Abstract
Macrophages constitute a heterogeneous population of myeloid cells that are essential for maintaining homeostasis and as a first line of innate responders controlling and organizing host defenses against pathogens. Monocyte–macrophage lineage cells are among the most functionally diverse and plastic cells of the immune system. They undergo specific activation into functionally distinct phenotypes in response to immune signals and microbial products. In mammals, macrophage functional heterogeneity is defined by two activation states, M1 and M2, which represent two polar ends of a continuum exhibiting pro-inflammatory and tissue repair activities, respectively. While the ancient evolutionary origin of macrophages as phagocytic defenders is well established, the evolutionary roots of the specialized division of macrophages into subsets with polarized activation phenotypes is less well defined. Accordingly, this chapter focuses on recent advances in the understanding of the evolution of macrophage polarization and functional heterogeneity with a focus on ectothermic vertebrates.
1.1 Introduction
Monocyte–macrophage lineage cells are found across all vertebrate species and play critical roles in homeostasis, wound healing, and immune responses. In addition to providing a first line of defense against pathogens, macrophages also undergo molecular reprogramming in response to microbial-, environmental-, and immune-derived signals that influence their subsequent interactions with various lymphocyte subsets [reviewed in (Biswas and Mantovani 2010; Mantovani et al. 2002; Mosser and Edwards 2008)]. This ability of macrophages to impact the course of immune responses is, in a large part, due to the inherent and adaptable plasticity of these cells. Indeed, monocyte–macrophage lineage cells have long been recognized as highly plastic with polarized populations that differ in terms of effector functions, cell surface receptor expression, and cytokine production [reviewed in (Italiani and Boraschi 2014)]. Broadly speaking, two main types of activation or polarization states are defined in mammals: a classically activated/inflammatory (M1) type macrophage and an alternatively activated/regenerative (M2)-type macrophage (Gordon and Martinez 2010; Gordon and Taylor 2005; Mills 2012). From an evolutionary perspective, it is unclear how far this definition of macrophage function stretches outside mammals. However, there are multiple lines of evidence of functional diversification of macrophages in ectothermic vertebrates (Bystrom et al. 2008; Grayfer et al. 2014c; Grayfer and Robert 2014 2015; McKinney et al. 1986; Rieger et al. 2010). While these functional variations may not strictly adhere to the mammalian paradigms, they imply that the ability of macrophages to adapt their roles to species-specific physiological cues is an evolutionary ancient trait.
Importantly, while the high plasticity of macrophages allows them to modify and reprogram their effector functions in response to immune stimuli, this functional malleability can also be manipulated by invading pathogens. Although macrophage adaptability provides a selective advantage in host resistance to pathogen, this same plasticity can sometimes be exploited and subverted by pathogens to invade the host. Indeed, in mammals, it is not infrequent in an infectious disease setting to find pathology associated with dynamic changes in macrophage activation, where M1 macrophages are associated with initiating and sustaining inflammation and M2 macrophages are associated with either resolution or chronic infection (Cassetta et al. 2011; Herbein and Varin 2010; Labonte et al. 2014; Sang et al. 2015; Shaked et al. 2014). Interestingly, in mouse models, the polarized M1/M2 phenotypes can, to some extent, be experimentally reversed in vitro and in vivo, which makes macrophages interesting targets for immune-modulation and therapeutic applications (Guiducci et al. 2005; Saccani et al. 2006).
As the embryonic origin, lineage commitment and monopoiesis are considered in another chapter of this book, and amphibian myelopoiesis has recently been discussed in detail in (Grayfer and Robert 2016), this chapter will focus on the current understanding of the functional diversification potential of macrophages from an evolutionary perspective. We will particularly discuss comparative approaches aimed at defining macrophage plasticity in genetically distant species in bony fish and amphibians.
1.2 Macrophage Polarization in Mammals
The original definition of mammalian M1 and M2 macrophages is derived from the Th1 and Th2 cytokines associated with their respective polarization and stem from the observation that macrophage activated with either the cytokine interferon gamma (IFN-γ) or lipopolysaccharides (LPS) in mouse strains with T helper type 1 (Th1) or T helper type 2 (Th2) backgrounds differed in their arginine metabolism (Mills et al. 2000). M1-type macrophages utilize inducible nitric oxide synthase (iNOS) to convert l-arginine to l-citruline and nitric oxide (NO) whereas M2 macrophages utilize arginase to convert l-arginine to l-ornithine, which is a precursor for polyamines and proline components of collagen, an important component in tissue repair (Mills 2001). For a detailed review of the immunobiology and regulation of iNOS, see the chapter by Lee at al. Notably, byproducts derived from either the iNOS or arginase pathways inhibit the reciprocal enzymes, thus stabilizing the M1 or M2 macrophage polarization states, respectively (Morris 2009; Munder et al. 1999). In mammals, classical M1 activation is induced by intracellular pathogens, bacterial cell wall components, and hallmark Th1 cytokines such as IFN-γ. This activation results in: (i) the production of an array of pro-inflammatory mediators including interleukin-12 (IL-12), interleukin 1 beta (IL-1β), tumor necrosis factor alpha (TNFα), interleukin 18 (IL-18), and interleukin 23 (IL-23); (ii) the production of reactive oxygen intermediates (ROI) and nitric oxide synthase-2 (NOS-2/iNOS)-dependent reactive nitrogen intermediates; and (iii) high antigen presenting activity resulting in an effective pathogen-killing phenotype. Comparably, M2 macrophage activity, originally defined as an alternative pathway of macrophage activation characterized by increased mannose receptor activity, is induced by interleukin 4 (IL-4) (Stein et al. 1992). More recent studies have shown that in addition to the prototypical Th2 cytokines IL-4 and IL-13, M2-like activation can be further amplified by fungal pathogens, parasites, immune complexes, components of the complement system (i.e., small proteins that normally circulate as inactive components but can be activated in response to pathogens), apoptotic cells, interleukin 10 (IL-10), and transforming growth factor (TGF-β). M2 macrophages are highly phagocytic, produce immunosuppressive cytokines such as TGF-β and IL-10. These cytokines establish a positive feedback loop that increase M2 macrophage polarization and typically facilitate the resolution of inflammation. However, M2 macrophages can also be reservoirs for intracellular pathogens facilitating chronic infection, aid in the growth of tumors, and cause allergic inflammation (Sica and Mantovani 2012).
The M1/M2 macrophage activation paradigm originally proposed to reflect the Th1 and Th2 nomenclature still requires frequent updates and fine-tuning to take into account the wide functional plasticity and high heterogeneity of macrophage populations as well as to integrate novel findings. One should realize that this M1/M2 paradigm similar to the now revised Th1/Th2 paradigm (including Treg, Th17, Th22, Th9, and TFH cells) represents two polar ends of a spectrum that may not fully take into account all the different activation scenarios. A uniform terminology based on both the tissue source of macrophages and the activation stimuli have recently been proposed (Murray et al. 2014). For example, M2 macrophage responses, depending on the stimuli has lead to the characterization of several M2 subtypes including: M2a (induced by exposure to IL-4 and IL-13); M2b (macrophages activated by immune complexes, Toll-like receptors (TLRs), or apoptotic cells); and M2c (macrophages deactivated by glucocorticoids, TGF-β or IL-10) (Biswas and Mantovani 2010; Mantovani et al. 2004; Martinez and Gordon 2014). While M2a and M2b macrophages drive Th2 responses, M2c cells are primarily involved in immune suppression and tissue remodeling (Murray et al. 2014). It is important to remember that M1/M2 macrophage polarization in vivo is not absolute and likely reflects a less straightforward and more complex process. Indeed, the list of macrophage subpopulations is still growing and additional subsets of monocyte-derived macrophages are actively investigated including tumor-associated macrophages (TAM), CD169+ macrophages, and most recently TCR+ macrophages (Allavena et al. 2008a,b; Mantovani et al. 2002; Sica et al. 2002) and [reviewed in (Chavez-Galan et al. 2015)].
