Abstract
Objective
Understanding the pathogenesis of systemic sclerosis (SSc) is confounded by considerable disease heterogeneity. Animal models of SSc that recapitulate distinct subsets of disease at the molecular level have not been delineated. We applied interspecies comparative analysis of genomic data from multiple mouse models of SSc and patients with SSc to determine which animal models best reflect the SSc intrinsic molecular subsets.
Methods
Gene expression measured in skin from mice with sclerodermatous graft-versus-host disease (sclGVHD), bleomycin-induced fibrosis, Tsk1/+ or Tsk2/+ mice was mapped to human orthologs and compared to SSc skin biopsy-derived gene expression. TGFβ activation was assessed using a responsive signature in mouse and Tnfrsf12a expression measured in SSc and mouse skin.
Results
Gene expression in skin from mice with sclGVHD and bleomycin-induced fibrosis corresponded to that in SSc patients in the inflammatory molecular subset. In contrast, Tsk2/+ mice showed gene expression corresponding to the fibroproliferative SSc subset. Bleomycin and Tsk2/+ mice showed enrichment of a TGFβ-responsive signature. Expression of Tnfrsf12a (the Tweak-Receptor / Fn14) is elevated in skin from fibroproliferative SSc patients and skin of Tsk2/+ mice.
Conclusion
This study reveals similarities in cutaneous gene expression between distinct mouse models of SSc and specific molecular subsets of the disease. Different pathways underlie the intrinsic subsets including TGF-β, IL13 and IL4. We identify a novel target, Tnfrsf12a that is elevated in skin from fibroproliferative patients and Tsk2/+ mice. The information will serve to inform mechanistic and translational pre-clinical studies in SSc.
Keywords: systemic sclerosis, skin, human intrinsic gene expression subsets, murine models, Tsk2/+, Tsk/+, sclGVHD, Bleomycin, fibrosis, DNA microrrays, interspecies comparison, TGF-beta, IL13, Tnfrsf12a (Tweak-Receptor)
A lack of clear genetic associations and the heterogeneity in clinical presentation of systemic sclerosis (SSc; scleroderma) have limited the development of a single agreed-upon animal model (1). We have demonstrated that molecular subsets of SSc exist (2-4) and that different signaling pathways underlie each subsets (5-7). Given the distinct molecular features of SSc subsets, it is possible that one or more mouse models might most accurately represent each. Identification of appropriate models for each subset is necessary to effectively develop therapeutics that specifically target each of these groups of patients. We conducted an integrated interspecies analysis of gene expression in skin of four commonly used mouse models of SSc to identify those models in which skin gene expression and deregulated pathways most accurately reflects that of the molecular subsets of SSc.
Many different signaling pathways have been implicated in SSc (8, 9). TGFβ and PDGF signatures have been demonstrated to underlie the fibroproliferative subset of patients (5, 7), whereas IL4 and IL13 signaling is predominantly enriched in the inflammatory subset of patients (6). Recent analyses have demonstrated that TGFβ signaling can span the fibroproliferative and inflammatory subsets (7, 10). Here we have directly compared the similarities and differences in the molecular events underlying pathogenesis of SSc, and in mouse models of the disease. To our knowledge this is the first integrative and comparative analysis of genome-wide expression in animal models for a heterogeneous human autoimmune disease.
Materials and Methods
Study Design and human subjects
Microarray data from human SSc samples were obtained from Milano et al ((2); GEO accession GSE9285). SSc patients met the ACR criteria for systemic sclerosis and included both diffuse and limited systemic sclerosis patients as well as a subset of patients with morphea. We performed biological rather than technical replicates for the different mouse samples. The study was designed to identify the best mouse models to study each of the human SSc subsets. This is a major unmet need since heterogeneity in the disease has confounded basic science studies as well as clinical trials. Our data show that different mouse models represent different subsets of SSc disease.
Mouse models of SSc
Mouse skin samples were obtained from different laboratories (Table 1). All animal protocols were institutionally approved by the Animal Care and Use Committee at University of Connecticut Health Center (UCONN), Brigham and Women's Hospital (BWH), Boston University School of Medicine (BUMC), Northwestern Feinberg School of Medicine (NW) and Drexel University College of Medicine (DUCM). Each biopsy included the hypodermal layer and fibrosis was confirmed at the site of biopsy in all models. The sclGVHD model was generated at BWH and data have been described previously (6). Depilated Tsk2/+ (11) mouse back skin samples from UCONN were stored in RNAlater (ThermoFisher Scientific, Waltham MA). These mice are heterozygous for the Tsk2 mutation on chromosome 1 and were maintained in an inbred line developed by backcrossing onto C57BL/6J (>N10). Tsk1/+ back skin samples were obtained from six week old mice (12) at BUMC. C57BL/6-FbnTsk+/+Pldnpa mice were obtained from Jackson Laboratories (Bar Harbor, ME) and maintained by breeding with C57BL/6 mice. Tsk1/+ and Tsk2/+ heterozygous mice were identified by assessment of skin tightness over the back and by genotyping as described (12). The bleomycin-induced skin fibrosis model was generated at the NW. The protocol chosen has been used previously to study skin fibrosis (13). In bleomycin-induced fibrosis, six- to eight-week-old female BALB/cJ mice were given daily subcutaneous injections of bleomycin (1 mg/kg) and sacrificed at either 5 days or 21 days. Skin at the site of injection was biopsied. All skin tissue was stored in RNAlater for shipping and storage.
Table 1. Eleven groups were specified for intrinsic gene analysis.
Four goups of human samples were specified based on the intrinsic subsets of SSc (2) and seven groups of mouse models were defined based on the model type and different timepoints were specified. (s.c. subcutaneous).
| Intrinsic Group | Description | Species | # of Microarrays |
|---|---|---|---|
| 1 | Fibroproliferative (previously diffuse-proliferation) | human | 27 |
| 2 | inflammatory | human | 17 |
| 3 | limited | human | 9 |
| 4 | normal-like | human | 22 |
| 5 | cGVHD 2 weeks | mouse | 9 |
| 6 | cGVHD 5 weeks | mouse | 4 |
| 7 | s.c. bleomycin 5 days | mouse | 3 |
| 8 | s.c bleomycin 21 days | mouse | 4 |
| 9 | Tsk2/+ 4-weeks | mouse | 4 |
| 10 | Tsk2/+ 16-weeks | mouse | 5 |
| 11 | Tsk1/+ 6-weeks | mouse | 4 |
In vivo TGFβ-responsive gene signature
Total RNA samples from the back skins of mice treated with subcutaneous pumps containing TGFβ were generated at BUMC. Prior to surgery, mice were injected with buprenorphine. For pump insertion, C57BL/6 mice were placed under general anesthesia using isoflourane by inhalation. After complete anesthesia was achieved, the mice were placed on a warm towel, their backs shaved, the surgical area sterilized with betadine and a 1 cm incision made through the skin over the interscapular region. The sterile pumps containing 50, 250 or 1250ng of TGFβ or PBS were inserted into a subcutaneous pocket made by gently teasing apart the fascia layer. Skin was closed with 1-2 staples. Mice were sacrificed 7 days after surgery and the skin from around the pump insertion site harvested for total RNA preparation.
