Abstract
In 2012, an unprecedented number of four distinct, partially overlapping filovirus-associated viral hemorrhagic fever outbreaks were detected in equatorial Africa. Analysis of complete virus genome sequences confirmed the reemergence of Sudan virus and Marburg virus in Uganda, and the first emergence of Bundibugyo virus in the Democratic Republic of the Congo.
Keywords: Filoviridae, Bundibugyo virus, Sudan virus, Marburg virus, Ebola virus
Results and discussion
Several viruses within the Filoviridae family, including Ebola virus (EBOV), Sudan virus (SUDV), Taï Forest virus (TAFV), Bundibugyo virus (BDBV), Marburg virus (MARV) and Ravn virus (RAVV), cause severe viral hemorrhagic fevers (VHFs) with high case-fatality (Hartman et al., 2010) in several equatorial African countries. A surveillance program to detect VHFs in Uganda was formally established in 2010 by the Centers for Disease Control and Prevention (CDC), Atlanta, USA, in collaboration with the Uganda Virus Research Institute (UVRI) and the Uganda Ministry of Health (UMoH) (MacNeil et al., 2011). Routine serologic and molecular diagnostic tests for various causative agents of VHF are performed on suspected case specimens submitted to the VHF reference laboratory located at UVRI, Entebbe. Between July and October 2012, alert cases were identified in Uganda or reported from the Democratic Republic of the Congo (DRC), and specimens were received by the CDC/UVRI VHF Program (Table 1). Four different filovirus disease outbreaks were initially identified by testing clinical samples via antigen capture, IgM ELISA, and/or qRT-PCR, using methods that were described before (Shoemaker et al., 2011; Towner et al., 2006, 2008). Subsequent virus isolation and complete viral genome sequencing studies were carried out at the CDC in Atlanta. Samples from patients with fatal and nonfatal outcomes that tested positive by qRT-PCR and showed low Ct values were chosen randomly for sequencing full viral genomes.
Table 1.
Locations of outbreaks, dates outbreaks were confirmed and number of confirmed cases.
| Location | Virus | Date of laboratory confirmation |
Last positive laboratory test (RT-PCR) |
# of days from detection of 1st case to last case |
# of laboratory confirmed cases (# of deaths) |
|---|---|---|---|---|---|
| Kibaale, UGA | SUDV | July 26th, 2012 | Aug 5th, 2012 | 11 | 11 (4) |
| Isiro, DRC | BDBV | Aug 16th, 2012 | Oct 11th, 2012 | 57 | 36 (13) |
| Kabale, UGA; Ibanda, UGA | MARV | Oct 18th, 2012 | Nov 7th, 2012 | 21 | 15 (4) |
| Luwero, UGA | SUDV | Nov 13th, 2012 | Nov 17th, 2012 | 5 | 6 (3) |
After each filovirus disease outbreak was identified, many partner organizations were involved in outbreak response, including UVRI, UMoH, CDC, Médecins Sans Frontières (MSF), DRC Ministry of Health (DRCMoH), Public Health Agency of Canada (PHAC) and World Health Organization (WHO). These organizations collaborated to establish epidemiological investigations, contact tracing, case management, logistical support, and patient isolation facilities. A mobile laboratory was established by CDC for the Ebola VHF outbreak in Isiro, DRC (this laboratory was maintained in the late stages of the outbreak by the staff from the Public Health Agency of Canada). Similarly, CDC established a mobile laboratory in Kabale, Uganda, during the Marburg virus disease outbreak. These field laboratories were necessary because of the considerable distance between the outbreak epicenters and the VHF laboratory in Entebbe. The mobile laboratories performed daily molecular testing on acute suspect cases and on confirmed cases in isolation wards to ensure patients were not viremic when they were discharged. The dates of laboratory confirmation of outbreaks, and time to the last acute case being laboratory confirmed are shown in Table 1. Once identified, these outbreaks were kept relatively small and quickly brought under control, relative to many of the past filovirus disease outbreaks. The ability to rapidly identify acute cases due to the availability of in-country filovirus diagnostics likely contributed to these successful control efforts. Complete reports on the epidemiologic and clinical aspects of the outbreaks will be presented elsewhere.