An additional challenge in defining activated macrophage subsets comes from species-specific variations. While relatively consistent molecular signatures is found between human and mouse macrophage cell lines, there is a variability in markers used to define M1 and M2 polarization between the two species in vivo (Martinez et al. 2013). For instance, there are no human homologs of the mouse M2 marker chitinase-3-like protein 3 (Chi313), also known as Ym1, highlighting the mterspecies differences between human and murine macrophages (Raes et al. 2005; Scotton et al. 2005). Nevertheless, the M1/M2 model provides a convenient framework, even though it is probably more accurate to view macrophage polarization as a continuum with overlapping cell surface expressions, cytokine secretions, and transcriptional regulators. In this context, studies aimed at elucidating the functional diversification of macrophages and the mechanisms underlying their activation in other evolutionary distant species may reveal useful understanding how macrophages adapt their function in responses to physiological and microbial cues.
1.3 Evolutionary Conservation of M1/M2-Like Functional Heterogeneity
Phagocytic cells with monocytic morphology are among the most ancestral immune-like cell types existing. We should not forget that the very first description of a macrophage by Élie Metchnikoff in 1905 was from the larvae of starfish, an echinoderm belonging to the class asteroida, a sea invertebrate (Metchnikoff 1905). However, while the evolutionary ancient origin of macrophages is well established, the origin and possible mechanisms governing macrophage functional heterogeneity in nonmammalian species is less well defined. Invertebrates such as insects, nematodes, echinoderms, mollusks, tunicates, or sponges possess numerous types of phagocytic leucocytes, collectively named immunocytes [i.e., hemocyte, coelomocyte, amebocyte, and plasmatocyte, reviewed in (Ottaviani 2011)]. These cells can respond to foreign material by secreting a variety of biologically active pathogen-binding and pathogen-killing substances including NO, reactive oxygen species, and hydrolytic enzymes (Dzik 2014; Franchini et al. 1995; Ottaviani and Franceschi 1997). Such a response is reminiscent of an M1-like phenotype. Conversely, invertebrate immunocytes can also exhibit some M2-like traits including wound-healing activity and to some extent involvement in tissue metabolism. For example, following activation, in the grey slug (Limax maximus), hemocytes have been shown to change their phenotype and transition into a collagen-fibrobast-like phenotype (Franchini and Ottaviani 2000). In addition, gene orthologs encoding some M2 markers such as mannose receptor proteins, chitinase-like proteins, and glycoproteins that share homology with Ym1 are also found in invertebrate phagocytes, which suggests that hemocytes are involved in wound healing and extracellular matrix synthesis (Badariotti et al. 2007; Kirkpatrick et al. 1995; Sricharoen et al. 2005). However, as stated above, microbial infections elicit both M1-like and M2-like transcriptional changes in invertebrate hemocytes, which implies that a strict mammalian type M1/M2 separation is probably lacking in invertebrates [reviewed in (Roszer 2015)].
Besides prototypical M1 and M2 functions, macrophage specialization into mammalian M1 - and M2 activation states can, at least partly, be inferred by tracing the molecular evolution of important receptors and effector molecules. As previously mentioned, the mammalian M1/M2 dichotomy is related to the cellular use of arginine (Mills et al. 2000). This amino acid can, through the activity of arginase, be converted into ornithine or alternatively be converted into nitric oxide by the activity of iNOS. The latter reaction makes the macrophage capable of killing invading pathogens, whereas the former is more tuned to tissue repair. Thus, the origin of macrophage specialization into mammalian M1- and M2 types can be considered to stem from two ancient molecular mechanisms: the cytotoxic activity of iNOS and the healing functions of arginase (Dzik 2014). While nitric oxide synthase functions similarly in invertebrates as in vertebrates, nitric oxide primarily serves as a signaling rather than a cytotoxic molecule in invertebrates. Hence, it has been argued that the ability of M1-like macrophages to produce large amounts of nitric oxide in response to microbial infections has emerged with vertebrates. Comparably arginase-1, which since the initial discovery of M2 macrophage activation is considered a prototypic murine M2 marker (Stempin et al. 2010), is ubiquitously found in prokaryote and eukaryotes. From an evolutionary perspective, arginase-1 has been proposed to have primarily been a wound healing protein, transcriptionally induced by proteins of the TGF family (Dzik 2014). However, most microorganisms and invertebrates possess a single arginase gene not involved in the ornithine–urea cycle (Samson 2000). In contrast, two genes encoding distinct, arginase isoforms have been identified in mammals: the cytosolic arginase-1 that is induced by IL-4 and IL-13 and the mitochondrial-associated arginase-2 that is upregulated by IL-10 and LPS (Lang et al. 2002; Munder et al. 1999). Moreover, two arginase genes have been described amphibians (Patterton and Shi 1994) and bony fish (Wright et al. 2004). Thus, based on molecular evidence, arginase, one of the key components driving the molecular mechanisms involved in macrophage polarization, can be traced as far back as bony fish, suggesting that M1/M2-like macrophage specialization arose during early vertebrate evolution.
1.3.1 Bony Fish
In bony fish, the best characterized macrophage phenotype is reminiscent of the mammalian pro-inflammatory M1 state [reviewed in (Hodgkinson et al. 2015)]. Studies have demonstrated that following exposure to various activation stimuli, bony fish macrophages display increased phagocytosis, increased production of reactive oxygen intermediates, elevated expression of inducible nitric oxide synthase (iNOS/NOS2), phagolysosomal acidification, and nutrient deprivation (Grayfer et al. 2014c; Neumann et al. 2000; Rieger et al. 2010). In addition, bony fish macrophages respond as mammalian macrophages to M1-inducing stimuli and microbial challenge by upregulating a prototypic M1 cytokine repertoire including TNFα, IL-lβ, interleukin 6 (IL-6), IL-12, interleukin 15 (IL-15), and IL-23 (Arts et al. 2010; Joerink et al. 2006a; Wang and Secombes 2013). While the classical M1-like activation of bony fish macrophages have been relatively well described, the functional occurrence of alternatively activated/M2 macrophages has proven more challenging to define. Efforts to characterize M2-like macrophages in fish have largely focused on the biology of typical M2 stimuli, the best characterized of which being interleukin IL-4 and IL-13. To date, two genes have been identified in bony fish that share homology with both the mammalian IL-4 and IL-13 cytokines (IL-4/13A and IL-4/13B) (Ohtani et al. 2008; Wang et al. 2016). Using a rainbow trout (Oncorhynchus mykiss) in vitro head kidney leucocyte culture system (in fish, myelopoiesis occurs in the head kidney), recombinant forms of both IL-4/13A and IL-4/13B were shown to be anti-inflammatory via upregulation of IL-10 and downregulation of IL-1β and IFN-γ (Wang et al. 2016). However, whether or not these cytokines directly act to polarize macrophages towards an M2-like state remains to be determined.
