TMZ resistance in GB patients. In this respect, a substantial body of evidence gained mostly in preclinical models supports the idea that many miRNAs interfere with the response of the cells to TMZ.
As discussed above, miR-21 is consistently upregulated in astrocytic tumors (grade II–IV) [154] and downmodulates an entire set of oncosuppressor genes [41, 155, 156]. Indeed, miR-21 has antiapoptotic activity in glioblastoma cells [40] and treatment of GB cells with TMZ results in miR-21 overexpression while its inhibition with specific anti-miR-21 results in high apoptotic levels upon treatment with TMZ [157].
The AEG-1 (astrocyte elevated gene-1), overexpressed in GB tumor samples, favors the infiltration capabilities of established GB cell lines [158], and its downmodulation by siRNA sensitizes the cells to TMZ. AEG-1 is directly targeted by miR-136 that, when exogenously overexpressed, increases the cytotoxic activity of TMZ [159]. In principle, the expression of miR-136 could be utilized as an indicator of drug response in GB patients.
Direct targeting of genes controlling the apoptotic pathway is another mechanism capable to modulate TMZ resistance in GB cells. For example, miR-139 inhibits the expression of the antiapoptotic gene Mcl-1, a member of the Bcl-2 family, and sensitizes GB cells to the effect of TMZ [160]. Similarly, miR-143 targets several genes involved in the pathogenesis of cancer like K- and N-RAS, Bcl-2, and IGF-IR. The overexpression of miR-143 sensitizes GB cells to apoptosis induced by TMZ and inhibits invasion and proliferation, and this effect has been attributed to the direct targeting of N-RAS and, indirectly, to the dephosphorylation of AKT and to the downmodulation of HIF and VEGF as a result of N-RAS inhibition [161].
A more direct link with TMZ resistance is attributed to miRNA targeting directly MGMT. The inhibition of MGMT through different mechanisms besides DNA methylation silencing may at least partly explain the positive response to treatment in patients without methylation of MGMT. In this respect, miR-603 and miR-181d directly target and independently coregulate MGMT inducing sensitivity to TMZ [162].
As mentioned in a previous section of this review, miR-29c is a direct inhibitor of the de novo DNA methyltransferases DNMT3a/DNMT3b and is an indirect suppressor of MGMT via silencing of Sp1, a MGMT transcription factor. Interestingly, forced expression of miR-29c, which is downmodulated in glioblastoma, sensitizes cells to TMZ [163].
Along with chemotherapy, radiotherapy is an integral part of the clinical management of GB and different miRNAs are involved in radiosensitization or radioresistance.
Low levels of ATM protein are a major determinant of radiosensitivity in glioblastoma, and ATM is the target of different miRNAs such as miR-100 and miR-26a. High level of miR-100 expression was found in the radiosensitive glioma cell line; on the other hand, its ectopic expression in radioresistant cells downmodulates ATM and sensitizes the cells to ionizing radiation [164]. Ionizing radiation induces ATM expression (and radioresistance), and miR-26a restores radiosensitivity by targeting ATM [164]. It thus appears that drug and radioresistance in GB are controlled by an array of miRNA that directly or indirectly interferes with multiple pathways involved in drug and radiation response.
9. miRNA and Innovative Therapies in Glioblastoma
The development of multiomics strategies has led to impressive advancements of the knowledge on the mechanisms behind cell transformation and has opened the possibility of selectively targeting cancer cells in many types of tumors including GB [165–168].
In principle, a drug-based “biologic therapy” is aimed at changing the cell phenotype through the use of molecules capable of blocking well-defined pathways. This can be achieved either through the functional inhibition of the enzymatic activities of a given protein or through the ablation of the protein itself. The first strategy leaves the protein unmodified while the second acts on the expression of the target protein and, in theory, should be more effective.
Transcriptional inhibition of a given gene can be obtained by RNA interference, a mechanism originally described in worms [169] and later in higher organisms [170–172]. In practice, it was observed that double-stranded RNA delivered into the cells caused the degradation of the target mRNA and this system is now widely employed for the transient or stable gene inactivation. MicroRNA, because of their hairpin and partially complementary structure, can be considered as an endogenous form of interfering RNA that depending on the extent of complementarity with their targets can either stop the translation or promote the degradation of the mRNA.
A major question to be answered is if miRNA modulation of gene transcription is powerful enough to have a therapeutic consequence in glioma also in view of the necessity of obtaining an adequate delivery at the tumor site. While in vitro assays demonstrated the feasibility of this approach, the in vivo translation of these studies appears a much more complex task. The partial knowledge of the miRNA networks, pathways, target genes, and of their interplay in healthy and diseased cells adds further difficulties to the short-term therapeutic utilization of these strategies.
One of the questions that need to be answered is if miRNA has the potential to enter the routine clinical practice. Along with this review, we have seen that suppressing certain oncomiR (i.e., miR-21) or inducing the expression of tumor suppressor miRNA like let-7 has dramatic effects on cell behavior and suppress GBM viability. Nevertheless, many major issues still remain, first of all, the problem of delivery, and also the choice between monospecific synthetic siRNA and polyspecific miRNA mimics or miRNA antagonists. siRNAs have the obvious advantage to selectively target specific pathway components while miRNA can interfere with multiple pathways at once. However, the off-target effects of the miRNA have to be carefully evaluated. Furthermore, if a siRNA cocktail seems a reasonable tool, the utilization of a miRNA cocktail seems more complex also because of the conspicuous off-target effect of this cocktail and because of the interactions between different miRNAs [114].
9.1. Biological Therapies in GB: The Delivery Issue
In Glioblastoma, the presence of the BBB represents a major challenge to the utilization of miRNA in therapy because if the BBB is damaged and permeable at the tumor site, its integrity is maintained at the infiltrating tumor areas that are those responsible for tumor relapse after initial surgery and radiochemotherapy [83]. Nevertheless, some preliminary results support the use of antago-miR or miRNA mimics in the therapy of glioma although the issue of the active concentration that can be achieved at the tumor site needs to be taken into consideration.
