Abstract
Background
We investigated the single nucleotide polymorphism (SNP) context of a previously identified periodontitis-associated locus and examined its association with microbial, biological and periodontal disease clinical parameters.
Methods
We annotated a 200Kb-spanning region of 1q12 previously highlighted in a genome-wide association scan among 4,910 European American individuals (SNP rs1633266). Two haplotype blocks were selected. We examined the association of these polymorphisms with data on microbial plaque composition, gingival crevicular fluid (GCF)-interleukin (IL)-1β levels and clinical parameters of periodontal disease. Descriptive analysis of IFI16 and AIM2 protein expression in gingival tissues from healthy (n=2) and chronic periodontitis individuals (n=2) was done via immunohistochemistry.
Results
The highlighted locus is a 100Kb region containing the interferon gamma-inducible protein 16 (IFI16) and absent in melanoma 2 (AIM2) genes. Two haplotype blocks rs6940 and rs1057028 were significantly associated with increased extent bleeding on probing and levels of microorganisms Porphyromonas gingivalis, Tannerella forsythia and Campylobacter rectus (p≤0.05). Haplotype block rs1057028 was also significantly associated with pathogens Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans, increased GCF-IL-1β levels and extent of probing depth≥4mm (p≤0.05). Prevalence of severe periodontitis (biofilm-gingival interface-P3 classification) was positively associated with haplotype block rs1057028. Similar trends were observed for haplotype block rs1057028. IFI16 and AIM2 protein expression was observed in multiple cell types of gingival tissues, including inflammatory cells.
Conclusion
This study found IFI16 and AIM2 SNPs associated with higher levels periodontal microorganisms and increased percentage of periodontal disease clinical parameters, suggesting the need for functional studies and additional fine-mapping of variants in the 1q12-locus.
Keywords: Pathogenesis of periodontal disease(s), genetics, host response
MeSH terms: Periodontitis, Innate Immune Response, Polymorphism, Genetic
INTRODUCTION
Periodontal disease is polygenic condition of the tooth supporting structures.1–4 Early studies among monozygotic and dizygotic twins showed that 33–48% of the variance in periodontal disease expression was attributable to genetics.1, 5 Notably, alterations in genes encoding proteins involved in the immune response are shown to influence the host microbiota and increase periodontal clinical parameters of disease. Individuals with variants in interleukin (IL)-1α, IL-1β and IL-6 are shown to have a unique periodontal microbiome with high levels of classical “red” and “orange” complex species that are known to be significantly associated with periodontal inflammation.6 IL-6 polymorphisms have also been moderately associated with the diagnosis of periodontitis.7 In addition, individuals with single gene mutations of β2 integrins leading to Leukocyte Adhesion Deficiency-1 show increased bacterial loads, decreased complexity of biofilms and severe periodontal bone loss.8 Together, the evidence supports the concept that genetic alterations controlling the immune response of the host can lead to alterations of microbial communities and predispose individuals to periodontal disease.
Genome-wide association studies (GWAS) and candidate gene studies have been used in attempt to identify single nucleotide polymorphisms (SNP)s that either contribute to the pathogenesis and/or risk of developing periodontal disease. To date, 4 studies have conducted GWAS analysis to identify SNPs associated with the American Academy of Periodontology (AAP)9 definition of chronic periodontal disease.10–13 No single marker met the genome-wide significance criteria, although four loci [ninein (NIN), abhydrolase domain containing 12B (ABD12B), WAS protein homolog associated with actin, golgi membranes and microtubules (WHAMM), adaptor-related protein complex-3 beta-2 subunit (AP3B2)] met gene-centric statistical significance criteria.12 Therefore, a new approach was utilized in order to identify SNPs that were relevant to the pathogenesis of periodontal disease. This approach utilized a combination of the levels of 8 classical periodontal pathogens and gingival crevicular fluid (GCF) IL-1β to derive periodontal complex traits (PCTs) via principal component analysis.14 The objective of this approach is for identifying loci related to the biological background and pathogenesis of periodontal disease. Approximately 2.5 million markers were evaluated among 975 European American adults. Several traits were derived by this analysis, with each trait having different eigenvalues (loadings) of the 8 microorganisms and GCF-IL-1β. PCT1 (named the Socransky Trait) was defined by a microbial community structure with high positive loadings of all periodontal pathogens, and correlated with clinical measurements of periodontal disease.15 Six loci were associated with PCT1, including C-Type Lectin Domain Family 19 Member A (CLEC19A), T-Cell Receptor Alpha Locus (TRA), Glycoprotein, Alpha-Galactosyltransferase 2, Pseudogene (GGTA2P), Transmembrane 9 Superfamily Member 2 (TM9SF2), RNA Binding Motif Single Stranded Interacting Protein 3 (RBMS3) and interferon (IFN) gamma-inducible protein 16 (IFI16)/absent in melanoma (AIM)2.14 The clinical, microbial and biological characterization of individuals with SNP variants in these 6 individual loci is currently unknown. This present study further investigates IFI16/AIM2 loci using bioinformatics, clinical, microbial and biological data.