As previously stated, another important hallmark of M2 activation is arginase activity. Using an in vitro macrophage model in the common carp (Cyprinus carpio L.), it was demonstrated that macrophages derived from head kidney leukocytes could be polarized into two different states by incubation with either dibutyryl cyclic adenosine mono phosphate (cAMP) or LPS. In this system, cAMP treatment resulted in highly upregulated arginase-2 gene expression, indicative of an M2-like phenotype (Joerink et al. 2006b,c). This is intriguing since cAMP stimulation has been shown to transform human M1 macrophages into resolution-phase macrophages (rMs) (Bystrom et al. 2008). These rMs exhibit an M2-like phenotype but with elevated M1 cell markers such as nitric oxide synthase (iNOS) and, thus, are neither classical M1 nor M2 macrophages. Rather, rMs Have a role in restoring tissue homeostasis following resolution of a inflammatory response. Finally, glucocorticoids, immune complexes, and IL-10 have been shown to be immune-suppressive on bony fish macrophages.
More recently, M1-like and M2-like polarization was demonstrated in vivo using a transgenic zebrafish (Danio rerio) larval model (Nguyen-Chi et al. 2015). This model uses TNFα gene expression as a marker for M1 macrophages and takes advantage of double transgenic fish lines in which macrophages express mCherry under the control of the macrophage-specific mpeg1 promoter and eGPF under the control of the tnfa promoter Tg (mpeg1 :mCherryF/tnfa:eGFP-F) in combination with wound-induced inflammation. By examining the gene expression profiles of tnfa− and tnfa+ macrophage populations isolated during early and late phases of inflammation, well-known markers of mammalian M1 macrophages including TNFα, IL1-β, and IL-6 were expressed in the tnfa+ macrophages, whereas tnfa− macrophages expressed TGF-β, C-C chemokine receptor type 2 (CCR2), and C-X-C chemokine receptor type 4 (CXCR4), which are markers of M2 activation in mammals (Beider et al. 2014; Hao et al. 2012; Mantovani et al. 2002; Martinez et al. 2006). Of note, neither Arginase 1 (Arg1) nor IL-10 expression was detected in zebrafish tnfa− macrophages. Collectively, these observations strongly argue for an evolutionary conservation of both M1-like and M2-like activation states in zebrafish. However, it is important to remember that both similarities and differences have been documented for the regulation of bony fish macrophage antimicrobial defenses, as compared to what has been described in mammals.
1.3.2 Amphibians
Anuran amphibians like Xenopus present the peculiarity of developmentally distinct macrophage populations in tadpoles and adults frogs, which can be further polarized into distinct functional subsets. Tadpole and adult macrophages elicited by intraperitoneal injection with heat-killed E. coli, and subsequently allowed to adhere in vitro in culture plates, are morphologically distinct. Compared to adult macrophages, a subset of tadpole macrophages is enlarged with a highly vacuolated morphology (Grayfer et al. 2012). Although hematopoiesis in Xenopus is localized to the subcapsular liver, macrophage precursors in X. laevis, unlike other vertebrates, are derived from the bone marrow [(Grayfer and Robert 2013) and reviewed in (Grayfer and Robert 2016)]. Since the bone marrow is absent in tadpoles, it is currently unknown how tadpole macrophage development and differentiation occurs.
Although M1/M2 is not fully defined in Xenopus, there is clear evidence in both tadpole and adult X. laevis of functional specialization driven by the two different colony stimulating factor-1 receptor (CSF-1R) ligands: colony-stimulating factor-1 (CSF-1) and interleukin-34 (IL-34) (Grayfer and Robert 2014, 2015). Across most vertebrate species, the survival, proliferation, differentiation, and functionality of cells of the monocytic lineage require binding of CSF-1 to its receptor CSF-1R (Dai et al. 2002), which is a marker for committed myeloid precursors and phagocyte populations (Guilbert and Stanley 1980; Lichanska et al. 1999; Tagoh et al. 2002). Although CSF-1 and CSF-1R genes display relatively low sequence conservation across evolutionary distant species, important hallmark sequence features as well as the intracellular signal-binding sites are highly conserved suggesting a shared function [(Garceau et al. 2010; Grayfer et al. 2014b; Grayfer and Robert 2013; Hanington et al. 2007; Wang et al. 2008) and reviewed in (Grayfer and Robert 2016)]. In support of the conserved function of CSF-1R, adult X. laevis bone marrow cells have been shown to respond to X. laevis recombinant CSF-1 and differentiate into primarily large mononuclear cells with characteristic macrophage morphology (Grayfer and Robert 2013). Similarly, bony fish CSF-1 is a strong inducer of macrophage growth and differentiation (Grayfer et al. 2009; Hanington and Belosevic 2007). Interestingly, the interleukin-34 (IL-34) cytokine, which is found across evolutionary distant species also binds to and activates CSF-1R in mammals resulting in the initiation of distinct biological activity and signal activation (Chihara et al. 2010; Liu et al. 2012; Wei et al. 2010). It is becoming increasingly evident that in X. laevis, the two types of macrophages elicited by either CSF-1 or IL-34 cytokines have distinct polarizing roles during infections both in tadpoles (Grayfer and Robert 2014) and adult frogs (Grayfer and Robert 2015) as discussed in the next section.
1.3.2.1 Macrophage Polarization During X. laevis Antiviral Responses
There is ample evidence implicating amphibian macrophages as important components in evasion, persistence, and dissemination during infections with the ranavirus, frog virus 3 (FV3), which is a large pox-like icosahedral dsDNA virus reviewed in (Chinchar et al. 2011; Grayfer et al. 2012) and more recently in (Grayfer and Robert 2016)]. Recent findings indicate that FV3 infects and likely uses macrophages to persist and disseminate throughout the host, suggesting that macrophage functions are modulated by the virus (Morales et al. 2010; Robert et al. 2007). In general, tadpoles are more susceptible and succumb to FV3 infections within 1–2 months, whereas adults are more resistant and typically clear the infection within a few weeks (Bayley et al. 2013; De Jesus Andino et al. 2012; Grayfer et al. 2014a; Hoverman et al. 2010).
Notably, the administration of X. laevis recombinant CSF-1 increases the susceptibility of X. laevis tadpole to FV3 infections (Grayfer and Robert 2014). In contrast; recombinant IL-34 increases tadpole resistance to FV3 infection (Grayfer and Robert 2014). As previously suggested (Grayfer and Robert 2014, 2015) this is possibly due to a CSF/IL-34-mediated expansion and biased polarization towards macrophage subpopulations with distinct antimicrobial properties. Consistent with these observations, in vitro studies have demonstrated that IL-34-elicited peritoneal macrophages express higher baseline levels of type I interferon, which following FV3 infection was further enhanced compared to CSF-elicited peritoneal macrophages (Grayfer and Robert 2014). IL-34-elicited peritoneal macrophages also express elevated levels of the NADPH oxidase components p67phox and gp91phox and display an increase in the major histocompatibility complex class I and II as well as β2-microglobulin gene expression. This is consistent with an enhanced antigen presentation capacity, which would contribute to enhance T cell-mediated anti-viral response (Grayfer and Robert 2014).