The ideal goals of the delivery across the BBB are as follows:
to increase the local drug concentration
to increase the possibility of using drugs that do not pass through the BBB
to increase the possibility of reaching the tumor niches surrounded by integral BBB that are responsible for tumor relapse
to increase the possibility of using antitumor drugs in low-grade glioma protected by a functional BBB
Delivery systems can be passive or active. The objective of the passive methods is the permeabilization of the BBB with hyperosmotic agents, surfactant, ultrasounds, and electromagnetic waves to transiently open the tight cell-cell junctions of the BBB [173]. In this respect, a randomized phase III clinical trial showed that the combined treatment of TMZ and pulsed electric fields is superior to the standard TMZ treatment [174]. The direct infusion of drugs or other bioactive molecules at the site of the lesion after craniotomy, even if highly selective, was found of limited utility because of the poor diffusion in the perilesional area where the tumor niches are [175].
Active transport toward the lesion is considered, in general, a more efficient mode to selectively deliver drugs or other molecules within the brain. The most promising active delivery systems are those based on nanoparticles of less than 200 nm [176] that carry on their surface molecules that can be recognized by specific receptors on the BBB, like transferrin, lactoferrin, transferrin receptor, and glutathione [177].
The most commonly utilized carriers for drug delivery in the CNS are liposomes at a single or double layer of approximately 100 nm of diameter that are engineered with molecules for tumor targeting [178, 179]. Some liposomal formulations have entered into the clinical practice, and others are being tested in clinical trials [179–183].
Other utilized delivery systems are the polymeric colloids (PDP) [184–186] or other colloidal formulations (LNC) [187–189] that can be modified to pass the BBB and to target the tumor utilizing two ligands [185]. The delivery systems based on nanoparticles are highly promising but their toxicity, biocompatibility, and payload retention must be carefully evaluated [190, 191].
9.2. Targeting Glioblastoma Cells with miRNA
Convention-enhanced delivery, a drug delivery method based on catheters stereotactically implanted to infuse the treatments directly to the tumor site, was utilized to deliver let-7a into the brain of mice xenografted with an aggressive GB. This treatment was well tolerated and was effective in reducing the expression of HMGA2, one of the targets of let-7a [192].
Although direct delivery of miRNA into the brain seems to be effective, intuitively non- or minimally invasive drug delivery methods may be preferable. In this respect, nanoparticles seem a very promising strategy and were exploited to deliver at the tumor site not only a variety of drugs but also miRNA [193]. In principle, nanoparticles should overcome the poor systemic stability of oligonucleotides and improve their delivery; as said above, different nanoparticle formulations are available each with advantages and disadvantages but they can all be engineered to target the tumor site and, in the case of brain tumors, to transit the BBB. To date, the most common carriers are targeted liposomes of 100 nm [178, 194] that are being tested in animal models [180, 181].
Several types of nanoparticles have been utilized to carry a number of miRNA and to test their biological effects. For example, antago-miR-21 carried by RNP were utilized to successfully rescue the expression of antioncogenic PTEN and of PDCD4 and to promote tumor regression in a model system [195]. Similarly, antago-miR-21, delivered by poly(lactic-co-glycolic acid) (PLGA), sensitizes the effect of TMZ in vitro [196].
Another interesting example of cooperative treatment in glioblastoma is provided by a multifunctional delivery system MSNPs (mesoporous silica nanoparticle) charged with TMZ molecules and decorated by an anti-miR-221 PNA-octaarginine conjugate (R8-PNA221) that increases the biological effect of TMZ in drug-resistant cells [197]. A similar effect was seen with miR-34a encapsulated in a polyglycerol scaffold [198].
Mesenchymal stem cells (MSC) are an interesting and potentially very effective method to target sites of injury or of inflammation and tumors for therapeutic purposes [199, 200], and it was demonstrated that functional miRNA can be conveyed to neural progenitor cells by cocultivation with appropriately engineered MSC [201].
As mentioned above, miR-10b is involved in tumor invasion and is an optimal therapeutic target because of its high and generalized expression in all GB subtypes [86–88]. A preclinical in vivo study focused on the inhibition of miR-10b in an orthotopic GB xenograft model compared the results of different delivery methods utilizing as endpoint the inhibition of the tumor growth [202]. Brain injections, systemic injections, and intracranial osmotic pumps were compared, and each one showed weak and strong points. The antagonist of miR-10b administered by the three routes resulted in the inhibition of miR-10b and in turn reactivated its target genes, attenuated tumor growth, and prolonged survival. Considering the possible translation from the bench to the bedside, the systemic injections of miR-10b inhibitor were less invasive compared to the other routes and had minimal or no side effects on extracranial tissues and with a good delivery through the BBB.
miRNA “sponges” are oligonucleotide sequences that contain many binding sites for a specific miRNA or miRNA family and act as competitive inhibitors of the binding of the miRNA to their targets [203]. The utility of these “sponges” in GB was recently demonstrated for miR-23b in an orthotopic in vivo model and showed the reduction of angiogenesis, migration and invasion, and in turn the malignancy of the tumor [204]. Circular RNAs (circRNA) are natural examples of sponges that are highly resistant to degradation and that are now subject of in-depth investigations because of their strong regulatory activity on miRNA [205].
10. Conclusions
MicroRNAs are epigenetic regulatory molecules that possessing multiple targets have a profound impact on cell physiology and pathology. MicroRNAs are players of the “epigenetic orchestra” that fine-tune the coordinate transcription of the genetic information. It is quite clear that control exerted by miRNA is extraordinary complex, that indeed a single miRNA can bind many genes, and that each gene can be recognized by many miRNAs in an extremely complex direct and indirect regulatory circuitry. Obviously, mastering this network could have dramatic effects on cell behavior.
Therefore, it is not surprising that although our knowledge of the complex effects and interactions between miRNA and genome is still incomplete, the potential implications of miRNA for the diagnosis and prognosis, for the patients' stratification and for their personalized therapy, were not overlooked. However, in order to translate the impressive basic knowledge so far gained on miRNA onto the clinical practice, several issues urgently need to be addressed. Besides the technicisms of delivery and targeting, the major problem remains that of understanding the miRNA effects not anymore at the level of single miRNA-target interaction, but utilizing a “holistic” approach to fully appreciate the balance between miRNA and target genes of opposite functions.
It is quite clear that this will be a highly demanding, but exciting, task for the scientists of the immediate future.