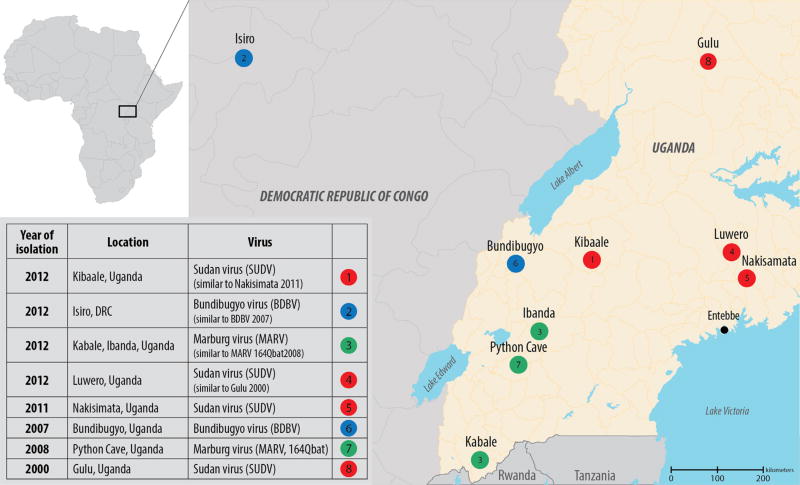
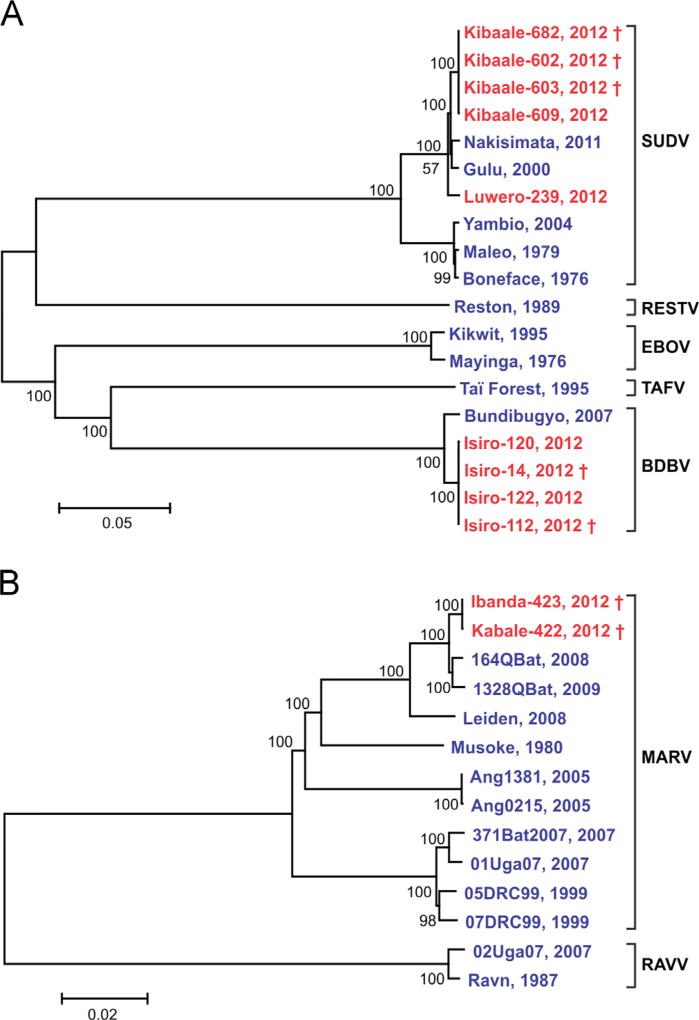
Samples from VHF alert cases in the Kibaale District of western Uganda were received at the CDC/UVRI laboratory in July 2012 (Fig. 1). Differential molecular testing of clinical samples identified the outbreak as Ebola virus disease associated with SUDV. Precise identification of the virus as SUDV was achieved at CDC in Atlanta by sequencing the complete genome of the virus directly from the clinical samples. Genomic analysis showed ~99.9% sequence identity among the virus genomes detected in the four acute case serum samples. Finding of nearly identical virus sequences is compatible with a single spillover event from the virus reservoir into the human population, with subsequent limited waves of human-to-human transmission. Interestingly, these sequences also showed a high identity (~99.2%) with the SUDV Nakisimata isolate (Shoemaker et al., 2011), detected in 2011 in the Luwero District, Uganda, and with the SUDV Gulu isolate (Towner et al., 2004) found in the Gulu District of northern Uganda in 2000 (Fig. 2). By the end of the outbreak (officially declared on October 4th, 2012) a total of 11 cases had been confirmed by molecular and serological testing.