Similar studies focused on adult peritoneal macrophages have confirmed that X. laevis possess functionally distinct macrophage populations (Grayfer and Robert 2015). Similar to tadpoles, IL-34-elicited adult peritoneal macrophages exhibit more potent antiviral activity against FV3 than CSF-elicited macrophages, which is indicated by a higher gene expression of NADPH oxidases components, a greater respiratory burst response (i.e., release of reactive oxygen species) and reactive oxygen production (Grayfer and Robert 2015) All these features are consistent with a M1-like phenotype. Contrasting with the classical M1 mammalian definition, however, IL-34 elicited macrophages also possess an elevated arginase-1 gene expression, which in mammals and fish is typically associated with the alternatively activated M2 phenotype. Characteristics of CSF-1- and IL-34-elicited X. laevis macrophage are summarized in Table 1.1.
Table 1.1.
Characteristics of CSF-1- and IL-34-derived X. laevis peritoneal macrophages
| Effector molecules expressed | Increased effector functions | Proposed function | References | |
|---|---|---|---|---|
| CSF-1-derived | iNOSa | Phagocytosis, NO production Microbicidal activity | Antibacterial | Grayfer and Robert (2014, 2015) |
| IL-34-derived | Arginase-l NADPH oxidase Type I IFNb | ROS production Antiviral activity | Antiviral | Grayfer and Robert (2014, 2015) |
Significantly higher iNOS expression in CSF-1-derived than IL-34-derived adult peritoneal macrophages
Significantly higher Type I IFN gene expression response in tadpole but not adult IL-34- derived peritoneal macrophages compared to respective CSP-derived peritoneal macrophages
1.3.2.2 Macrophage Polarization During X. laevis Antibacterial Responses
As in the case of viral pathogens, macrophages that are essential immune effector cells are also used by certain intracellular bacteria to survive and thrive. An example in humans highlighting this paradoxical role of macrophages is Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB). Mtb preferentially infects macrophages, replicates and persists inside the cell, and causes chronic infections. A hallmark feature of the immune response to Mtb infections is the formation of compact aggregates or granulomas primarily made up of blood-derived macrophages and epithelioid cells (i.e., activated macrophages that often merge together via tightly interdigitated cell membranes that link adjacent cells) surrounded by additional immune cells including neutrophils, dendritic cells, T cells, and B cells [reviewed in (Silva Miranda et al. 2012)]. It has been postulated that granulomas are formed in an attempt to wall off and sequester Mtb. However, granulomas also provide mycobacteria with a niche in which it can modulate the immune response, survive over long periods of time, and under certain conditions reactivate and cause clinic disease (Adams 1976; Martin et al. 2016). This ability of Mtb to survive inside macrophages is postulated to result in part from the Mtbs ability to subvert pro-inflammatory M1 functions (Cronan and Tobin 2014; Huang et al. 2015; Marino et al. 2015) To understand how Mtb can modulate macrophage polarization and functions especially during early stage of granuloma formations, nonmammalian alternative animal models have proven valuable (O’Toole 2010). For example, important insight into the dynamic structures of granulomas has been gathered from studies using Mycobacterium marinum (Mm) infected zebrafish models (Cronan and Tobin 2014; Davis et al. 2002; Davis and Ramakrishnan 2009; Meijer 2016; Meijer et al. 2005; Stinear et al. 2008). Mm is a natural pathogen of ectothermic vertebrates causing a systemic disease with formation of macrophage aggregates and containment of bacteria in granulomas that show strong similarity with human TB granulomas. Studies in zebrafish not only confirmed the importance of macrophage-mediated phagocytosis and initial pro-inflammatory signaling in controlling Mm growth, but also provided a novel insight into host macrophage–mycobacteria interaction (Cronan and Tobin 2014; Davis et al. 2002; Davis and Ramakrishnan 2009; Meijer et al. 2005). Notably early granuloma formation, which was thought to be a relatively static host defense mechanism, was shown in zebrafish to be a dynamic structure actively utilized by Mm to, via the Mm virulence factor RD-1, recruit nascent macrophages to the site of the primary granuloma where they phagocytose the bacillus (Davis and Ramakrishnan 2009). Subsequently newly infected macrophages depart the primary granuloma, initiate new granulomas at distal locations, and thus facilitate Mm dissemination (Cronan and Tobin 2014; Davis et al. 2002; Davis and Ramakrishnan 2009; Meijer et al. 2005). In addition, using the zebrafish model, it has been suggested that Mm manipulate macrophage recruitment to preferentially recruit a subpopulation of iNOS-deficient macrophages. This would selectively enhance Mm phagocytosis by macrophages with reduced bactericidal activity and thus provide a niche for bacterial growth (Cambier et al. 2014).
Moreover, genetically engineered X. laevis transgenic lines expressing fluorescent reporter genes in macrophages are likely to prove useful for further in vivo characterization of macrophage activity and polarization during a complex infection in living animals. In addition, unlike zebrafish, T cells develop within 2 weeks of age in X. laevis tadpoles. Furthermore, the majority of these T cells are so-called innate T cells expressing semi-invariant T cell receptors that are restricted by MHC class-I-like molecules [reviewed in (Edholm et al. 2014; Robert and Edholm 2014)]. Specifically, deep-sequencing has revealed that 80% of CD8 intermediate and CD8 negative T cells express 6 invariant TCRα rearrangements (Edholm et al. 2013). The polarization potential of one or more of these iT cell population on macrophages open new avenues of investigation. Indeed, preliminary evidence using loss-of-function reverse genetic combined with transgenesis has identified an iT cell subset critical for anti-Mm host resistance (i.e., transgenic tadpoles lacking this iT cell population are more susceptible to Mm infection; Edholm, Rhoo and Robert, unpublished).
As in zebrafish, several transgenic lines with different subsets of myeloid lineages labeled with different colored fluorescent proteins have been characterized (Paredes et al. 2015). These lines include: a xLurp1:EGFP Tg line in which myeloid cells (e.g., granulocytes and monocytic leukocytes) express EGFP under the xLurp (Ly-6/uPAR-related protein) promoter (Paredes et al. 2015; Smith et al. 2002); a xmpeg:mCherry Tg line in which only mononuclear phagocytes (i.e., mainly macrophages) express the red fluorescence label mCherry under the zebrafish macrophage-specific mpeg promoter (Ellett et al. 2011); and a double mpeg1: mCherry and xlurp1:GFP line (Paredes et al. 2015).