Acknowledgments
This study was supported by the Italian Ministry of Health, Core Grant “Ricerca Corrente” to the IRCCS AOU San Martino-IST and by the grant from the “Fondazione Compagnia di San Paolo, Torino”: “Terapie innovative per il glioblastoma” (2016–2018).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Crocetti E., Trama A., Stiller C., et al. Epidemiology of glial and non-glial brain tumours in Europe. European Journal of Cancer. 2012;48(10):1532–1542. doi: 10.1016/j.ejca.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Louis D. N., Perry A., Reifenberger G., et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Chen R., Cohen A. L., Colman H. Targeted therapeutics in patients with high-grade gliomas: past, present, and future. Current Treatment Options in Oncology. 2016;17(8):p. 42. doi: 10.1007/s11864-016-0418-0. [DOI] [PubMed] [Google Scholar]
- 4.Kelly A. D., Issa J. J. The promise of epigenetic therapy: reprogramming the cancer epigenome. Current Opinion in Genetics & Development. 2017;42:68–77. doi: 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Biswas S., Rao C. M. Epigenetics in cancer: fundamentals and beyond. Pharmacology & Therapeutics. 2017;173:118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Gosden R. G., Feinberg A. P. Genetics and epigenetics — nature’s pen-and-pencil set. The New England Journal of Medicine. 2007;356(7):731–733. doi: 10.1056/NEJMe068284. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noushmehr H., Weisenberger D. J., Diefes K., et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturm D., Witt H., Hovestadt V., et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 10.Hegi M. E., Diserens A. C., Gorlia T., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England Journal of Medicine. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Stupp R., Mason W. P., van den Bent M. J., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England Journal of Medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 12.Toyota M., Ahuja N., Ohe-Toyota M., Herman J. G., Baylin S. B., Issa J. P. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller B. F., Sanchez-Vega F., Elnitski L. The emergence of pan-cancer CIMP and its elusive interpretation. Biomolecules. 2016;6(4) doi: 10.3390/biom6040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Rijnsoever M., Elsaleh H., Joseph D., McCaul K., Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clinical Cancer Research. 2003;9(8):2898–2903. [PubMed] [Google Scholar]
- 15.Kelly A. D., Kroeger H., Yamazaki J., et al. A CpG island methylator phenotype in acute myeloid leukemia independent of IDH mutations and associated with a favorable outcome. Leukemia. 2017 doi: 10.1038/leu.2017.12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang F., Turcan S., Rimner A., et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Science Translational Medicine. 2011;3(75, article 75ra25) doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe M., Ohira M., Kaneda A., et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Research. 2005;65(3):828–834. [PubMed] [Google Scholar]
- 18.Banelli B., Merlo D. F., Allemanni G., Forlani A., Romani M. Clinical potentials of methylator phenotype in stage 4 high-risk neuroblastoma: an open challenge. PLoS One. 2013;8(5, article e63253) doi: 10.1371/journal.pone.0063253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanemura A., Terando A. M., Sim M. S., et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clinical Cancer Research. 2009;15(5):1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyupoglu I. Y., Savaskan N. E. Epigenetics in brain tumors: HDACs take center stage. Current Neuropharmacology. 2016;14(1):48–54. doi: 10.2174/1570159x13666151030162457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banelli B., Carra E., Barbieri F., et al. The histone demethylase KDM5A is a key factor for the resistance to temozolomide in glioblastoma. Cell Cycle. 2015;14(21):3418–3429. doi: 10.1080/15384101.2015.1090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liau B. B., Sievers C., Donohue L. K., et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20(2):233–246.e7. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sareddy G. R., Nair B. C., Krishnan S. K., et al. KDM1 is a novel therapeutic target for the treatment of gliomas. Oncotarget. 2013;4(1):18–28. doi: 10.18632/oncotarget.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 25.Kinnaird A., Zhao S., Wellen K. E., Michelakis E. D. Metabolic control of epigenetics in cancer. Nature Reviews. Cancer. 2016;16(11):694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 26.Coolen M. W., Stirzaker C., Song J. Z., et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nature Cell Biology. 2010;12(3):235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banelli B., Daga A., Forlani A., et al. Small molecules targeting histone demethylase genes (KDMs) inhibit growth of temozolomide-resistant glioblastoma cells. Oncotarget. 2017;8(21):34896–34910. doi: 10.18632/oncotarget.16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helin K., Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502(7472):480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 29.Moller H. G., Rasmussen A. P., Andersen H. H., Johnsen K. B., Henriksen M., Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Molecular Neurobiology. 2013;47(1):131–144. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson S. M., Grosshans H., Shingara J., et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Sampson V. B., Rong N. H., Han J., et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Research. 2007;67(20):9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 32.Lee S. T., Chu K., Oh H. J., et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. Journal of Neuro-Oncology. 2011;102(1):19–24. doi: 10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 33.Papagiannakopoulos T., Friedmann-Morvinski D., Neveu P., et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31(15):1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuchty S., Arjona D., Li A., et al. Prediction of associations between microRNAs and gene expression in glioma biology. PLoS One. 2011;6(2, article e14681) doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Chao T., Li R., et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. Journal of Molecular Medicine. 2009;87(1):43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 36.Sun F., Fu H., Liu Q., et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Letters. 2008;582(10):1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 37.Silber J., Lim D. A., Petritsch C., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Medicine. 2008;6:p. 14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Dai L. X., Zhang S., et al. Regulation of epidermal growth factor receptor signaling by plasmid-based microRNA-7 inhibits human malignant gliomas growth and metastasis in vivo. Neoplasma. 2013;60(3):274–283. doi: 10.4149/neo_2013_036. [DOI] [PubMed] [Google Scholar]