RNA isolation and microarray hybridization
Total RNA was isolated from mouse skin samples using standard Trizol (Invitrogen, Carlsbad, CA) procedures. Samples were manually homogenized as previously described (2). Samples were further purified using RNA cleanup with RNeasy mini-columns (Qiagen, Valencia, CA). RNA was quantified on a NanoDrop ND-1000 Spectrophotometer (Agilent Technologies, Santa Clara, CA) and integrity assessed by gel electrophoresis.
Samples were amplified and labeled using the Agilent Low Input Linear Amplification kit (Agilent Technologies, Santa Clara, CA) and were hybridized against Universal Mouse Reference (UMR) (Strategene, La Jolla, CA) to Agilent Whole Mouse Genome arrays (G4122F) (Agilent Technologies, Santa Clara, CA) in a common reference based design as previously described (2, 5).
Microarray data processing and analysis
Human gene expression data from Milano et al (2) were lowess-normalized log2 Cy5/Cy3 ratios, filtered for intensity/background ratio ≥1.5 in one or both channels and for which at least 80% of the data of sufficient quality were present. This resulted in 28,495 probes for analysis, which were collapsed to 14,276 gene symbols.
Mouse gene expression data were lowess-normalized log2 Cy5/Cy3 ratios, filtered for intensity/background ratio ≥1.5 in one or both channels and for which at least 80% of the data was of sufficient quality were present. This resulted in 13,773 probes selected from Agilent Mouse Whole Genome microarray. These probes were then collapsed by gene symbol, resulting in 9,776 genes.
Both human and mouse data tables were multiplied by negative one, thereby converting the log2 Cy5/Cy3 ratios to log2 Cy3/Cy5 ratios for all analyses.
For interspecies comparisons mouse genes were matched to their human orthologs using the Mouse Genome Database (MGD) at Jackson Laboratories (14). Mouse genes were matched to their human orthologs based on Mouse/Human Orthology with Phenotype Annotations table (MGI 5.22). 62.53% of mouse genes mapped to a human ortholog. This resulted in 6,113 genes. Missing data values were imputed using KNN-nearest neighbors (k=10) function in the bioinformatics toolbox of Matlab R2008b. Systematic bias was corrected using distance weighted discrimination (DWD) in Matlab (15).
Intrinsic genes were selected using an intrinsic gene identifier algorithm (2, 16) was performed (Table 1). 1,217 genes that demonstrated a weight intrinsic score threshold below 0.30 were selected for further analysis (2, 16). Each column was standardized using the root mean square (RMS) for each array, calculated by taking the sum of the square of the expression values for each gene, divided by the number of genes, and taking the square root of this value. The values of the genes on each array were divided by its RMS. All human and mouse data were median centered and clustered using Cluster 3.0 (17) and visualized in TreeView 1.0.13 (17). The p-values for branches on the dendrogram were calculated using R SigClust package with default settings (* in Figure 2 indicates p < 0.001). Consensus Clustering was performed using GenePattern ConsensusClustering with 2,000 iterations and K=3,4 and 5, which clearly shows 4 stable clusters (Figure 2; Supplemental Figure S1).
Figure 2. Gene expression patterns are shared in human SSc skin and in the mouse models.
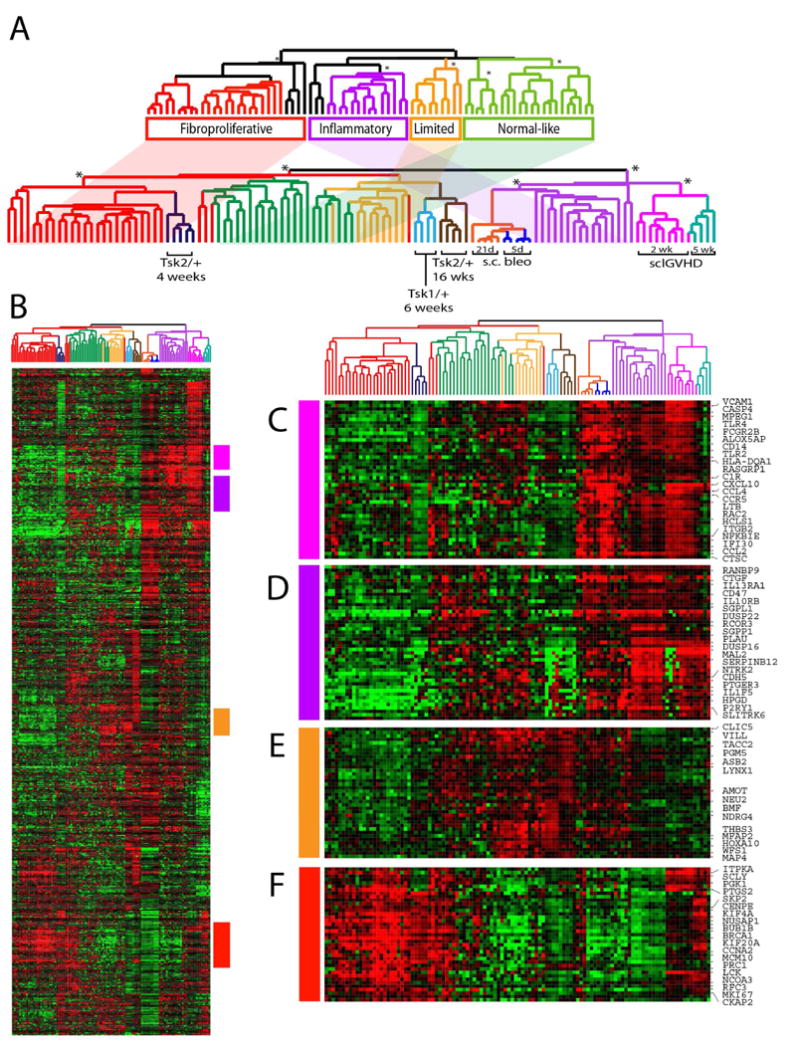
(A) Integrated human mouse expression data clustering. The upper dendrogram shows the organization of the molecular subsets of SSc. The shading indicates the placement of these subsets in the integrated dataset cluster analysis. Microarrays from the mouse models are interspersed among the human subsets. Notably, Tsk2/+ at 4-weeks cluster on the same branch as the fibroproliferative subset while sclGVHD mice at 2- and 5-weeks are clustered with the inflammatory subset. Samples from the bleomycin mouse model show are most closely associated with inflammatory subset but show aspects of inflammatory and fibroproliferative gene expression. (B) 1217 intrinsic genes were clustered in the gene and array dimensions. The sample dendrogram is that from Figure 1. Selected clusters of interest are shown. Gene expression features shared by the sclGVHD mice and the inflammatory subset include those induced by IL13 (C) and IFN-signaling (D). Genes associated with proliferation were up-regulated in Tsk2/+ mice at 1 month and in the fibroproliferative subset (F). A cluster of genes was found up-regulated in the inflammatory and limited subsets as well as the sclGVHD and other models (E).
Pair-wised Pearson correlations for all human and mouse models of SSc were calculated using the R package corrplot to generate the heatmap showing the Pearson product-moment correlation coefficient (Pearson's r) between human SSc and mouse models of SSc (Supplemental Figure S2).