Using this double transgenic line in combination with fluorescently labeled Mm, the diversity and plasticity of X. laevis macrophage subset during infection has begun to be further examined in vivo in adult and tadpoles (Fig. 1.1). Using intravital imaging, it is possible to visualize in real time the migration, accumulation. and granulomatous formation in response to Mm infection (Rhoo and Robert, unpublished data). With regard to IL-34/CSF driven macrophage polarization during Mm infection in Xenopus, distinct expression kinetics of IL-34 and CSF gene have been observed in pilot experiments in adult frogs. Notably, IL-34 is highly expressed during the early stages of infection peaking at 6 dpi followed by decreased expression at later time points. In contrast, CSF-1 gene expression continues to increase until 12 days post-Mm infection suggesting a polarization from M1-like to M2-like phenotype during Mm infection highlighting the importance of macrophage effector choices in the context of Mm infection progression. Although still preliminary, these observations further implicate CSF-1 and IL-34 as important factors in macrophage polarization and function during mycobacteria infection (Table 1.1).
Fig. 1.1.
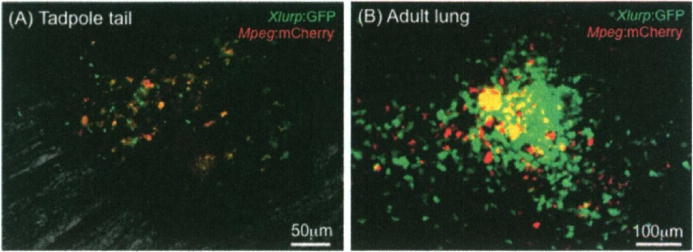
Visualization of macrophage involvement in granuloma formation during Mm infection in xlurp:GFP/xmpeg:mCherry double transgenic X. laevis. (a) Recruitement and accumulation of lurp+/mpeg+ (red-orange) macrophages at the site of infection in the tadpole’s tail 8 days after Mm infection. The infection was done by intramuscular injection of 1000 forming unit (CFU) in a volume of 100 nl in the middle section of the tail of a 3 weeks old tadpole. (b) Small granuloma-like accumulation of lurp+/mpeg− leukocytes (green) and lurp+/mpeg+ macrophages (red-orange) in the lung of a young adult 19 days after Mm infection by intraperitoneal injection of 1 × 106 CFU in a volume of 100 μl. Images were taken on unfixed whole mount organ for adult and live tadpole under narcosis using a Leica DMIRB inverted fluorescence microscope
Concerning the role of CSF and IL-34 in macrophage polarization, it is noteworthy that CSF-1 can alter the magnitude of M1/M2 polarized phenotypes in mammals (Verreck et al. 2004, 2006). For example, human macrophages cultured in presence of recombinant CSF-1 poorly respond to LPS ± IFNγ stimulation. However, while these CSF-1 treated cells are unable to generate the pro-inflammatory cytokines, IL-12 or IL-23, they can produce significant amounts of IL-10 in response to the same stimuli (i.e., LPS ± IFNγ). Thus, although CSF-1 stimulation alone does not recapitulate a full M1/M2 phenotype when compared with prototypic polarizing stimuli (e.g., IFNγ, TLRs, IL-4, IL-13, etc.), CSF-1 stimulation has impacts on macrophage polarizing sensitivity. These studies suggest that CSF-1 predisposes monocyte–macrophage to exhibit a differential M2 phenotype (Verreck et al. 2004, 2006). Notably, CSF-l-derived human monocytes are more susceptible to infection with an attenuated strain of Mycobacterium bovis (Bacille de Calmette et Guérin), the causative agent of tuberculosis in cattle, as determined by increased phagocytosis and enhanced bacterial outgrowth (Verreck et al. 2004, 2006). Although the modulating potential and putative suppressive role of CSF-1 in macrophage polarization is partially recapitulated in mice (Fleetwood et al. 2007) and to some extent in Xenopus, in bony fish CSF-1 stimulation appears to skew macrophages towards an M1-like state (Grayfer et al. 2009) and reviewed in (Hodgkinson et al. 2015).
Interestingly, the single CSF-1 gene in mammals and birds exhibits alternatively splicing (Garceau et al. 2010; Manos 1988; Rettenmier and Roussel 1988), whereas in amphibians the single CSF-1 gene does not appear to be alternatively spliced (Grayfer and Robert 2013) and bony fish possess two distinct CSF-1 genes (Wang et al. 2008). It is presently unknown whether the distinct fish molecules encoded by these two CSF-1 genes have distinct biological roles and, whether, as previously suggested (Grayfer and Robert 2016) these gene products may recapitulate the mammalian CSF-1 splice variants.
Thus, it appears that bony fish and amphibian macrophages, similar to their mammalian counterparts, exhibit a variety of functional roles and an ability to become polarized towards either an M1-like inflammatory type or an M2-like resolution functional state (summarized in Table 1.2). However, more extensive comparative research will be required for elucidating mechanisms governing macrophage functional heterogeneity and for understanding macrophage evolution. Also, given the heterogeneity in macrophage populations, it is likely that the evolution of macrophage polarization is more complex, reflecting the species adaptations, the environment and, the source of pathogenic stimuli. For example, anuran amphibians not only exhibit two quite distinctive developmental stages: tadpoles and adult frogs, but these life stages also exhibit different antigen receptor repertoires for both B and T cells and occupy different ecological niches (e.g., different diets and biotopes) that are likely confronted to different pathogens. As such tadpole and adult frog macrophages are likely exposed to different stimuli.
Table 1.2.
Summary of prototypical M1 and M2 macrophage functions and hallmark genes
| Animal taxa | M1-like characteristics | M2-like | ||||||
|---|---|---|---|---|---|---|---|---|
| Antimicrobial activity | NO, ROI iNOS | M1 polarizing cytokines (IFNγ TNFα) | M1-cytokine profilesa | Wound healing | Arginaseb | M1 polarizing cytokines (IL-4, IL-3) | M1-cytokine profilesc | |
| Invertebrates (insects, snails) | + | + | ? | ? | + | + Arginase-2 | – | ? |
| Teleost (Cyprinids, salmonids) | + | + | + | + | + | + Arginase-1/2 | +d | + |
| Amphibians (Xenopus) | + | + | + | + | + | Arginase-1/2 | +e | + |
| Mammals (Human, mice) | + | + | + | + | + | Arginase-1/2 | + | + |
Up-regulation of pro-inflammatory cytokines in response to microbial challenge, including TNF-α, IL-1β), IL-12 and IL-23, has been demonstrated in teleost and amphibians
Arginase is encoded by a single gene in invertebrates, whereas in vertebrates two genes code for the cytosolic arginase-1 and the mitochondria arginase-2 (postulated to represent the ancestral gene). Mammalian M2 macrophage express arginase-1, while alternative macrophage activation in teleost (carp) coincides with increased arginase-2 transcript levels
M2-type cytokines such as IL-10 and TGF-β have been described in teleost and amphibians
In teleost fish, two genes share homology with both IL-4 and Il-13 (IL4/13A and IL4/13B)
To date in Xenopus, a single gene shares homology with both IL-4 and IL-13 (IL4/13)
1.4 Conclusion
Functional heterogeneity and adaptable plasticity are hallmarks of monocyte–macrophage lineage cells, highlighting the essential roles of these cells in maintaining homeostasis, as well as effector functions during pro-inflammatory and anti-inflammatory immune responses. Similar to their mammalian counterparts, macrophages of ectothermic vertebrates are now recognized to have the ability to adapt their functional roles to species-specific physiological cues suggesting that macrophage functional polarization towards distinct activation states is an evolutionary ancient trait. However, although broadly defined M1-like and M2-like macrophages have been demonstrated in nonmammalian species, this terminology covers an array of functionally disparate groups of macrophages. As such the full spectrum of macrophage activation, polarization, and functions in vivo is less straightforward and not strictly adhering to their mammalian counterparts. In addition, although T and B cell-deficient mice do possess the potential for macrophage polarization (Mills et al. 2000), the immune system consists of a spectrum of immune cell populations clearly influencing and being influenced by macrophage polarization. Indeed, it is likely that macrophages encounter both M1-like and M2-like polarizing stimuli simultaneously within an inflamed tissue microenvironment, which may in part explain the wide spectrum of macrophage-activated phenotypes in vivo. Furthermore, it is well recognized that depending on anatomical and physiologic settings, macrophages exhibit different capacities for polarization. Thus, further research into the molecular cues and mechanisms regulating mammalian and nonmammalian macrophages will further the understanding of macrophage functional regulation.