- 39.Kefas B., Godlewski J., Comeau L., et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Research. 2008;68(10):3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 40.Chan J. A., Krichevsky A. M., Kosik K. S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Research. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 41.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papagiannakopoulos T., Shapiro A., Kosik K. S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Research. 2008;68(19):8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 43.Qi J. H., Ebrahem Q., Moore N., et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nature Medicine. 2003;9(4):407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- 44.Corsten M. F., Miranda R., Kasmieh R., Krichevsky A. M., Weissleder R., Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Research. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 45.Fornari F., Gramantieri L., Ferracin M., et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 46.Gillies J. K., Lorimer I. A. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6(16):2005–2009. doi: 10.4161/cc.8.17.9489. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C. Z., Zhang J. X., Zhang A. L., et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Molecular Cancer. 2010;9:p. 229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu M., Zheng X., Wu S., et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Molecular Cancer. 2011;10:p. 59. doi: 10.1186/1476-4598-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia H., Yan Y., Hu M., et al. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κB activity. Neuro-Oncology. 2013;15(4):413–422. doi: 10.1093/neuonc/nos296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nan Y., Han L., Zhang A., et al. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Research. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 51.Chen L., Zhang J., Han L., et al. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncology Reports. 2012;27(3):854–860. doi: 10.3892/or.2011.1535. [DOI] [PubMed] [Google Scholar]
- 52.Gwak H. S., Kim T. H., Jo G. H., et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7(10, article e47449) doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Comincini S., Allavena G., Palumbo S., et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biology & Therapy. 2013;14(7):574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dirks P. B. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Molecular Oncology. 2010;4(5):420–430. doi: 10.1016/j.molonc.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abou-Antoun T. J., Hale J. S., Lathia J. D., Dombrowski S. M. Brain cancer stem cells in adults and children: cell biology and therapeutic implications. Neurotherapeutics. 2017;14(2):372–384. doi: 10.1007/s13311-017-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavon I., Zrihan D., Granit A., et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro-Oncology. 2010;12(5):422–433. doi: 10.1093/neuonc/nop061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ambs S., Prueitt R. L., Yi M., et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Research. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volinia S., Calin G. A., Liu C. G., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uziel T., Karginov F. V., Xie S., et al. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandres E., Cubedo E., Agirre X., et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Molecular Cancer. 2006;5:p. 29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Q., Gumireddy K., Schrier M., et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biology. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 62.Yu J., Wang F., Yang G. H., et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochemical and Biophysical Research Communications. 2006;349(1):59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 63.Zhang L., Volinia S., Bonome T., et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(19):7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heng H. H., Bremer S. W., Stevens J. B., et al. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastasis Reviews. 2013;32(3-4):325–340. doi: 10.1007/s10555-013-9427-7. [DOI] [PubMed] [Google Scholar]
- 65.Peng S., Dhruv H., Armstrong B., et al. Integrated genomic analysis of survival outliers in glioblastoma. Neuro-Oncology. 2017;19(6):833–844. doi: 10.1093/neuonc/now269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godek K. M., Venere M., Wu Q., et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discovery. 2016;6(5):532–545. doi: 10.1158/2159-8290.CD-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castellano L., Dabrowska A., Pellegrino L., et al. Sustained expression of miR-26a promotes chromosomal instability and tumorigenesis through regulation of CHFR. Nucleic Acids Research. 2017;45(8):4401–4412. doi: 10.1093/nar/gkx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hell M. P., Thoma C. R., Fankhauser N., Christinat Y., Weber T. C., Krek W. miR-28-5p promotes chromosomal instability in VHL-associated cancers by inhibiting Mad2 translation. Cancer Research. 2014;74(9):2432–2443. doi: 10.1158/0008-5472.CAN-13-2041. [DOI] [PubMed] [Google Scholar]
- 69.Choi Y. E., Pan Y., Park E., et al. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. eLife. 2014;3, article e02445 doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z., Yin H., Zhang Y., et al. miR-214-mediated downregulation of RNF8 induces chromosomal instability in ovarian cancer cells. Cell Cycle. 2014;13(22):3519–3528. doi: 10.4161/15384101.2014.958413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin Y., Zhang S., Deng S., et al. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia. 2017;31(5):1123–1135. doi: 10.1038/leu.2016.325. [DOI] [PubMed] [Google Scholar]
- 72.Dal Bello M. G., Alama A., Coco S., Vanni I., Grossi F. Understanding the checkpoint blockade in lung cancer immunotherapy. Drug Discovery Today. 2017;22(8):1266–1273. doi: 10.1016/j.drudis.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 73.Schadendorf D., Fisher D. E., Garbe C., et al. Melanoma. Nature Reviews Disease Primers. 2015;1, article 15003 doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 74.Preusser M., Lim M., Hafler D. A., Reardon D. A., Sampson J. H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nature Reviews Neurology. 2015;11(9):504–514. doi: 10.1038/nrneurol.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wainwright D. A., Chang A. L., Dey M., et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clinical Cancer Research. 2014;20(20):5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garg A. D., Vandenberk L., Van Woensel M., et al. Preclinical efficacy of immune-checkpoint monotherapy does not recapitulate corresponding biomarkers-based clinical predictions in glioblastoma. OncoImmunology. 2017;6(4, article e1295903) doi: 10.1080/2162402X.2017.1295903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smolle M. A., Calin H. N., Pichler M., Calin G. A. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. The FEBS Journal. 2017;284(13):1952–1966. doi: 10.1111/febs.14030. [DOI] [PubMed] [Google Scholar]