Both IFI16 and AIM2 are members of the IFN-inducible PYHIN protein family that contain C-terminal DNA-binding hematopoietic expression, interferon-inducible nature, and nuclear localization (HIN) domain(s) and an N-terminal Pyrin domain (PYD) that belongs to the death domain superfamily of signaling molecules.16, 17 Both IFI16 and AIM2 are intracellular recognition sensors that trigger inflammatory responses against DNA from the host and microorganisms.17 Increased expression of AIM2 has been reported in a number of inflammatory diseases, including psoriasis, atopic dermatitis, venous ulcers, inflammatory disease and periodontitis suggesting involvement with inflammation.18–22 Expression of IFI16 in inflammatory diseases is less explored but increased expression is reported in inflammatory bowel disease.22 To our knowledge, no study has explored the expression of IFI16 in periodontal tissues. Because of the critical role of these proteins in innate immunity, the purpose of this study was to evaluate the relationship between SNPs in the IFI16 and AIM2 loci with periodontal microorganisms, levels of GCF-IL-1β and clinical parameters of periodontal disease. Meanwhile, descriptive analysis of IFI16 and AIM2 protein expression in gingival samples derived from healthy and individuals with periodontal disease showed expression in multiple cells, including inflammatory cells. Conclusively, our previous14 and present study supports that variants in IFI16/AIM2 are associated with increased loads of periodontal pathogens and increased parameters of clinical disease.
MATERIALS AND METHODS
GWAS population
A total of 4,910 Northern European descendants were enrolled in the Dental Atherosclerosis Risk in Communities (DARIC) Cohort as described.23, 24 Blood was collected as described for genotyping for ~2.5 million markers.10 GCF was collected at 4 gingival sampling areas from the mesio-buccal region of each first molar from each individual and stored for further analysis of IL-1β levels.23 Plaque samples were collected from the subgingival mesio-buccal site of the maxillary right first molar, and stored for further DNA whole chromosomal checkerboard for the 8 periodontal pathogens. Periodontal measurements (n=4,766) in all teeth at 6 sites per tooth were collected and included number of missing teeth, gingival index (GI), plaque index (PI), probing depths (PD), clinical attachment level (CAL) and bleeding on probing (BOP). All sites were examined by trained and calibrated examiners with >90% agreement.
Bioinformatic approaches
Initial analysis of genome-wide imputed SNP data was done with a software package.25 The results of that analysis revealed one genome-wide significant SNP in the IFI16 region (lead SNP rs1633266). We then identified and visualized markers in linkage disequilibrium with this polymorphism.14,26,*The criteria used to prioritize and select SNPs of interest for this analysis were: a) a statistical significance criterion (p<5×10−5 considered as ‘suggestive’ evidence of association), b) biological plausibility of genes in the region, c) functional significance and d) linkage disequilibrium, the non-random association in the occurrence of alleles at two loci, represented by the square of the correlation coefficient between two indicator variables (r2). SNPs with suggestively association (p<5×10−5) with PCT1 were selected and were then carried forward for screening of missense SNPs with predicted functional protein damage.27–29 Using these criteria, we identified 2 SNPs of interest, rs6940 and rs1057028 located in the IFI16 region. We then searched and evaluated SNPs in perfect linkage disequilibrium (Northern and Western European ancestry panel, r2=1, D′=1 cutoff) with these 2 markers as previously described.30,† We further gathered protein information and performed alignment of the different IFI16 isoforms [clustal 0 (1.1.1) multiple sequence alignment].31,‡ The SNPs within these loci were carried forward to tests of association with clinical and biological parameters, including plaque microorganisms, GCF-IL-1β levels, periodontal clinical measurements and biofilm-gingival interface (BGI)-periodontal disease classification.32 The BGI classification was selected because it defines biological phenotypes based on 8 periodontal pathogens, serum immunoglobulin (Ig)G (17 bacteria), 16 GCF-mediators, PD and BOP, representing the individual’s current disease activity and inflammatory condition. Previous GWAS studies from our group have already shown that traditional AAP/ADA periodontal disease classification9 that utilize CAL, a measurement of the loss of tissue or history disease, does not allow identification of groups with similar biological characteristics.10, 14
Plaque microbial analysis
A subset of 909 participants of the DARIC cohort was evaluated for plaque microbial composition using DNA-DNA checkerboard as previously described.32 One plaque sample was used from each individual and assayed by DNA checkerboard for the 8 periodontal pathogens. Microorganism levels were expressed as counts using known microbial standards for Porphyromonas gingivalis (Pg), Prevotella intermedia (Pi), Treponema denticola (Td), Tannerella forsythia (Tf), Campylobacter rectus (Cr), Fusobacterium nucleatum (Fn), Aggregatibacter actinomycetemcomitans (Aa) and Prevotella nigrescens (Pn). The total counts reflect a sum of these targeted pathogens for each individual.