In this regard, the growing genetic resources in particular the availability of ectothermic vertebrates transgenic lines combined with live-imaging techniques will be useful in the further deciphering of macrophage polarization. Indeed, nonmammalian animal models such as zebrafish and Xenopus present several attractive features for in vivo studies and intravital microscopy. Because of external fertilization, early developmental stages are accessible to experimentation. In addition, zebrafish larvae and Xenopus tadpoles have a relatively small size, are transparent, and do not require temperature control or extensive aseptic conditions. All these attributes are convenient for intravital microscopy. The availability of inbred lines allowing adoptive cell transfer as well as transgenic lines with fluorescently labeled macrophage and other immune cell populations further empower these nonmammalian animal models. Owing to the additional possibilities offered by loss-of-function approaches using genome editing technology, ectothermic vertebrates models are in position to significantly advance our understanding of the plasticity and “raison d’être” of macrophage activation.
Acknowledgments
We would like to thank Ms. Maureen Banach for critical reading of the manuscript. Funding support R24-AI-059830 from the National Institute of Allergy and Infectious Diseases (NIH/NIAID) and IOS-1456213 from the National Science Foundation.
References
- Adams DO. The granulomatous inflammatory response. A review. Am J Pathol. 1976;84:164–192. [PMC free article] [PubMed] [Google Scholar]
- Allavena P, Sica A, Garlanda C, Mantovani A. The Yin-Yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol Rev. 2008a;222:155–161. doi: 10.1111/j.1600-065X.2008.00607.x. [DOI] [PubMed] [Google Scholar]
- Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008b;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Arts JA, Tijhaar EJ, Chadzinska M, Savelkoul HF, Verburg-van Kemenade BM. Functional analysis of carp interferon-gamma: evolutionary conservation of classical phagocyte activation. Fish Shellfish Immunol. 2010;29:793–802. doi: 10.1016/j.fsi.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Badariotti F, Lelong C, Dubos MP, Favrel P. Characterization of chitinase-like proteins (Cg-Clp1 and Cg-Clp2) involved in immune defence of the mollusc Crassostrea gigas. FEBS J. 2007;274:3646–3654. doi: 10.1111/j.1742-4658.2007.05898.x. [DOI] [PubMed] [Google Scholar]
- Bayley AE, Hill BJ, Feist SW. Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Dis Aquat Organ. 2013;103:171–183. doi: 10.3354/dao02574. [DOI] [PubMed] [Google Scholar]
- Beider K, Bitner H, Leiba M, Gutwein O, Koren-Michowitz M, Ostrovsky O, Abraham M, Wald H, Galun E, Peled A, Nagler A. Multiple myeloma cells recruit tumor-supportive macrophages through the CXCR4/CXCL12 axis and promote their polarization toward the M2 phenotype. Oncotarget. 2014;5:11283–11296. doi: 10.18632/oncotarget.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Bystrom J, Evans I, Newson J, Stables M, Toor I, van Rooijen N, Crawford M, Colville-Nash P, Farrow S, Gilroy DW. Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood. 2008;112:4117–4127. doi: 10.1182/blood-2007-12-129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetta L, Cassol E, Poli G. Macrophage polarization in health and disease. ScientificWorldJoumal. 2011;11:2391–2402. doi: 10.1100/2011/213962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Galan L, Olleros ML, Vesin D, Garcia I. Much more than M1 and M2 macrophages, there are also CD169(+) and TCR(+) macrophages. Front Immunol. 2015;6:263. doi: 10.3389/fimmu.2015.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, Kimura F, Okada S. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010;17:1917–1927. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Yu KH, Jancovich JK. The molecular biology of frog vims 3 and other iridoviruses infecting cold-blooded vertebrates. Viruses. 2011;3:1959–1985. doi: 10.3390/v3101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan MR, Tobin DM. Fit for consumption: zebrafish as a model for tuberculosis. Dis Model Mech. 2014;7:777–784. doi: 10.1242/dmm.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- De Jesus Andino F, Chen G, Li Z, Grayfer L, Robert J. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus Frog-Virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology. 2012;432:435–443. doi: 10.1016/j.virol.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzik JM. Evolutionary roots of arginase expression and regulation. Front Immunol. 2014;5:544. doi: 10.3389/fimmu.2014.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Albertorio Saez LM, Gill AL, Gill SR, Grayfer L, Haynes N, Myers JR, Robert J. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proc Natl Acad Sci U S A. 2013;110:14342–14347. doi: 10.1073/pnas.1309840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Grayfer L, Robert J. Evolution of nonclassical MHC-dependent invariant T cells. Cell Mol Life Sci. 2014;71:4763–4780. doi: 10.1007/s00018-014-1701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117(4):e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- Franchini A, Ottaviani E. Repair of molluscan tissue injury: role of PDGF and TGF-beta1. Tissue Cell. 2000;32:312–321. doi: 10.1054/tice.2000.0118. [DOI] [PubMed] [Google Scholar]
- Franchini A, Conte A, Ottaviani E. Nitric oxide: an ancestral immunocyte effector molecule. Adv Neuroimmunol. 1995;5:463–478. doi: 10.1016/0960-5428(95)00029-1. [DOI] [PubMed] [Google Scholar]
- Garceau V, Smith J, Paton IR, Davey M, Fares MA, Sester DP, Burt DW, Hume DA. Pivotal advance: avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J Leukoc Biol. 2010;87:753–764. doi: 10.1189/jlb.0909624. [DOI] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Colony-stimulating factor-l-responsive macrophage precursors reside in the amphibian (Xenopus laevis) bone marrow rather than the hematopoietic subcapsular liver. J Innate Immun. 2013;5:531–542. doi: 10.1159/000346928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. J Leukoc Biol. 2014;96:1143–1153. doi: 10.1189/jlb.4A0614-295R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Distinct functional roles of amphibian (Xenopus laevis) colony-stimulating factor-1- and interleukin-34-derived macrophages. J Leukoc Biol. 2015;98:641–649. doi: 10.1189/jlb.4AB0315-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Robert J. Amphibian macrophage development and antiviral defenses. Dev Comp Immunol. 2016;58:60–67. doi: 10.1016/j.dci.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Hanington PC, Belosevic M. Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol. 2009;26:406–413. doi: 10.1016/j.fsi.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Grayfer L, Andino Fde J, Chen G, Chinchar GV, Robert J. Immune evasion strategies of ranaviruses and innate immune responses to these emerging pathogens. Viruses. 