Fig. 1.
Map of the Democratic Republic of the Congo (DRC) and Uganda showing the locations of filovirus-caused viral hemorrhagic fever (VHF) outbreaks in 2012. Isolation sites of the most genetically similar viruses from previous years are also shown.
Fig. 2.
Phylogenetic trees comparing representative full-length genomes of ebolavirus (A) and marburgvirus (B). Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011), using the neighbor-joining method. Bootstrap values listed at the nodes provide statistical support for 500 replicates. Scale bar indicates the number of substitutions per site.
Genomes from 2012 are in red; † indicates fatal cases. Genbank accession numbers used in A are: FJ968794, EU338380, NC_014373, FJ217162, NC_004161, JN638998, NC_006432, AF086833, AY354458, and KC242783. SUDV sequences from 2012 outbreaks are: KC545389, KC545390, KC545391, KC545392 and KC589025. BDBV sequences from 2012 outbreaks are: KC545393, KC545394, KC545395, and KC545396. Genbank accession numbers used in B are: DQ447654, JX458858, FJ750958, JX458853, JN408064, FJ750957, DQ447649, DQ217792, DQ447658, DQ447650, DQ447651, and FJ750953. MARV sequences from 2012 outbreaks are: KC545387 and KC545388.
In mid-August 2012, samples from VHF alert cases in Isiro and Dungu Health Zones, Province Orientale in northeastern DRC, were dispatched to CDC/UVRI by MSF Switzerland staff working in that region (Fig. 1). The initial diagnostics and subsequent full genome sequence analysis confirmed the presence of BDBV in the clinical samples from the Isiro Health Zone, but not the Dungu area. The analysis showed that the virus genomes present in the Isiro clinical samples were ~98.6% identical to those of the original BDBV isolated in the Bundibugyo District of western Uganda in 2007 (Towner et al., 2008). The high degree of sequence identity (~99.2%) found among the viruses detected in four serum samples from acute cases was again suggestive of a single spillover event into the human population with subsequent human-to-human transmission. A total of 36 cases were confirmed by the end of the outbreak (officially declared on November 26th, 2012).
In October 2012, samples were received by CDC/UVRI from VHF alert cases in the Kabale and Ibanda Districts of southwestern Uganda (Fig. 1). Initial testing at CDC/UVRI identified this as a Marburg virus disease outbreak. During the following 3 weeks, a total of 15 additional cases were confirmed in the districts of Kabale, Ibanda, Mbarara, and Kampala (Fig. 1, Table 1) based on testing at the CDC mobile laboratory in Kabale and/or serology at the VHF laboratory at CDC/UVRI. Preliminary epidemiological investigations identified two chains of transmission: (1) a case infected in Ibanda and spread to persons in Kabale, some of whom sought care in Kampala, and (2) cases in Ibanda, some of whom sought care in Mbarara. No epidemiologic data could be found to link these two clusters, however, sequence analysis of complete viral genomes in serum samples from one acute case from each cluster showed nearly identical sequences (~99.9%) indicating that these cases were likely part of the same human-to-human transmission chain. Moreover, these viral genome sequences were highly similar (~99.3%) to two MARV isolates previously found in Rousettus aegyptiacus bats (Fig. 2) captured in 2008 and 2009 in the nearby Python Cave in Queen Elizabeth National Park, southwest Uganda (Amman et al., 2012). UMoH officially declared the end of the outbreaks in Kabale and Ibanda Districts on December 21st, 2012.