- 78.Charles N. A., Holland E. C., Gilbertson R., Glass R., Kettenmann H. The brain tumor microenvironment. Glia. 2011;59(8):1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 79.Yang I., Han S. J., Kaur G., Crane C., Parsa A. T. The role of microglia in central nervous system immunity and glioma immunology. Journal of Clinical Neuroscience. 2010;17(1):6–10. doi: 10.1016/j.jocn.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tili E., Michaille J. J., Wernicke D., et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prabowo A. S., van Scheppingen J., Iyer A. M., et al. Differential expression and clinical significance of three inflammation-related microRNAs in gangliogliomas. Journal of Neuroinflammation. 2015;12:p. 97. doi: 10.1186/s12974-015-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iyer A., Zurolo E., Prabowo A., et al. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One. 2012;7(9, article e44789) doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schlageter K. E., Molnar P., Lapin G. D., Groothuis D. R. Microvessel organization and structure in experimental brain tumors: microvessel populations with distinctive structural and functional properties. Microvascular Research. 1999;58(3):312–328. doi: 10.1006/mvre.1999.2188. [DOI] [PubMed] [Google Scholar]
- 84.Li Y., Wang Y., Yu L., et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Letters. 2013;339(2):260–269. doi: 10.1016/j.canlet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 85.Gabriely G., Wurdinger T., Kesari S., et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and Cellular Biology. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun L., Yan W., Wang Y., et al. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Research. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 87.Lin J., Teo S., Lam D. H., Jeyaseelan K., Wang S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death & Disease. 2012;3, article e398 doi: 10.1038/cddis.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu D. G., Wang Y. Y., Fan L. G., et al. MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chinese Medical Journal. 2011;124(17):2616–2621. [PubMed] [Google Scholar]
- 89.Wang S., Olson E. N. AngiomiRs—key regulators of angiogenesis. Current Opinion in Genetics & Development. 2009;19(3):205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wurdinger T., Tannous B. A., Saydam O., et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14(5):382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agrawal R., Pandey P., Jha P., Dwivedi V., Sarkar C., Kulshreshtha R. Hypoxic signature of microRNAs in glioblastoma: insights from small RNA deep sequencing. BMC Genomics. 2014;15:p. 686. doi: 10.1186/1471-2164-15-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang L., Deng Z., Shatseva T., et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30(7):806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 93.Kosaka N., Iguchi H., Yoshioka Y., Hagiwara K., Takeshita F., Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. The Journal of Biological Chemistry. 2012;287(2):1397–1405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Skog J., Wurdinger T., van Rijn S., et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han X., Wang J., Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics, Proteomics & Bioinformatics. 2017;15(2):59–72. doi: 10.1016/j.gpb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liang S. B., Fu L. W. Application of single-cell technology in cancer research. Biotechnology Advances. 2017;35(4):443–449. doi: 10.1016/j.biotechadv.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 97.Ellingson B. M., Chung C., Pope W. B., Boxerman J. L., Kaufmann T. J. Pseudoprogression, radionecrosis, inflammation or true tumor progression? Challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. Journal of Neuro-Oncology. 2017 doi: 10.1007/s11060-017-2375-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boisselier B., Gallego Perez-Larraya J., Rossetto M., et al. Detection of IDH1 mutation in the plasma of patients with glioma. Neurology. 2012;79(16):1693–1698. doi: 10.1212/WNL.0b013e31826e9b0a. [DOI] [PubMed] [Google Scholar]
- 99.Capper D., Simon M., Langhans C. D., et al. 2-hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. International Journal of Cancer. 2012;131(3):766–768. doi: 10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]
- 100.Contreras-Naranjo J. C., Wu H. J., Ugaz V. M. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab on a Chip. 2017 doi: 10.1039/c7lc00592j. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kucharzewska P., Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. Journal of Extracellular Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morishita M., Takahashi Y., Nishikawa M., et al. Quantitative analysis of tissue distribution of the B16BL6-derived exosomes using a streptavidin-lactadherin fusion protein and iodine-125-labeled biotin derivative after intravenous injection in mice. Journal of Pharmaceutical Sciences. 2015;104(2):705–713. doi: 10.1002/jps.24251. [DOI] [PubMed] [Google Scholar]
- 103.Shao H., Chung J., Lee K., et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nature Communications. 2015;6:p. 6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muller L., Muller-Haegele S., Mitsuhashi M., Gooding W., Okada H., Whiteside T. L. Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect antitumor immune activity and might predict survival. OncoImmunology. 2015;4(6, article e1008347) doi: 10.1080/2162402X.2015.1008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Regazzo G., Terrenato I., Spagnuolo M., et al. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. Journal of Experimental & Clinical Cancer Research. 2016;35(1):p. 124. doi: 10.1186/s13046-016-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Q., Li P., Li A., et al. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. Journal of Experimental & Clinical Cancer Research. 2012;31:p. 97. doi: 10.1186/1756-9966-31-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao H., Shen J., Hodges T. R., Song R., Fuller G. N., Heimberger A. B. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Molecular Cancer. 2017;16(1):p. 59. doi: 10.1186/s12943-017-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Z., Yao H., Lin S., et al. Transcriptional and epigenetic regulation of human microRNAs. Cancer Letters. 2013;331(1):1–10. doi: 10.1016/j.canlet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 109.Ha M. J., Baladandayuthapani V., Do K. A. DINGO: differential network analysis in genomics. Bioinformatics. 2015;31(21):3413–3420. doi: 10.1093/bioinformatics/btv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doecke J. D., Wang Y., Baggerly K. Co-localized genomic regulation of miRNA and mRNA via DNA methylation affects survival in multiple tumor types. Cancer Genetics. 2016;209(10):463–473. doi: 10.1016/j.cancergen.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 111.Lopez-Bertoni H., Lal B., Li A., et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene. 2015;34(30):3994–4004. doi: 10.1038/onc.2014.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jha P., Agrawal R., Pathak P., et al. Genome-wide small noncoding RNA profiling of pediatric high-grade gliomas reveals deregulation of several miRNAs, identifies downregulation of snoRNA cluster HBII-52 and delineates H3F3A and TP53 mutant-specific miRNAs and snoRNAs. International Journal of Cancer. 2015;137(10):2343–2353. doi: 10.1002/ijc.29610. [DOI] [PubMed] [Google Scholar]