2012;4:1075–1092. doi: 10.3390/v4071075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, De Jesus Andino F, Robert J. The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. J Virol. 2014a;88:5766–5777. doi: 10.1128/JVI.00223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Edholm ES, Robert J. Mechanisms of amphibian macrophage development: characterization of the Xenopus laevis colony-stimulating factor-1 receptor. Int J Dev Biol. 2014b;58:757–766. doi: 10.1387/ijdb.140271jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Hodgkinson JW, Belosevic M. Antimicrobial responses of teleost phagocytes and innate immune evasion strategies of intracellular bacteria. Dev Comp Immunol. 2014c;43:223–242. doi: 10.1016/j.dci.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–3446. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- Guilbert U, Stanley ER. Specific interaction of murine colony-stimulating factor with mononuclear phagocytic cells. J Cell Biol. 1980;85:153–159. doi: 10.1083/jcb.85.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington PC, Belosevic M. Interleukin-6 family cytokine M17 induces differentiation and nitric oxide response of goldfish (Carassius auratus L.) macrophages. Dev Comp Immunol. 2007;31:817–829. doi: 10.1016/j.dci.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Hanington PC, Wang T, Secombes CJ, Belosevic M. Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J Biol Chem. 2007;282:31865–31872. doi: 10.1074/jbc.M706278200. [DOI] [PubMed] [Google Scholar]
- Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol. 2012;2012:948098. doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson JW, Grayfer L, Belosevic M. Biology of bony fish macrophages. Biology (Basel) 2015;4:881–906. doi: 10.3390/biology4040881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoverman JT, Gray MJ, Miller DL. Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis Aquat Organ. 2010;89:97–107. doi: 10.3354/dao02200. [DOI] [PubMed] [Google Scholar]
- Huang Z, Luo Q, Guo Y, Chen J, Xiong G, Peng Y, Ye J, Li J. Mycobacterium tuberculosis-induced polarization of human macrophage orchestrates the formation and development of tuberculous granulomas in vitro. PLoS One. 2015;10:e0129744. doi: 10.1371/journal.pone.0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs functional differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerink M, Forlenza M, Ribeiro CM, de Vries BJ, Savelkoul HF, Wiegertjes GF. Differential macrophage polarisation during parasitic infections in common carp (Cyprinus carpio L) Fish Shellfish Immunol. 2006a;21:561–571. doi: 10.1016/j.fsi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Joerink M, Ribeiro CM, Stet RJ, Hermsen T, Savelkoul HF, Wiegertjes GF. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol. 2006b;177:61–69. doi: 10.4049/jimmunol.177.1.61. [DOI] [PubMed] [Google Scholar]
- Joerink M, Savelkoul HF, Wiegertjes GF. Evolutionary conservation of alternative activation of macrophages: structural and functional characterization of arginase 1 and 2 in carp (Cyprinus carpio L) Mol Immunol. 2006c;43:1116–1128. doi: 10.1016/j.molimm.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick RB, Matico RE, McNulty DE, Strickler JE, Rosenberg M. An abundantly secreted glycoprotein from Drosophila melanogaster is related to mammalian secretory proteins produced in rheumatoid tissues and by activated macrophages. Gene. 1995;153:147–154. doi: 10.1016/0378-1119(94)00756-i. [DOI] [PubMed] [Google Scholar]
- Labonte AC, Tosello-Trampont AC, Hahn YS. The role of macrophage polarization in infectious and inflammatory diseases. Mol Cells. 2014;37:275–285. doi: 10.14348/molcells.2014.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- Lichanska AM, Browne CM, Henkel GW, Murphy KM, Ostrowski MC, McKercher SR, Maki RA, Hume DA. Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.l. Blood. 1999;94:127–138. [PubMed] [Google Scholar]
- Liu H, Leo C, Chen X, Wong BR, Williams LT, Lin H, He X. The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochim Biophys Acta. 2012;1824:938–945. doi: 10.1016/j.bbapap.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos MM. Expression and processing of a recombinant human macrophage colony-stimulating factor in mouse cells. Mol Cell Biol. 1988;8:5035–5039. doi: 10.1128/mcb.8.11.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovam A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Marino S, Cilfone NA, Mattila JT, Linderman JJ, Flynn JL, Kirschner DE. Macrophage polarization drives granuloma outcome during Mycobacterium tuberculosis infection. Infect Immun. 2015;83:324–338. doi: 10.1128/IAI.02494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CJ, Carey AF, Fortune SM. A bug’s life in the granuloma. Semin Immunopathol. 2016;38:213–220. doi: 10.1007/s00281-015-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S. The Ml and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, Thomas B, Fabbri M, Crawshaw A, Ho LP, Ten Hacken NH, Cobos Jimenez V, Kootstra NA, Hamann J, Greaves DR, Locati M, Mantovani A, Gordon S. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–e69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- McKinney EC, Haynes L, Droese AL. Macrophage-like effector of spontaneous cytotoxicity from the shark. Dev Comp Immunol. 1986;10:497–508. doi: 10.1016/0145-305x(86)90171-0. [DOI] [PubMed] [Google Scholar]
- Meijer AH. Protection and pathology in TB: learning from the zebrafish model. Semin Immunopathol. 2016;38:261–273. doi: 10.1007/s00281-015-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Verbeek FJ, Salas-Vidal E, Corredor-Adamez M, Bussman J, van der Sar AM, Otto GW, Geisler R, Spaink HP. Transcriptome profiling of adult zebrafish at the late stage of chronic tuberculosis due to Mycobacterium marinum infection. Mol Immunol. 2005;42:1185–1203. doi: 10.1016/j.molimm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Metchnikoff EM. Immunity in infective disease. Cambridge University Press; Cambridge: 1905. [Google Scholar]
- Mills CD. Macrophage arginine metabolism to ornithine/urea or nitric oxide/citrulline: a life or death issue. Crit Rev Immunol. 2001;21:399–425. [PubMed] [Google Scholar]
- Mills CD. M1 and M2 macrophages: oracles of health and disease. Crit Rev Immunol. 2012;32:463–488. doi: 10.1615/critrevimmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. [Google Scholar]
- Morales HD, Abramowitz L, Gertz J, Sowa J, Vogel A, Robert J. Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J Virol. 2010;84:4912–4922. doi: 10.1128/JVI.02486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM., Jr Recent advances in arginine metabolism: roles and regulation of the arginases. Br J Pharmacol. 2009;157:922–930. doi: 10.1111/j.1476-5381.2009.00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder M, Eichmann K, Moran JM, Centeno F, Soler G, Modolell M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol. 1999;163:3771–3777. [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann NF, Stafford JL, Belosevic M. Biochemical and functional characterisation of macrophage stimulating factors secreted by mitogen-induced goldfish kidney leucocytes. Fish Shellfish Immunol. 2000;10:167–186. doi: 10.1006/fsim.1999.0236. [DOI] [PubMed] [Google Scholar]