Pathway module map analysis was performed in Genomica (18). Gene expression data for the merged datasets was matched to the appropriate Entrez Gene Identifier corresponding to the human gene annotation. Statistically significantly enriched GO terms were selected (p < 0.05; FDR 0.05; hypergeometric distribution) and clustered according to their enrichment scores. Samples were organized as per the sample clustering of the 1,217 intrinsic genes (Figure 3).
Figure 3. Module map of coordinately regulated GO terms in human SSc and in mouse models.
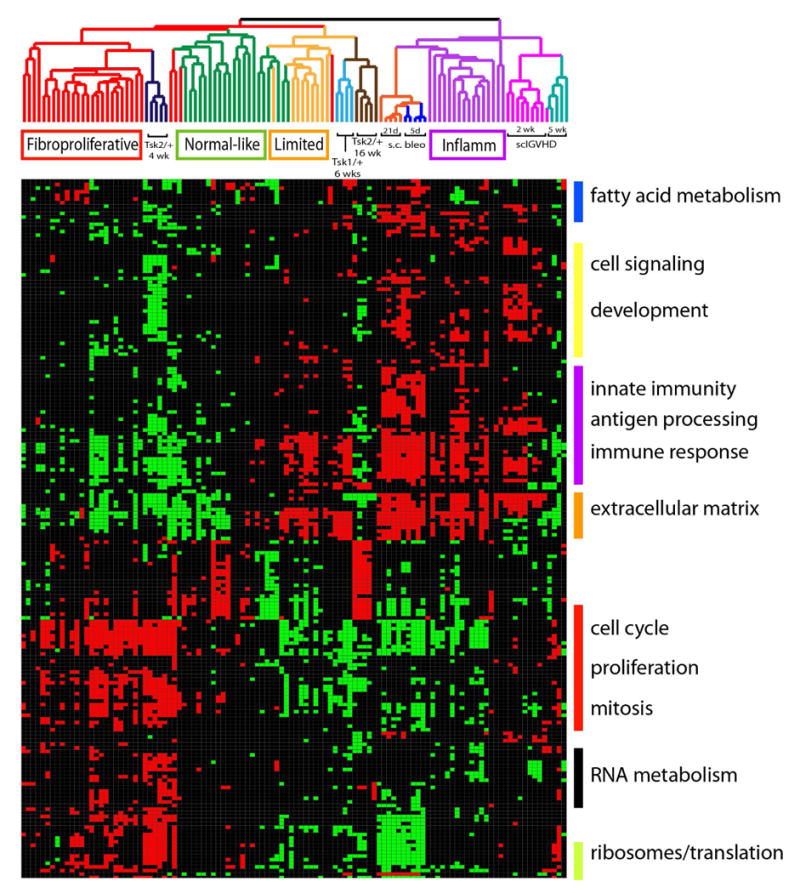
A module map of enriched GO terms was created using gene expression from the integrated interspecies microarray dataset. Modules significantly enriched (p < 0.05, FDR 0.05, hypergeometric distribution) in at least 15 of the 109 arrays were selected and are displayed. Clusters of select GO terms are shown to the right of the module map. Each column represents a microarray and each row is a GO term. The arrays have been ordered as per the intrinsic clustering shown in Figure 1. Positively enriched and negatively enriched modules are shown by red and green squares respectively.
Gene expression data from this study are available from NCBI GEO at accession number GSE71999 (Tsk1/+ Tsk2/+, and bleomycin-induced skin fibrosis). The data for the sclGVHD mouse model ((6); GSE24410) and human SSc skin data ((2); GSE9285) were published previously.
Immunohistochemistry
To visualize TWEAK-R positive and Ki-67 positive cells, paraffin-embedded sections were deparaffinized with two washes of xylene and two washes of ethanol. Antigens were unmasked with 10 mM citrate buffer at 100 oC for 25 min, then blocked with 5% goat serum. For mouse biopsies, rabbit anti-TWEAK-R monoclonal antibody at 1:100 (ab109365, Abcam, Cambridge, MA), or rabbit anti-Ki67 polyclonal antibody at 1:250 (ab9260, Millipore, Darmstadt, Germany) in blocking buffer was applied to the sections overnight at 4°C. Slides were washed three times with PBS and incubated with Cy3-conjugated goat anti-rabbit IgG at 1:200 (TWEAK-R) or at 1:500 (Ki-67, JacksonImmuno-111-165-006, Jackson ImmunoResearch Laboratories, West Grove) for 40 min at RT. Nuclei were visualized with DAPI (Life Technologies, Carlsbad CA). An IgG isotype negative control was used to control for background staining.
Staining of human SSc skin biopsies followed the procedure above, except the mouse anti-TWEAK-R monoclonal antibody was used (1:20; BioLegend, Clone ITEM-4). Sections were washed to remove unbound antibody and then incubated with goat-anti-mouse-Cy3 (1:400; Jackson ImmunoResearch West Grove PA) for 40 min at room temperature. A mouse IgG2b isotype negative control (ThermoFisher, Waltham MA) was used and analyzed in parallel. Eleven biopsies from Milano et al. (2) were analyzed using intrinsic subset assignments from (7), which were identical to Milano except that dSSc7 forearm, which was assigned to fibroproliferative, and dSSc2 forearm, which was assigned to normal-like. The biopsies stained were fibroproliferative (dSSc1, dSSc7, dSSc11), inflammatory (dSSc6, lSSc7), limited (lSSc1, lSSc4, and lSSc5 where two independent skin samples were analyzed) and normal-like (lSSc7, dSSc2, Nor2, Nor3, Nor5). Positive cells in the dermis and epidermal skin appendages were counted by a blinded expert; epidermal staining was not quantified. Statistical significance was calculated using a Mann-Whitney test. Variable increased staining was observed in fibroproliferative and inflammatory patients, and due to the low (n=2) patients analyzed in the inflammatory subset these data were graphed together with those of the fibroproliferative subset.
Statistical Analysis
Pearson correlations were calculated and plotted in Microsoft Office Excel 2007. Positive cells in the sections were counted at 40X magnification from at least 6 fields of view per sample, and four different mice per genotype. The numbers per field were compared for statistical significance by 1-way ANOVA using GraphPad Prism software.
Results
To our knowledge no direct comparisons have been made between the bleomycin-induced SSc mouse model or the Tsk2/+ model with human SSc; although both mouse models share features with human SSc, such as immune cell filtration, ECM deposition, and skin fibrosis (19-22). The datasets and analyses for human SSc skin and the sclGVHD mouse model gene expression have been previously published (2, 6). We used DWD analysis methods (15) to remove platform and species biases, and to integrate the human SSc biopsy and the mouse model gene expression datasets (Figure 1). Groupings in the human and mouse tissue datasets were specified based on intrinsic subset classification (2) or by mouse model (Table 1). 1,217 genes were selected from the merged dataset; samples and genes were organized by hierarchical clustering.
Figure 1. Implementation of integrated interspecies analysis for systemic sclerosis.