In November 2012, the UVRI/CDC laboratory received samples from VHF alert cases in the Luwero District, Uganda and identified this as another SUDV outbreak. Over the next few weeks, a total of six cases were confirmed from the relatively close districts of Luwero, Jinja, and Nakasongola. There were only 5 days between the laboratory diagnosis of the first and last acute cases (Table 1). Nearly identical (~100%) virus NP gene sequences were found for the viruses detected in three serum samples, suggesting a single chain of human-to-human transmission. Moreover, the full-length genomic sequence obtained from a viral isolate showed a closest identity to the SUDV Gulu strain isolated in Uganda in 2000 (Towner et al., 2004), with the SUDV Kibaale isolates identified earlier in 2012 being more distant. The end of the outbreak was officially declared on Jan 16th, 2013.
Conclusions
In summary, continuous VHF surveillance, established by CDC, UVRI, and UMoH, combined with rapid laboratory testing, allowed the identification and confirmation of four filovirus disease outbreaks during the second half of 2012. Following initial filovirus identification by conventional serological methods and qRT-PCR, sequence characterization of complete viral genomes from clinical samples confirmed the reemergence of SUDV and MARV in Uganda, and the first emergence of BDBV in DRC. Prior to 2012, BDBV had only been detected in a single outbreak in western Uganda in 2007 (Towner et al., 2008).
Several lines of evidence strongly indicate that bats are a likely natural reservoir for EBOV and MARV (Amman et al., 2012; Leroy et al., 2005; Pourrut et al., 2007; Towner et al., 2007, 2009). Indeed, MARV and Ravn viruses have been repeatedly isolated from the common African fruit bat, R. aegyptiacus (Amman et al., 2012; Towner et al., 2009). Moreover, the seasonal pattern of MARV circulation in R. aegyptiacus bats has been tentatively linked to a high risk of infection in humans (Amman et al., 2012). Interestingly, the timing of this Marburg virus disease outbreak in Uganda coincides with the second annual virus pulse in bat populations. Ongoing ecological investigations are currently underway to examine the potential origins of the virus spillover events initiating these recent outbreaks. Furthermore, our sequence analysis indicated the reemergence of SUDV as two single, but independent chains of transmission events in July and October 2012.
Acknowledgments
We thank all people who participated in the international multi-agency outbreak response in Uganda and DRC in 2012, including the staff from MSF Spain, Belgium, Switzerland, and France; WHO; UMoH; DRCMoH; Public Health Agency of Canada, and CDC-Atlanta. We also thank Edmund Newman (Health Protection Agency, Porton Down) for supporting the mobile laboratory in Kabale. We acknowledge the medical staff that provided care for patients. We also thank Tatyana Klimova for her assistance during the editing process of this manuscript, Craig Manning for the map design, and Joel Vincent for the assistance during the sequence assembly.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8(10):e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Towner JS, Nichol ST. Ebola and Marburg hemorrhagic fever. Clin. Lab. Med. 2010;30(1):161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–5761. doi: 10.1038/438575a. 438 (7068) 575–576. [DOI] [PubMed] [Google Scholar]
- MacNeil A, Farnon EC, Morgan OW, Gould P, Boehmer TK, Blaney DD, et al. Filovirus outbreak detection and surveillance: lessons from Bundibugyo. J. Infect. Dis. 2011;204(Suppl 3):S761–S767. doi: 10.1093/infdis/jir294. [DOI] [PubMed] [Google Scholar]
- Pourrut X, Delicat A, Rollin PE, Ksiazek TG, Gonzalez JP, Leroy EM. Spatial and temporal patterns of Zaire ebolavirus antibody prevalence in the possible reservoir bat species. J. Infect. Dis. 2007;196(Suppl 2):S176–S183. doi: 10.1086/520541. [DOI] [PubMed] [Google Scholar]
- Shoemaker T, MacNeil A, Balinandi S, Campbell S, Wamala JF, McMullan LK, et al. Reemerging Sudan Ebola virus disease in Uganda. Emerg. Infect. Dis. 2011;18(9):1480–1483. doi: 10.3201/eid1809.111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 2004;78(8):4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 2006;80(13):6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Pourrut X, Albarino CG, Nkogue CN, Bird BH, Grard G, et al. Marburg virus infection detected in a common African bat. PLoS ONE. 2007;2(1):e764. doi: 10.1371/journal.pone.0000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4(11):e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5(7):e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]