- 113.Maugeri M., Barbagallo D., Barbagallo C., et al. Altered expression of miRNAs and methylation of their promoters are correlated in neuroblastoma. Oncotarget. 2016;7(50):83330–83341. doi: 10.18632/oncotarget.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parodi F., Carosio R., Ragusa M., et al. Epigenetic dysregulation in neuroblastoma: a tale of miRNAs and DNA methylation. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2016;1859(12):1502–1514. doi: 10.1016/j.bbagrm.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 115.Barbagallo D., Condorelli A., Ragusa M., et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fabbri M., Garzon R., Cimmino A., et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garzon R., Liu S., Fabbri M., et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duursma A. M., Kedde M., Schrier M., le Sage C., Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yin Y., Qiu S., Peng Y. Functional roles of enhancer of zeste homolog 2 in gliomas. Gene. 2016;576, 1, Part 2:189–194. doi: 10.1016/j.gene.2015.09.080. [DOI] [PubMed] [Google Scholar]
- 120.Vire E., Brenner C., Deplus R., et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 121.Crea F., Hurt E. M., Farrar W. L. Clinical significance of polycomb gene expression in brain tumors. Molecular Cancer. 2010;9:p. 265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ren G., Baritaki S., Marathe H., et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Research. 2012;72(12):3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 123.Kim S. H., Joshi K., Ezhilarasan R., et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Reports. 2015;4(2):226–238. doi: 10.1016/j.stemcr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan T. Y., Wang H., Xiang P., et al. Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. International Journal of Clinical and Experimental Pathology. 2014;7(10):6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 125.de Vries N. A., Hulsman D., Akhtar W., et al. Prolonged Ezh2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Reports. 2015;10(3):383–397. doi: 10.1016/j.celrep.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 126.Allfrey V. G., Mirsky A. E. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science. 1964;144(3618):p. 559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 127.Dali-Youcef N., Froelich S., Moussallieh F. M., et al. Gene expression mapping of histone deacetylases and co-factors, and correlation with survival time and 1H-HRMAS metabolomic profile in human gliomas. Scientific Reports. 2015;5, article 9087 doi: 10.1038/srep09087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Noonan E. J., Place R. F., Pookot D., et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 129.Nohata N., Hanazawa T., Kinoshita T., et al. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. British Journal of Cancer. 2013;108(8):1648–1658. doi: 10.1038/bjc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen J. F., Mandel E. M., Thomson J. M., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sandhu S. K., Volinia S., Costinean S., et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(49):20047–20052. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bronisz A., Wang Y., Nowicki M. O., et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Research. 2014;74(3):738–750. doi: 10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu X., Wang Y., Yu T., et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro-Oncology. 2017;19(9):1195–1205. doi: 10.1093/neuonc/nox017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roman-Gomez J., Agirre X., Jimenez-Velasco A., et al. Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. Journal of Clinical Oncology. 2009;27(8):1316–1322. doi: 10.1200/JCO.2008.19.3441. [DOI] [PubMed] [Google Scholar]
- 135.Lee H. K., Finniss S., Cazacu S., et al. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget. 2013;4(2):346–361. doi: 10.18632/oncotarget.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Asuthkar S., Velpula K. K., Chetty C., Gorantla B., Rao J. S. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3(11):1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ying Z., Li Y., Wu J., et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Research. 2013;73(2):990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Geng J., Luo H., Pu Y., et al. Methylation mediated silencing of miR-23b expression and its role in glioma stem cells. Neuroscience Letters. 2012;528(2):185–189. doi: 10.1016/j.neulet.2012.08.055. [DOI] [PubMed] [Google Scholar]
- 139.Lee H. K., Bier A., Cazacu S., et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8(2, article e54652) doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Speranza M. C., Frattini V., Pisati F., et al. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget. 2012;3(7):723–734. doi: 10.18632/oncotarget.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bier A., Giladi N., Kronfeld N., et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4(5):665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Szulwach K. E., Li X., Smrt R. D., et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. The Journal of Cell Biology. 2010;189(1):127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schliesser M. G., Claus R., Hielscher T., et al. Prognostic relevance of miRNA-155 methylation in anaplastic glioma. Oncotarget. 2016;7(50):82028–82045. doi: 10.18632/oncotarget.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Y., Sun E., Li X., et al. miR-155 contributes to Df1-induced asthma by increasing the proliferative response of Th cells via CTLA-4 downregulation. Cellular Immunology. 2017;314:1–9. doi: 10.1016/j.cellimm.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 145.Ruan J., Lou S., Dai Q., Mao D., Ji J., Sun X. Tumor suppressor miR-181c attenuates proliferation, invasion, and self-renewal abilities in glioblastoma. Neuroreport. 2015;26(2):66–73. doi: 10.1097/WNR.0000000000000302. [DOI] [PubMed] [Google Scholar]
- 146.Ayala-Ortega E., Arzate-Mejia R., Perez-Molina R., et al. Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines. BMC Cancer. 2016;16:p. 226. doi: 10.1186/s12885-016-2273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.D'Atri S., Graziani G., Lacal P. M., et al. Attenuation of O 6-methylguanine-DNA methyltransferase activity and mRNA levels by cisplatin and temozolomide in Jurkat cells. The Journal of Pharmacology and Experimental Therapeutics. 2000;294(2):664–671. [PubMed] [Google Scholar]
- 148.Buckner J. C., Ballman K. V., Michalak J. C., et al. Phase III trial of carmustine and cisplatin compared with carmustine alone and standard radiation therapy or accelerated radiation therapy in patients with glioblastoma multiforme: North Central Cancer Treatment Group 93-72-52 and Southwest Oncology Group 9503 Trials. Journal of Clinical Oncology. 2006;24(24):3871–3879. doi: 10.1200/JCO.2005.04.6979. [DOI] [PubMed] [Google Scholar]