- Nguyen-Chi M, Laplace-Builhe B, Travnickova J, Luz-Crawford P, Tejedor G, Phan QT, Duroux-Richard I, Levraud JP, Kissa K, Lutfalla G, Jorgensen C, Djouad F. Identification of polarized macrophage subsets in zebrafish. Elife. 2015;4:e07288. doi: 10.7554/eLife.07288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole R. Experimental models used to study human tuberculosis. Adv Appl Microbiol. 2010;71:75–89. doi: 10.1016/S0065-2164(10)71003-0. [DOI] [PubMed] [Google Scholar]
- Ohtani M, Hayashi N, Hashimoto K, Nakanishi T, Dijkstra JM. Comprehensive clarification of two paralogous interleukin 4/13 loci in teleost fish. Immunogenetics. 2008;60:383–397. doi: 10.1007/s00251-008-0299-x. [DOI] [PubMed] [Google Scholar]
- Ottaviani E. Immunocyte: the invertebrate counterpart of the vertebrate macrophage. Invertebr Surviv J. 2011;8:1–4. [Google Scholar]
- Ottaviani E, Franceschi C. The invertebrate phagocytic immunocyte: clues to a common evolution of immune and neuroendocrine systems. Immunol Today. 1997;18:169–174. doi: 10.1016/s0167-5699(97)84663-4. [DOI] [PubMed] [Google Scholar]
- Paredes R, Ishibashi S, Borrill R, Robert J, Amaya E. Xenopus: an in vivo model for imaging the inflammatory response following injury and bacterial infection. Dev Biol. 2015;408:213–228. doi: 10.1016/j.ydbio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterton D, Shi YB. Thyroid hormone-dependent differential regulation of multiple arginase genes during amphibian metamorphosis. J Biol Chem. 1994;269:25328–25334. [PubMed] [Google Scholar]
- Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174:6561. doi: 10.4049/jimmunol.174.11.6561. author reply 6561-2. [DOI] [PubMed] [Google Scholar]
- Rettenmier CW, Roussel MF. Differential processing of colony-stimulating factor 1 precursors encoded by two human cDNAs. Mol Cell Biol. 1988;8:5026–5034. doi: 10.1128/mcb.8.11.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger AM, Hall BE, Barreda DR. Macrophage activation differentially modulates particle binding, phagocytosis and downstream antimicrobial mechanisms. Dev Comp Immunol. 2010;34:1144–1159. doi: 10.1016/j.dci.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Robert J, Edholm ES. A prominent role for invariant T cells in the amphibian Xenopus laevis tadpoles. Immunogenetics. 2014;66:513–523. doi: 10.1007/s00251-014-0781-6. [DOI] [PubMed] [Google Scholar]
- Robert J, Abramowitz L, Gantress J, Morales HD. Xenopus laevis: a possible vector of Ranavirus infection? J Wildl Dis. 2007;43:645–652. doi: 10.7589/0090-3558-43.4.645. [DOI] [PubMed] [Google Scholar]
- Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani A, Schioppa T, Porta C, Biswas SK, Nebuloni M, Vago L, Bottazzi B, Colombo MP, Mantovani A, Sica A. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- Samson ML. Drosophila arginase is produced from a nonvital gene that contains the elav locus within its third intron. J Biol Chem. 2000;275:31107–31114. doi: 10.1074/jbc.M001346200. [DOI] [PubMed] [Google Scholar]
- Sang Y, Miller LC, Blecha F. Macrophage polarization in virus-host interactions. J Clin Cell Immunol. 2015;6:311. doi: 10.4172/2155-9899.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotton CJ, Martinez FO, Smelt MJ, Sironi M, Locati M, Mantovani A, Sozzani S. Transcriptional profiling reveals complex regulation of the monocyte IL-1 beta system by IL-13. J Immunol. 2005;174:834–845. doi: 10.4049/jimmunol.174.2.834. [DOI] [PubMed] [Google Scholar]
- Shaked I, Hanna DB, Gleissner C, Marsh B, Plants J, Tracy D, Anastos K, Cohen M, Golub ET, Karim R, Lazar J, Prasad V, Tien PC, Young MA, Landay AL, Kaplan RC, Ley K. Macrophage inflammatory markers are associated with subclinical carotid artery disease in women with human immunodeficiency virus or hepatitis C vims infection. Arterioscler Thromb Vasc Biol. 2014;34:1085–1092. doi: 10.1161/ATVBAHA.113.303153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Saccani A, Mantovani A. Tumor-associated macrophages: a molecular perspective. Int Immunopharmacol. 2002;2:1045–1054. doi: 10.1016/s1567-5769(02)00064-4. [DOI] [PubMed] [Google Scholar]
- Silva Miranda M, Breiman A, Allain S, Deknuydt F, Altare F. The tuberculous granuloma: an unsuccessful host defence mechanism providing a safety shelter for the bacteria? Clin Dev Immunol. 2012;2012:139127. doi: 10.1155/2012/139127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Kotecha S, Towers N, Latinkic BV, Mohun TJ. XPOX2-peroxidase expression and the XLURP-l promoter reveal the site of embryonic myeloid cell development in Xenopus. Mech Dev. 2002;117:173–186. doi: 10.1016/s0925-4773(02)00200-9. [DOI] [PubMed] [Google Scholar]
- Sricharoen S, Kim JJ, Tunkijjanukij S, Soderhall I. Exocytosis and proteomic analysis of the vesicle content of granular hemocytes from a crayfish. Dev Comp Immunol. 2005;29:1017–1031. doi: 10.1016/j.dci.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempin CC, Dulgerian LR, Garrido VV, Cerban FM. Arginase in parasitic infections: macrophage activation, immunosuppression, and intracellular signals. J Biomed Biotechnol. 2010;2010:683485. doi: 10.1155/2010/683485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagoh H, Himes R, Clarke D, Leenen PJ, Riggs AD, Hume D, Bonifer C. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 2002;16:1721–1737. doi: 10.1101/gad.222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff TH. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci U S A. 2004;101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreck FA, de Boer T, Langenberg DM, van der Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-gamma- and CD40L-mediated costimulation. J Leukoc Biol. 2006;79:285–293. doi: 10.1189/jlb.0105015. [DOI] [PubMed] [Google Scholar]
- Wang T, Secombes CJ. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 2013;35:1703–1718. doi: 10.1016/j.fsi.2013.08.030. [DOI] [PubMed] [Google Scholar]
- Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol. 2008;181:3310–3322. doi: 10.4049/jimmunol.181.5.3310. [DOI] [PubMed] [Google Scholar]
- Wang T, Johansson P, Abos B, Holt A, Tafalla C, Jiang Y, Wang A, Xu Q, Qi Z, Huang W, Costa MM, Diaz-Rosales P, Holland JW, Secombes CJ. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget. 2016;7:10917–10946. doi: 10.18632/oncotarget.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, Williams LT, Lin H, Stanley ER. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J Leukoc Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PA, Campbell A, Morgan RL, Rosenberger AG, Murray BW. Dogmas and controversies in the handling of nitrogenous wastes: expression of arginase type I and II genes in rainbow trout: influence of fasting on liver enzyme activity and mRNA levels in juveniles. J Exp Biol. 2004;207:2033–2042. doi: 10.1242/jeb.00958. [DOI] [PubMed] [Google Scholar]