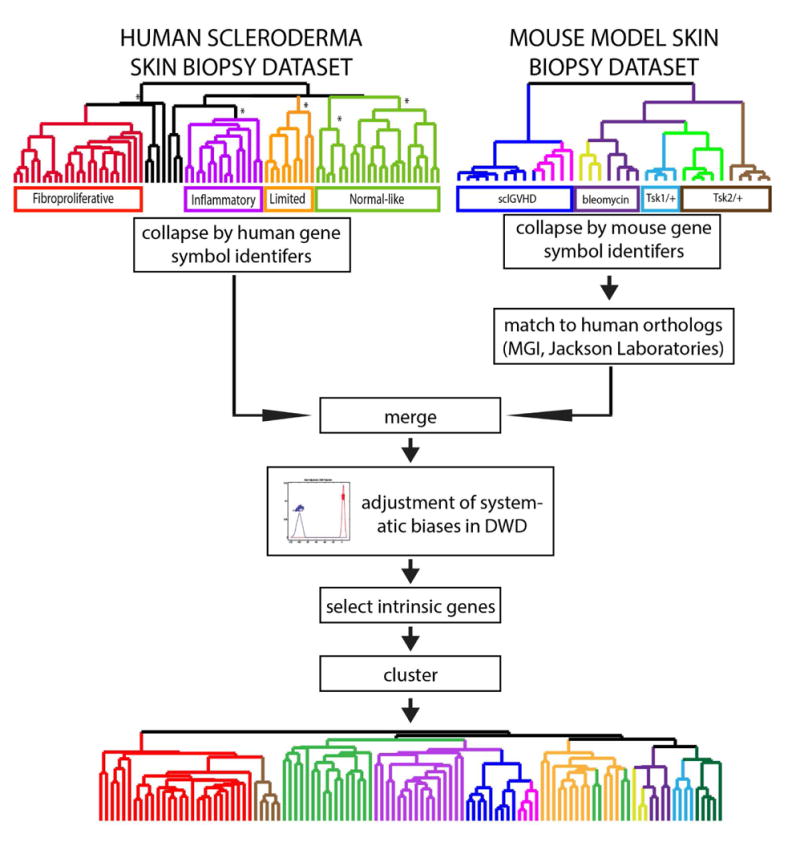
The schematic of the analysis strategy based on that of Herschkowitz et al. (49) is shown. Human and mouse datasets were merged as described in the text, the biases removed using DWD, and the intrinsic genes selected and clustered for further analysis.
As expected, after hierarchical clustering, SSc skin biopsy samples clustered into the same groupings of the original molecular subsets (2). We were specifically interested in understanding how gene expression in different mouse models resembles specific subsets of human SSc. The sclGVHD samples clustered adjacent to the inflammatory patient samples, consistent with prior findings (6), and the Tsk2/+ mice at 4-weeks of age shared a dendrogram branch with the fibroproliferative subset, suggesting that the skin of these mice share gene expression features with the skin from patients in this subset (Figure 2A). Bleomycin-injected mice at 5 days and at 21 days clustered together, and shared a branch with the inflammatory subset. Samples from Tsk1/+ mice and the Tsk2/+ mice at 16-weeks clustered on a branch with the limited subset of patients. Each cluster containing both mouse and human samples is statistically significant (Figure 2; p < 0.001). We performed 2,000 iterations of K-means clustering using consensus cluster, each using 2/3 of the data that showed four stable clusters (Supplemental Figure S1). We plotted the pair-wised Pearson correlation values for all human samples and mouse models (Supplemental Figure S2), which supports the clustering results in Figure 2.
We have previously demonstrated enrichment of IL13-driven gene expression patterns in both the inflammatory subset and in sclGVHD mice (6). As previously demonstrated, membrane-associated IL13-specific receptor subunit IL13RA1 has coordinately high relative expression in both groups along with AIF1 and CCL2, both of which are IL13-regulated; all show high expression again in this interspecies comparison (Figure 2C) (23). Expression of many of these genes is similarly increased in the bleomycin-induced fibrosis model, consistent with the characterized role of IL13 signaling in fibrosis in these mice (24, 25).
Despite similarities between sclGVHD and bleomycin-induced skin fibrosis, a prominent cluster of interferon genes distinguishes these two models. This group of genes is not induced by bleomycin, but is highly upregulated in early sclGVHD. Many of the genes in this cluster are expressed at intermediate levels in SSc inflammatory skin and are targets associated with interferon signaling, such as STAT1, IRF1, CCL5, IFI30, and JAK3 (Figure 2D).
A large number of the genes found coordinately regulated in the fibroproliferative subset and in Tsk2/+ mice at 4-weeks of age are associated with cell proliferation, including KI67, CENPE, KIF20A, MCM7, and POLE2 (Figure 2F) (26). Skin samples from Tsk2/+ mice and their wild-type littermates were stained for the key signature molecule of the proliferative subset, KI67. The results (shown in Figure 5F-G) confirmed the microarray findings that showed up-regulated KI67 mRNA transcripts, which are expressed in significantly more cells, especially in hair follicle locations, primarily in male Tsk2/+ mice in which the disease is more severe; KI67 was not analyzed in female Tsk2/+ mice. Interestingly, gene expression in skin samples from Tsk2/+ mice at 16-weeks and Tsk1/+ at 6-weeks of age did not show a strong resemblance to the genes differentially expressed in diffuse SSc, however, they do show some resemblance to limited SSc. In our mechanistic studies of sclGVHD and Tsk2/+ (6, 27), we found a strong correlation between age of the mice, the gene expression in the model and their resemblance of the human SSc subsets.
Figure 5. Tweak R (TNFRSF12A) is differentially expressed in fibroproliferative SSc patients and Tsk2/+.