- 149.Brandes A. A., Basso U., Reni M., et al. First-line chemotherapy with cisplatin plus fractionated temozolomide in recurrent glioblastoma multiforme: a phase II study of the Gruppo Italiano Cooperativo di Neuro-Oncologia. Journal of Clinical Oncology. 2004;22(9):1598–1604. doi: 10.1200/JCO.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 150.Shen D. W., Pouliot L. M., Hall M. D., Gottesman M. M. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacological Reviews. 2012;64(3):706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guo Y., Yan K., Fang J., Qu Q., Zhou M., Chen F. Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. Journal of Experimental & Clinical Cancer Research. 2013;32:p. 41. doi: 10.1186/1756-9966-32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen X., Zhang Y., Shi Y., et al. MiR-873 acts as a novel sensitizer of glioma cells to cisplatin by targeting Bcl-2. International Journal of Oncology. 2015;47(4):1603–1611. doi: 10.3892/ijo.2015.3143. [DOI] [PubMed] [Google Scholar]
- 153.Messaoudi K., Clavreul A., Lagarce F. Toward an effective strategy in glioblastoma treatment. Part I: resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discovery Today. 2015;20(7):899–905. doi: 10.1016/j.drudis.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 154.Conti A., Aguennouz M., La Torre D., et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. Journal of Neuro-Oncology. 2009;93(3):325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- 155.Asangani I. A., Rasheed S. A., Nikolova D. A., et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 156.Zhu S., Si M. L., Wu H., Mo Y. Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) The Journal of Biological Chemistry. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 157.Wong S. T., Zhang X. Q., Zhuang J. T., Chan H. L., Li C. H., Leung G. K. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Research. 2012;32(7):2835–2841. [PubMed] [Google Scholar]
- 158.Emdad L., Sarkar D., Lee S. G., et al. Astrocyte elevated gene-1: a novel target for human glioma therapy. Molecular Cancer Therapeutics. 2010;9(1):79–88. doi: 10.1158/1535-7163.MCT-09-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu H., Liu Q., Cai T., Chen Y. D., Liao F., Wang Z. F. MiR-136 modulates glioma cell sensitivity to temozolomide by targeting astrocyte elevated gene-1. Diagnostic Pathology. 2014;9:p. 173. doi: 10.1186/s13000-014-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li R. Y., Chen L. C., Zhang H. Y., et al. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neuroscience & Therapeutics. 2013;19(7):477–483. doi: 10.1111/cns.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang L., Shi Z. M., Jiang C. F., et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5(14):5416–5427. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kushwaha D., Ramakrishnan V., Ng K., et al. A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget. 2014;5(12):4026–4039. doi: 10.18632/oncotarget.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiao S., Yang Z., Qiu X., et al. miR-29c contribute to glioma cells temozolomide sensitivity by targeting O6-methylguanine-DNA methyltransferases indirectely. Oncotarget. 2016;7(31):50229–50238. doi: 10.18632/oncotarget.10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ng W. L., Yan D., Zhang X., Mo Y. Y., Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 2010;9(11):1170–1175. doi: 10.1016/j.dnarep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 165.Yoo B. C., Kim K. H., Woo S. M., Myung J. K. Clinical multi-omics strategies for the effective cancer management. Journal of Proteomics. 2017 doi: 10.1016/j.jprot.2017.08.010. in press. [DOI] [PubMed] [Google Scholar]
- 166.Uhm J. H., Porter A. B. Treatment of glioma in the 21st century: an exciting decade of postsurgical treatment advances in the molecular era. Mayo Clinic Proceedings. 2017;92(6):995–1004. doi: 10.1016/j.mayocp.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 167.Mangani D., Weller M., Roth P. The network of immunosuppressive pathways in glioblastoma. Biochemical Pharmacology. 2017;130:1–9. doi: 10.1016/j.bcp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 168.Polivka J., Jr., Polivka J., Holubec L., et al. Advances in experimental targeted therapy and immunotherapy for patients with glioblastoma multiforme. Anticancer Research. 2017;37(1):21–33. doi: 10.21873/anticanres.11285. [DOI] [PubMed] [Google Scholar]
- 169.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 170.Hammond S. M., Bernstein E., Beach D., Hannon G. J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 171.Shrivastava N., Srivastava A. RNA interference: an emerging generation of biologicals. Biotechnology Journal. 2008;3(3):339–353. doi: 10.1002/biot.200700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Paddison P. J. RNA interference in mammalian cell systems. Current Topics in Microbiology and Immunology. 2008;320:1–19. doi: 10.1007/978-3-540-75157-1_1. [DOI] [PubMed] [Google Scholar]
- 173.Karim R., Palazzo C., Evrard B., Piel G. Nanocarriers for the treatment of glioblastoma multiforme: current state-of-the-art. Journal of Controlled Release. 2016;227:23–37. doi: 10.1016/j.jconrel.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 174.Stupp R., Taillibert S., Kanner A. A., et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. Journal of the American Medical Association. 2015;314(23):2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 175.Pardridge W. M. Drug and gene delivery to the brain: the vascular route. Neuron. 2002;36(4):555–558. doi: 10.1016/S0896-6273(02)01054-1. [DOI] [PubMed] [Google Scholar]
- 176.Lu W., Zhang Y., Tan Y. Z., Hu K. L., Jiang X. G., Fu S. K. Cationic albumin-conjugated pegylated nanoparticles as novel drug carrier for brain delivery. Journal of Controlled Release. 2005;107(3):428–448. doi: 10.1016/j.jconrel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 177.Chen Y., Liu L. Modern methods for delivery of drugs across the blood-brain barrier. Advanced Drug Delivery Reviews. 2012;64(7):640–665. doi: 10.1016/j.addr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 178.Pakunlu R. I., Wang Y., Saad M., Khandare J. J., Starovoytov V., Minko T. In vitro and in vivo intracellular liposomal delivery of antisense oligonucleotides and anticancer drug. Journal of Controlled Release. 2006;114(2):153–162. doi: 10.1016/j.jconrel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 179.Chang H. I., Yeh M. K. Clinical development of liposome-based drugs: formulation, characterization, and therapeutic efficacy. International Journal of Nanomedicine. 2012;7:49–60. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Charest G., Sanche L., Fortin D., Mathieu D., Paquette B. Glioblastoma treatment: bypassing the toxicity of platinum compounds by using liposomal formulation and increasing treatment efficiency with concomitant radiotherapy. International Journal of Radiation Oncology, Biology, Physics. 2012;84(1):244–249. doi: 10.1016/j.ijrobp.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koukourakis M. I., Koukouraki S., Fezoulidis I., et al. High intratumoural accumulation of stealth liposomal doxorubicin (Caelyx) in glioblastomas and in metastatic brain tumours. British Journal of Cancer. 2000;83(10):1281–1286. doi: 10.1054/bjoc.2000.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sengupta S., Thaci B., Crawford A. C., Sampath P. Interleukin-13 receptor alpha 2-targeted glioblastoma immunotherapy. BioMed Research International. 2014;2014:8. doi: 10.1155/2014/952128.952128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang Y., Jeong Lee H., Boado R. J., Pardridge W. M. Receptor-mediated delivery of an antisense gene to human brain cancer cells. The Journal of Gene Medicine. 2002;4(2):183–194. doi: 10.1002/jgm.255. [DOI] [PubMed] [Google Scholar]