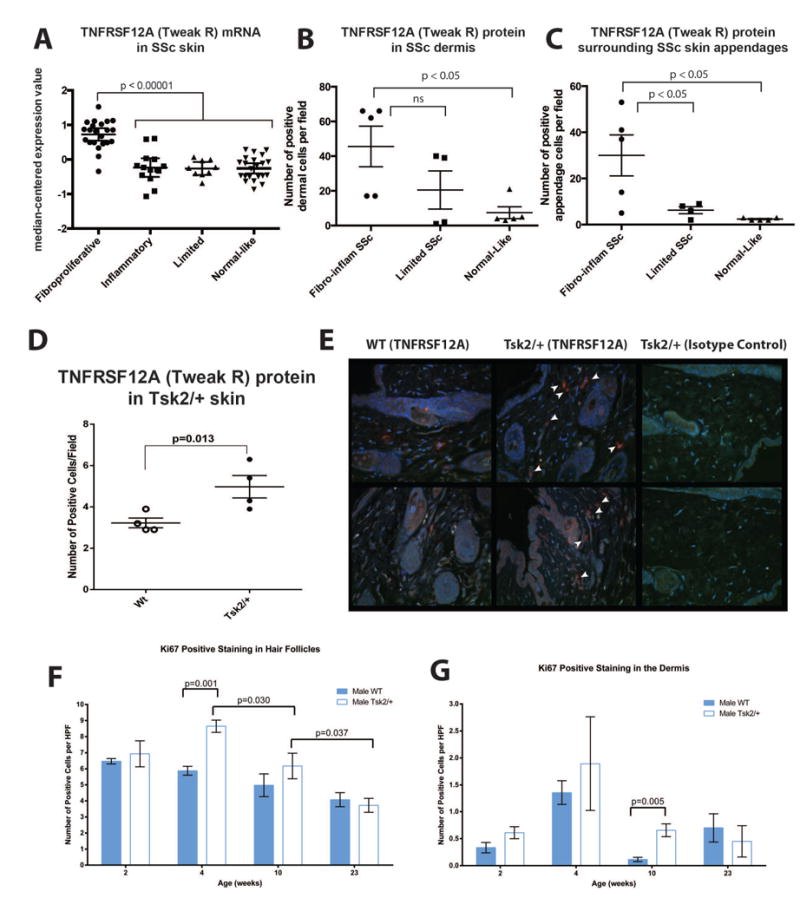
(A) Fibroproliferative SSc patients express significant more tnfrsf12a (Tweak-R; p < 0.00001). (B) Fibroproliferative and inflammatory SSc patients (n=5) have higher TNFRSF12A protein in the dermis and (C) surrounding epidermal skin appendages than patients in the limited intrinsic subset (n=3), or the normal/normal-like subset (n=3 healthy controls; n=2 SSc normal-like)(p < 0.05; one way ANOVA). (D) Tsk2/+ mice expressed 1.6-fold more TNFRSF12A (TWEAK-R) than wild type littermates (p=0.013). Four-week-old female mice were scored for the number of TWEAK-R (Fn14)+ cells per field of view (400X magnification). Significance was calculated with a paired t-test using at least eight fields of view per mouse and four mice per genotype. (E) Representative immunofluorescent images of two wild type mice (Left), two Tsk2/+ mice (center) and the isotype control staining (right). Skin samples were evaluated by immunofluorescence for the presence of TWEAK-R(red) and DAPI(blue). (F-G) KI67 staining for proliferating cells in the hair follicles and dermis of Tsk2/+ mice show significant increase in the 4- and 10-week samples.
To examine the shared features of gene expression in the molecular subsets and in the mouse models on a genomic scale, we created a module map of enriched GO terms in the integrated human SSc and mouse model gene expression dataset (Figure 3). Many GO terms associated with proliferation and mitosis such as mitotic checkpoint, cell division, regulation of mitosis and DNA replication initiation were similarly positively enriched in the Tsk2/+ mice at 4-weeks and in the fibroproliferative biopsies, consistent with the increased expression of genes involved in cell cycle progression. Other GO terms coordinately regulated in this subset and in 4-week Tsk2/+ mice include those associated with RNA metabolism (RNA splicing, mRNA processing and RNA binding), translation (protein-RNA complex assembly, ribosome and eukaryotic 43S initiation complex) and DNA repair (DNA-dependent DNA replication and DNA repair). Proteins involved in these processes such as topoisomerase I and RNA polymerase I are frequent antigenic targets for autoantibodies in SSc (28-32). GO terms associated with lipid biogenesis such as fatty acid metabolic process, lipid metabolic process and sterol biosynthetic process, are enriched and downregulated in a portion of fibroproliferative biopsies and similarly downregulated in the Tsk2/+ mouse skin biopsies. Loss of subcutaneous fat is a characteristic feature of both diffuse SSc (33) and Tsk2/+ model (22). Additionally, it has been demonstrated that lipid biogenesis and fibrotic processes in fibroblasts are mutually antagonistic signaling systems. Activation TGFβ-signaling inhibits lipid biogenesis via PPARγ pathways and vice versa (13, 34). Additionally, PPARγ ligands have been shown to suppress bleomycin-induced fibrosis in lungs (35).
Modules shared in skin from sclGVHD mice at two weeks and the inflammatory subset are predominantly associated with inflammatory processes, such as immune system process, inflammatory response, chemokine activity, and response to biotic stimulus. Increased expression of ECM-associated pathways is also evident in the inflammatory and limited subsets, as well as in the bleomycin-induced fibrosis and Tsk1/+ models, suggesting that pathways underlying fibrosis in these patients and these models may also be conserved.
Having established that skin from Tsk2/+ mice at 4-weeks shares gene expression features with the fibroproliferative subset, we specifically examined a role for TGFβ-signaling associated gene expression in this model. An in vivo TGFβ-responsive signature in mouse skin was generated and expression of these genes examined in skin from Tsk2/+ animals. Total RNA was prepared from skin samples of C57Bl/6 mice that had been surgically implanted with subcutaneous pumps containing 50, 250 or 1250ng of TGFβ, or phosphate-buffered saline (PBS) as a control, for 7 days. Gene expression data relative to the PBS treatment control is shown. We have termed the 719 probes that showed a 2-fold or more change in gene expression the mouse TGFβ-responsive signature (Figure 4). Canonical TGFβ targets found induced in this signature include TIMP1, PAI1, COL1A1, SPP1, LOX, THBS and SPARC, demonstrating that this signature is representative of a response to TGFβ in the skin of these animals. The data for the mouse TGFβ-responsive signature was extracted from the compendium of gene expression in skin of the models. Expression of TGFβ-responsive signature genes is enriched in skin from Tsk2/+ animals at 4-weeks (Figure 4B) suggesting that TGFβ-signaling is active in the skin of these mice. Since both the bleomycin-induced skin fibrosis and Tsk1/+ pathogenesis have been shown to involve TGFβ-signaling, we confirmed the activation of TGFβ-signaling in Tsk1/+ and bleomycin-induced skin fibrosis (Figure 4C). Analysis of the bleomycin-induced skin fibrosis model using GSEA confirms enrichment of the TGFβ-activated gene expression (Supplemental Figure S3).
Figure 4. TGFβ-responsive signature gene expression in SSc mouse models.
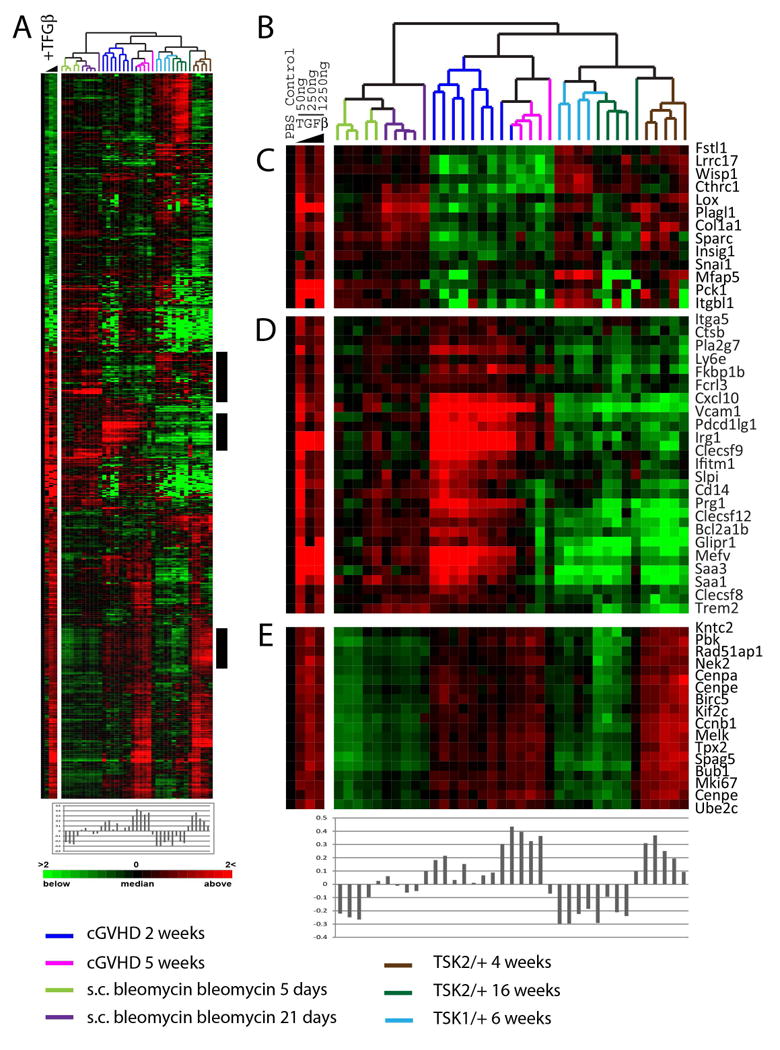
(A) Pumps containing PBS or 50, 250 or 1250ng of TGFβ were surgically inserted subcutaneously in B57/B6 mice for 7 days and skin analyzed by DNA microarray. 719 genes changed in expression >2-fold from the PBS control in at two doses of TGFβ. Data were T0 transformed against the PBS control and the blue wedge is indicative of increasing TGFβ concentrations. Data for the 719 TGFβ-responsive genes were extracted from the SSc mouse models and clustered in the array and gene dimensions. Pearson correlations of the 1250ng TGFβ dose and each microarray were calculated and are plotted directly beneath the heatmap. The TGFβ dose response is shown to the left of the heatmap. The highest TGFβ gene expression is observed in 5 week samples from the sclGVHD mouse and 4-week old Tsk2/+ mice. (B) Dendrogram of mouse samples analyzed colored-coded by model. (C) Canonical TGFβ targets COL1A1, WISP1, SPARC, and TIMP1 were found TGFβ-responsive. (D) TGFβ-induced genes highly expressed in the sclGVHD model. (E) Proliferation genes induced by the TGFβ treatment.
We recently demonstrated that PDGF signaling is also increased in the fibroproliferative subset of SSc patients (9). We analyzed the PDGF stimulated gene set from Johnson et al. and find PDGF stimulated gene expression increased in Tsk2/+ mice at 4-weeks of age and in the sclGVHD mouse model (data not shown). PDGF gene expression was not enriched in either the bleomycin-induced fibrosis model, Tsk1/+ or Tsk2/+ at 16-weeks of age.
To confirm the molecular similarities between the patients in the fibroproliferative subset and Tsk2/+ mice we analyzed the expression of the TGFβ-regulated gene tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a; also designated Tweak-R, fibroblast growth factor-inducible-14). Tweak-R is highly expressed in the fibroproliferative subset and was among a set of genes highly correlated to worse skin disease (2). Tweak-R mRNA levels were also found increased in the fibroproliferative subsets of Milano et al. (Figure 5A; p < 0.00001); gene expression differences for Tweak-R (Tnfrsf12a) were confirmed by qRT-PCR (2). Immunohistochemistry analysis of a subset of biopsies from the Milano et al. study (2) showed that TWEAK-R protein levels were increased in the dermis and around epidermal skin appendages of patients of the fibroproliferative and inflammatory subsets (Figure 5B-C; p < 0.05, Mann-Whitney test). The Tsk2/+ animal model has cycles throughout young life while the disease is becoming established. We used this information to select skin samples at peak disease activity for assessment of TWEAK-R expression. We performed immunohistochemistry on sections of skin taken from female Tsk2/+ and their wild-type littermates to detect the levels of TWEAK-R (Figure 5D - E). Tweak-R is induced by TGFβ (36) and there is an ∼1.5-fold increase in numbers of TWEAK-R positive cells at in skin of 4-week-old female Tsk2/+ mice compared to their wild-type littermates (Figure 5D - E; p = 0.013) and in male Tsk2/+ mice compared to their wild-type littermates (not shown). Taken together with the findings of enrichment of TGFβ-signaling primarily in the fibroproliferative skin biopsies of the Milano cohort (5), we believe that Tsk2/+ skin at 4-weeks reflects the biology of this subset of SSc.
Discussion
The identification of mouse models that share gene expression patterns with the fibroproliferative and inflammatory subsets of SSc provides the research community with valuable tools in which to closely examine the molecular mechanisms underlying disease in these subsets. The data presented here are consistent with our earlier findings that sclGVHD mice share underlying mechanistic pathways with the inflammatory subset of SSc (6). Here we have extended those findings by identifying Tsk2/+ mice at 4-weeks of age as a promising model of disease for the fibroproliferative subset. We used multiple approaches to demonstrate parallels in gene expression in this mouse and fibroproliferative skin biopsies, including analysis of individual genes and clusters, examination of global similarities based on co-expression of GO terms, and specifically testing for enrichment of TGFβ-responsive gene expression in both species. PDGF stimulated gene expression has also been shown to underlie the fibroproliferative subset and we find PDGF gene expression also increased in the Tsk2/+ mouse.
The role of TGFβ signaling in fibrosis and as a driver of disease in SSc is well established. An important new finding is the demonstration that Tsk2/+ mice have activation of TGFβ responsive gene expression. We have shown that Tsk2/+ results from an ENU-induced point mutation in the PIIINP fragment of the Col3a1 gene (27). We show that the TGFβ target, Tnfrsf12a (Tweak-R or Fn14) is highly expressed both in SSc patients of this subset and in Tsk2/+ mice at 4-weeks of age. Interestingly, mice deficient for Tweak-R have significantly reduced liver progenitor cell proliferation in response to chemical liver injury (37). Its ligand, Tweak / Tnfsf12, has been implicated as a driver of inflammation and proliferation of fibroblasts / epithelial cells and can contribute to kidney fibrosis (38, 39). The kidneys of Tweak KO mice show a decreased number of myofibroblasts with lower proliferation, and reduced ECM accumulation; mice over-expressing TWEAK have increased kidney fibrosis (38). We propose that a common mechanism underlying the fibroproliferative subset of SSc and the Tsk2/+ model is due in part to TGFβ signaling leading to activation of the Tweak / Fn14 signaling axis.
The deregulation of TGFβ signaling in the bleomycin model has been previously demonstrated (40, 41) and we observe activation of TGFβ signaling here (Supplemental Figure S3). The bleomycin-induced fibrosis model has also been used to advance knowledge of the roles for both TGFβ and IL13 in fibrosis and has been particularly informative for revealing the interactions between these signaling pathways in fibrotic processes (24, 25). The inflammatory gene expression signature dominates the Bleomycin fibrosis signature and the clustering results. This is consistent with data showing that mice lacking a functional inflammasome are not susceptible to bleomycin-induced fibrosis (42-45). Our data suggest the bleomycin-induced skin fibrosis model shows similarities to the early inflammatory response observed in patients and shows both TGFβ and IL13 signatures, both of which are implicated in SSc pathogenesis.