- 184.Ambruosi A., Gelperina S., Khalansky A., Tanski S., Theisen A., Kreuter J. Influence of surfactants, polymer and doxorubicin loading on the anti-tumour effect of poly(butyl cyanoacrylate) nanoparticles in a rat glioma model. Journal of Microencapsulation. 2006;23(5):582–592. doi: 10.1080/02652040600788080. [DOI] [PubMed] [Google Scholar]
- 185.Liu Y., Lu W. Recent advances in brain tumor-targeted nano-drug delivery systems. Expert Opinion on Drug Delivery. 2012;9(6):671–686. doi: 10.1517/17425247.2012.682726. [DOI] [PubMed] [Google Scholar]
- 186.Tosi G., Costantino L., Ruozi B., Forni F., Vandelli M. A. Polymeric nanoparticles for the drug delivery to the central nervous system. Expert Opinion on Drug Delivery. 2008;5(2):155–174. doi: 10.1517/17425247.5.2.155. [DOI] [PubMed] [Google Scholar]
- 187.Allard E., Huynh N. T., Vessieres A., et al. Dose effect activity of ferrocifen-loaded lipid nanocapsules on a 9L-glioma model. International Journal of Pharmaceutics. 2009;379(2):317–323. doi: 10.1016/j.ijpharm.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 188.Allard E., Passirani C., Garcion E., et al. Lipid nanocapsules loaded with an organometallic tamoxifen derivative as a novel drug-carrier system for experimental malignant gliomas. Journal of Controlled Release. 2008;130(2):146–153. doi: 10.1016/j.jconrel.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 189.Laine A. L., Clavreul A., Rousseau A., et al. Inhibition of ectopic glioma tumor growth by a potent ferrocenyl drug loaded into stealth lipid nanocapsules. Nanomedicine. 2014;10(8):1667–1677. doi: 10.1016/j.nano.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 190.Bhabra G., Sood A., Fisher B., et al. Nanoparticles can cause DNA damage across a cellular barrier. Nature Nanotechnology. 2009;4(12):876–883. doi: 10.1038/nnano.2009.313. [DOI] [PubMed] [Google Scholar]
- 191.Gou N., Onnis-Hayden A., Gu A. Z. Mechanistic toxicity assessment of nanomaterials by whole-cell-array stress genes expression analysis. Environmental Science & Technology. 2010;44(15):5964–5970. doi: 10.1021/es100679f. [DOI] [PubMed] [Google Scholar]
- 192.Halle B., Marcusson E. G., Aaberg-Jessen C., et al. Convection-enhanced delivery of an anti-miR is well-tolerated, preserves anti-miR stability and causes efficient target de-repression: a proof of concept. Journal of Neuro-Oncology. 2016;126(1):47–55. doi: 10.1007/s11060-015-1947-2. [DOI] [PubMed] [Google Scholar]
- 193.Ganju A., Khan S., Hafeez B. B., et al. miRNA nanotherapeutics for cancer. Drug Discovery Today. 2017;22(2):424–432. doi: 10.1016/j.drudis.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Goren D., Horowitz A. T., Tzemach D., Tarshish M., Zalipsky S., Gabizon A. Nuclear delivery of doxorubicin via folate-targeted liposomes with bypass of multidrug-resistance efflux pump. Clinical Cancer Research. 2000;6(5):1949–1957. [PubMed] [Google Scholar]
- 195.Lee T. J., Yoo J. Y., Shu D., et al. RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Molecular Therapy. 2017;25(7):1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ananta J. S., Paulmurugan R., Massoud T. F. Nanoparticle-delivered antisense microRNA-21 enhances the effects of temozolomide on glioblastoma cells. Molecular Pharmaceutics. 2015;12(12):4509–4517. doi: 10.1021/acs.molpharmaceut.5b00694. [DOI] [PubMed] [Google Scholar]
- 197.Bertucci A., Prasetyanto E. A., Septiadi D., et al. Combined delivery of temozolomide and anti-miR221 PNA using mesoporous silica nanoparticles induces apoptosis in resistant glioma cells. Small. 2015;11(42):5687–5695. doi: 10.1002/smll.201500540. [DOI] [PubMed] [Google Scholar]
- 198.Shatsberg Z., Zhang X., Ofek P., et al. Functionalized nanogels carrying an anticancer microRNA for glioblastoma therapy. Journal of Controlled Release. 2016;239:159–168. doi: 10.1016/j.jconrel.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 199.Amano S., Li S., Gu C., et al. Use of genetically engineered bone marrow-derived mesenchymal stem cells for glioma gene therapy. International Journal of Oncology. 2009;35(6):1265–1270. doi: 10.3892/ijo_00000443. [DOI] [PubMed] [Google Scholar]
- 200.Ahmed A. U., Alexiades N. G., Lesniak M. S. The use of neural stem cells in cancer gene therapy: predicting the path to the clinic. Current Opinion in Molecular Therapeutics. 2010;12(5):546–552. [PMC free article] [PubMed] [Google Scholar]
- 201.Lee H. K., Finniss S., Cazacu S., Xiang C., Brodie C. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells and Development. 2014;23(23):2851–2861. doi: 10.1089/scd.2014.0146. [DOI] [PubMed] [Google Scholar]
- 202.Teplyuk N. M., Uhlmann E. J., Gabriely G., et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: first steps toward the clinic. EMBO Molecular Medicine. 2016;8(3):268–287. doi: 10.15252/emmm.201505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ebert M. S., Sharp P. A. MicroRNA sponges: progress and possibilities. RNA. 2010;16(11):2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen L., Zhang K., Shi Z., et al. A lentivirus-mediated miR-23b sponge diminishes the malignant phenotype of glioma cells in vitro and in vivo. Oncology Reports. 2014;31(4):1573–1580. doi: 10.3892/or.2014.3012. [DOI] [PubMed] [Google Scholar]
- 205.Hansen T. B., Jensen T. I., Clausen B. H., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 206.Cai K., Wan Y., Sun G., Shi L., Bao X., Wang Z. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncology Reports. 2012;28(6):2101–2106. doi: 10.3892/or.2012.2027. [DOI] [PubMed] [Google Scholar]
- 207.Wang X. R., Luo H., Li H. L., et al. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro-Oncology. 2013;15(11):1491–1501. doi: 10.1093/neuonc/not107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yu L., Lu J., Zhang B., et al. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncology Letters. 2013;5(4):1223–1228. doi: 10.3892/ol.2013.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Smits M., Nilsson J., Mir S. E., et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1(8):710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xiaoping L., Zhibin Y., Wenjuan L., et al. CPEB1, a histone-modified hypomethylated gene, is regulated by miR-101 and involved in cell senescence in glioma. Cell Death & Disease. 2013;4, article e675 doi: 10.1038/cddis.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zheng F., Liao Y. J., Cai M. Y., et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61(2):278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 212.Qiu S., Huang D., Yin D., et al. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2013;1832(10):1697–1707. doi: 10.1016/j.bbadis.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 213.Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Molecular Cell. 2009;36(1):61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Guo P., Lan J., Ge J., Nie Q., Mao Q., Qiu Y. miR-708 acts as a tumor suppressor in human glioblastoma cells. Oncology Reports. 2013;30(2):870–876. doi: 10.3892/or.2013.2526. [DOI] [PubMed] [Google Scholar]