The resemblance of the Tsk1/+ mouse to the SSc subsets is not clear or consistent with prior findings (46). We found no significant parallels of gene expression in skin of these animals at 6-weeks of age with the molecular subsets of SSc. Detailed analysis of Tsk1/+ at other time points might show stronger similarities to gene expression in SSc skin. The Genomica module map analysis did not demonstrate any contribution to gene expression patterns by B-lymphocytes in Tsk1/+ as has been previously reported. It is possible that the reported reliance of the phenotype in these mice on functional IL4 signaling (47) is relevant to the inflammatory or limited subsets, however the GO term analysis implemented in these studies is not sensitive enough to draw definitive conclusions from these data.
Previous comparisons of human SSc and human cGVHD at the histological level show key similarities as well as distinct differences. Fleming et al. compared human cGVHD to human SSc histologically (48) and showed a similar extent of fibrosis, increased hyaluronan staining, myofibroblasts numbers and intimal hyperplasia of the microvasculature including increased smooth muscle cell layers. SSc and cGVHD showed positive dermal KI67 staining. The major difference was that SSc showed differences in the pattern of fibrotic change and a decrease in staining for endothelial cell markers indicating capillary rarefaction, not found in cGVHD.
This study has several limitations. The bleomycin-induced fibrosis model results are likely specific to the protocol used here. In addition, the TGFβ pump model captures only a single time point and this cytokine likely induces changes in a time dependent-manner. We note that the proliferating cells observed in SSc are mainly located in the epidermis with some proliferating cells around the vasculature and other epidermal skin appendages (2, 48). Since the structure of mouse skin, particularly the epidermis, is quite different in mice, this could limit interpretation of the Tsk2/+ data and mapping of our subsets.
With few tools available for studying disease mechanisms in this poorly understood disease, the benefits of the results of this study are clear. By using gene expression as a readily quantifiable phenotype in skin, we have shown that the Tsk/2+ mouse, the sclGVHD and bleomycin-induced fibrosis mouse models are similar to their respective human subsets not by gross morphology approximations, but by robust genome-scale molecular measures. This means it is the expression of thousands of genes, rather than any single gene, that drives these results. Additionally, identification of the pathways similarly deregulated in human SSc subsets and in mouse models, provides new urgently needed tools for optimizing development of therapeutics that specifically target each of the intrinsic subsets of scleroderma. The mechanisms in these mouse models can be used to identify drugs for SSc patients in the clinic and target their molecular subsets. The development of a routine diagnostic for SSc patients as well as drugs that target each subset will greatly facilitate treatment of this disease.
Supplementary Material
Acknowledgments
Funding Sources: This work has been supported by grants from: the Scleroderma Research Foundation (SRF www.srfcure.org) to MLW, AOA, and JV; National Institutes of Health (NIH; www.nih.gov) grants R01AR061384-01 to MLW, EPB, CA, NIH 2R01AR051089 and P30AR061271 to RL; and PR100338 from the Department of Defense (DoD; www.defense.gov; MLW, EPB, CA). Funding agencies have had no role in the interpretation of the data or in writing the manuscript.
Footnotes
The authors report no conflict of interest with regards to this work.
References
- 1.Christner PJ, Jimenez SA. Animal models of systemic sclerosis: insights into systemic sclerosis pathogenesis and potential therapeutic approaches. Curr Opin Rheumatol. 2004;16(6):746–52. doi: 10.1097/01.bor.0000137893.68929.86. [DOI] [PubMed] [Google Scholar]
- 2.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3(7):e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132(5):1363–73. doi: 10.1038/jid.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinchcliff ME, Huang CC, Wood TA, Mahoney JM, Martyanov V, Bhattacharya S, et al. Molecular Signatures in Skin Associated with Clinical Improvement During Mycophenolate Treatment in Systemic Sclerosis. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.130. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sargent JL, Milano A, Bhattacharyya S, Varga J, Connolly MK, Chang HY, et al. A TGFbeta-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2009;130(3):694–705. doi: 10.1038/jid.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenblatt MB, Sargent JL, Farina G, Tsang K, Lafyatis R, Glimcher LH, et al. Interspecies Comparison of Human and Murine Scleroderma Reveals IL-13 and CCL2 as Disease Subset-Specific Targets. Am J Pathol. 2012 doi: 10.1016/j.ajpath.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson ME, Mahoney JM, Taroni J, Sargent JL, Marmarelis E, Wu MR, et al. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS One. 2015;10(1):e0114017. doi: 10.1371/journal.pone.0114017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsumoto TR, Whitfield ML, Connolly MK. The pathogenesis of systemic sclerosis. Annual review of pathology. 2011;6:509–37. doi: 10.1146/annurev-pathol-011110-130312. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ME, Pioli PA, Whitfield ML. Gene expression profiling offers insights into the role of innate immune signaling in SSc. Seminars in immunopathology. 2015 doi: 10.1007/s00281-015-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahoney JM, Taroni J, Martyanov V, Wood TA, Greene CS, Pioli PA, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol. 2015;11(1):e1004005. doi: 10.1371/journal.pcbi.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barisic-Dujmovic T, Boban I, Clark SH. Regulation of collagen gene expression in the Tsk2 mouse. J Cell Physiol. 2008;215(2):464–71. doi: 10.1002/jcp.21319. [DOI] [PubMed] [Google Scholar]
- 12.Bayle J, Fitch J, Jacobsen K, Kumar R, Lafyatis R, Lemaire R. Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Invest Dermatol. 2008;128(4):871–81. doi: 10.1038/sj.jid.5701101. [DOI] [PubMed] [Google Scholar]
- 13.Wu M, Melichian DS, Chang E, Warner-Blankenship M, Ghosh AK, Varga J. Rosiglitazone abrogates bleomycin-induced scleroderma and blocks profibrotic responses through peroxisome proliferator-activated receptor-gamma. Am J Pathol. 2009;174(2):519–33. doi: 10.2353/ajpath.2009.080574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36(Database issue):D724–8. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20(1):105–14. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 16.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34(2):166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 19.Adamson IY, Bowden DH. The pathogenesis of bloemycin-induced pulmonary fibrosis in mice. Am J Pathol. 1974;77(2):185–97. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Shi W, Wang YL, Chen H, Bringas P, Jr, Datto MB, et al. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol. 2002;282(3):L585–93. doi: 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- 21.Christner PJ, Siracusa LD, Hawkins DF, McGrath R, Betz JK, Ball ST, et al. A high-resolution linkage map of the tight skin 2 (Tsk2) locus: a mouse model for scleroderma (SSc) and other cutaneous fibrotic diseases. Mammalian genome : official journal of the International Mammalian Genome Society. 1996;7(8):610–2. doi: 10.1007/s003359900181. [DOI] [PubMed] [Google Scholar]
- 22.Christner PJ, Peters J, Hawkins D, Siracusa LD, Jimenez SA. The tight skin 2 mouse. An animal model of scleroderma displaying cutaneous fibrosis and mononuclear cell infiltration. Arthritis Rheum. 1995;38(12):1791–8. doi: 10.1002/art.1780381212. [DOI] [PubMed] [Google Scholar]
- 23.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35(3):337–46. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita M, Yamamoto T, Nishioka K. Upregulation of interleukin-13 and its receptor in a murine model of bleomycin-induced scleroderma. Int Arch Allergy Immunol. 2004;135(4):348–56. doi: 10.1159/000082331. [DOI] [PubMed] [Google Scholar]
- 25.Aliprantis AO, Wang J, Fathman JW, Lemaire R, Dorfman DM, Lafyatis R, et al. Transcription factor T-bet regulates skin sclerosis through its function in innate immunity and via IL-13. Proc Natl Acad Sci U S A. 2007;104(8):2827–30. doi: 10.1073/pnas.0700021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 27.Long KB, Li Z, Burgwin CM, Choe SG, Martyanov V, Sassi-Gaha S, et al. The Tsk2/+ mouse fibrotic phenotype is due to a gain-of-function mutation in the PIIINP segment of the Col3a1 gene. J Invest Dermatol. 2015;135(3):718–27. doi: 10.1038/jid.2014.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greidinger EL, Flaherty KT, White B, Rosen A, Wigley FM, Wise RA. African-American race and antibodies to topoisomerase I are associated with increased severity of scleroderma lung disease. Chest. 1998;114(3):801–7. doi: 10.1378/chest.114.3.801. [DOI] [PubMed] [Google Scholar]
- 29.Hu PQ, Fertig N, Medsger TA, Jr, Wright TM. Correlation of serum anti-DNA topoisomerase I antibody levels with disease severity and activity in systemic sclerosis. Arthritis Rheum. 2003;48(5):1363–73. doi: 10.1002/art.10977. [DOI] [PubMed] [Google Scholar]
- 30.Steen VD. Autoantibodies in systemic sclerosis. Semin Arthritis Rheum. 2005;35(1):35–42. doi: 10.1016/j.semarthrit.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Hanke K, Dahnrich C, Bruckner CS, Huscher D, Becker M, Jansen A, et al. Diagnostic value of anti-topoisomerase I antibodies in a large monocentric cohort. Arthritis Res Ther. 2009;11(1):R28. doi: 10.1186/ar2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimer G, Rose KM, Scheer U, Tan EM. Autoantibody to RNA polymerase I in scleroderma sera. J Clin Invest. 1987;79(1):65–72. doi: 10.1172/JCI112809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleischmajer R, Damiano V, Nedwich A. Alteration of subcutaneous tissue in systemic scleroderma. Arch Dermatol. 1972;105(1):59–66. [PubMed] [Google Scholar]
- 34.Ghosh AK, Bhattacharyya S, Lakos G, Chen SJ, Mori Y, Varga J. Disruption of transforming growth factor beta signaling and profibrotic responses in normal skin fibroblasts by peroxisome proliferator-activated receptor gamma. Arthritis Rheum. 2004;50(4):1305–18. doi: 10.1002/art.20104. [DOI] [PubMed] [Google Scholar]
- 35.Aoki Y, Maeno T, Aoyagi K, Ueno M, Aoki F, Aoki N, et al. Pioglitazone, a peroxisome proliferator-activated receptor gamma ligand, suppresses bleomycin-induced acute lung injury and fibrosis. Respiration. 2009;77(3):311–9. doi: 10.1159/000168676. [DOI] [PubMed] [Google Scholar]
- 36.Milks MW, Cripps JG, Lin H, Wang J, Robinson RT, Sargent JL, et al. The role of Ifng in alterations in liver gene expression in a mouse model of fulminant autoimmune hepatitis. Liver Int. 2009;29(9):1307–15. doi: 10.1111/j.1478-3231.2009.02028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakubowski A, Ambrose C, Parr M, Lincecum JM, Wang MZ, Zheng TS, et al. TWEAK induces liver progenitor cell proliferation. J Clin Invest. 2005;115(9):2330–40. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ucero AC, Benito-Martin A, Fuentes-Calvo I, Santamaria B, Blanco J, Lopez-Novoa JM, et al. TNF-related weak inducer of apoptosis (TWEAK) promotes kidney fibrosis and Ras-dependent proliferation of cultured renal fibroblast. Biochim Biophys Acta. 2013;1832(10):1744–55. doi: 10.1016/j.bbadis.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 39.Zhu LX, Zhang HH, Mei YF, Zhao YP, Zhang ZY. Role of tumor necrosis factor-like weak inducer of apoptosis (TWEAK)/fibroblast growth factor-inducible 14 (Fn14) axis in rheumatic diseases. Chinese medical journal. 2012;125(21):3898–904. [PubMed] [Google Scholar]
- 40.Lakos G, Takagawa S, Chen SJ, Ferreira AM, Han G, Masuda K, et al. Targeted disruption of TGF-beta/Smad3 signaling modulates skin fibrosis in a mouse model of scleroderma. Am J Pathol. 2004;165(1):203–17. doi: 10.1016/s0002-9440(10)63289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago B, Gutierrez-Canas I, Dotor J, Palao G, Lasarte JJ, Ruiz J, et al. Topical application of a peptide inhibitor of transforming growth factor-beta1 ameliorates bleomycin-induced skin fibrosis. J Invest Dermatol. 2005;125(3):450–5. doi: 10.1111/j.0022-202X.2005.23859.x. [DOI] [PubMed] [Google Scholar]
- 42.Gasse P, Mary C, Guenon I, Noulin N, Charron S, Schnyder-Candrian S, et al. IL-1R1/MyD88 signaling and the inflammasome are essential in pulmonary inflammation and fibrosis in mice. J Clin Invest. 2007;117(12):3786–99. doi: 10.1172/JCI32285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Artlett CM, Sassi-Gaha S, Rieger JL, Boesteanu AC, Feghali-Bostwick CA, Katsikis PD. The inflammasome activating caspase 1 mediates fibrosis and myofibroblast differentiation in systemic sclerosis. Arthritis Rheum. 2011;63(11):3563–74. doi: 10.1002/art.30568. [DOI] [PubMed] [Google Scholar]
- 44.Artlett CM. The Role of the NLRP3 Inflammasome in Fibrosis. The open rheumatology journal. 2012;6:80–6. doi: 10.2174/1874312901206010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derk CT, Artlett CM, Jimenez SA. Morbidity and mortality of patients diagnosed with systemic sclerosis after the age of 75: a nested case-control study. Clin Rheumatol. 2006:1–4. doi: 10.1007/s10067-005-0177-y. [DOI] [PubMed] [Google Scholar]
- 46.Baxter RM, Crowell TP, McCrann ME, Frew EM, Gardner H. Analysis of the tight skin (Tsk1/+) mouse as a model for testing antifibrotic agents. Lab Invest. 2005;85(10):1199–209. doi: 10.1038/labinvest.3700331. [DOI] [PubMed] [Google Scholar]
- 47.Ong CJ, Ip S, Teh SJ, Wong C, Jirik FR, Grusby MJ, et al. A role for T helper 2 cells in mediating skin fibrosis in tight-skin mice. Cell Immunol. 1999;196(1):60–8. doi: 10.1006/cimm.1999.1537. [DOI] [PubMed] [Google Scholar]
- 48.Fleming JN, Shulman HM, Nash RA, Johnson PY, Wight TN, Gown A, et al. Cutaneous chronic graft-versus-host disease does not have the abnormal endothelial phenotype or vascular rarefaction characteristic of systemic sclerosis. PLoS One. 2009;4(7):e6203. doi: 10.1371/journal.pone.0006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