Gingival crevicular fluid IL-1β levels
Four GCF strips were eluted and analyzed separately for each individual (n=4,407). IL-1β levels were evaluated by enzyme-linked immune-absorbent assay (ELISA) according to the manufacturer’s instructions as previously described.33 GCF analyte concentration data were pooled to provide a mean value for each individual in ng/mL.
Sample collection for immunohistochemistry
To describe the tissue distribution of IFI16 and AIM2 in human gingival tissues, gingival biopsies were taken from 2 individuals with healthy periodontium and 2 with chronic periodontal disease according to the AAP/ADA classification.9 All enrolled participants into this study provided written informed consent, which was approved by the Institutional Review Board (IRB) of the University of North Carolina at Chapel Hill. Major exclusion criteria included symptom of any systemic disease, antibiotic use within 1 month prior to the examination, or medical treatment for any known disease that is associated with periodontal disease within the last 3 months. Gingival biopsies were harvested either during routine periodontal flap surgeries from participants clinically diagnosed with chronic periodontitis or crown lenthening surgeries and healthy volunteers. A tissue biopsy sample (~3mm×4mm) was removed from underneath the papillae, buccally or lingually, to include the col area of depth of the osseous crest. Upon removal, gingival tissues were fixed in 10% neutral buffered formalin overnight, dehydrated (70% alcohol), and embedded in paraffin for the immunohistochemical procedure.
Immunohistochemistry
Gingival tissue sections (5μm thick) were obtained in the sagittal direction including the epithelial and connective tissues. The slides were stained with rabbit polyclonal anti-IFI16* and rabbit polyclonal anti-AIM2†. Anti-rabbit horseradish peroxidase (HRP)-DAB staining‡ was used according to the manufacturer’s instructions and the slides were counterstained with hematoxylin. Photo images were captured using an Olympus BX61 microscope§.
Statistical analysis
General linear models (PROC GLM of SAS, version 9.4) were used to examine associations between the SNPs of interest and adjusted mean counts of microorganisms, GCF-IL-1β levels and clinical measurements, adjusting for microbial plaque levels (p≤0.05). Chi-square tests were used to examine SNP associations with periodontal disease category (BGI-classification) (p≤0.05).32
RESULTS
SNP identification and analysis
A sample of 4,910 Northern European descendants was genotyped and evaluated as periodontal complex traits (PCT), being PCT1 associated with periodontal clinical parameters of disease.14 To select variants correlation with clinical parameters, SNP prioritization was based upon statistical significance, linkage disequilibrium (r2), biological relevance, gene proximity, coding sequences and functional prediction. IFI16 rs1633266, an intron variant, was the lead SNP most significantly associated with the Socransky trait (p=3.1×10−8, Supplemental Figure 1). Nine additional SNPs were located in the IFI16 region and 1 in the AIM2 region (rs2793845) that were in high disequilibrium (r2≥0.8) with the lead SNP in IFI16, rs1633266; 1 SNP in the IFI16 region with an r2≥0.6 and 16 SNPs in the IFI16 region with r2>0.4 (Supplemental Figure 1). IFI16 and AIM2 are both localized in the 1q25.2 locus and are transcribed in opposite directions (Supplemental Figure 1). Among the SNPs that correlated with PCT1 (p≤5×10−5, suggestive evidence of association), 21 were localized in the IFI16 locus and 1 closest to AIM2 (within 2 kb downstream of the 3′ end of a transcript) (Supplemental table 1). Analysis with SNAP30 indicated the existence of 2 tight haplotype blocks in perfect linkage disequilibrium (r2=1 and D′=1), including IFI16 rs6940 (missense) with neighboring gene AIM2 rs2793845 and several additional intronic SNPs, while the second block identified by missense rs1057028 included rs1057027 and additional introns, all located in the IFI16 region (Table 1). Bioinformatic analysis of functional damages potentially caused by missense SNPs demonstrated that rs6940 (T>S) affected all 4 isoforms with a prediction of possibly damaging (score 0.584–0.782), rs1057027 as benign and rs1057028 (Y>N) as probably damaging to the protein function of isoform 3 (score 0.995) (Supplemental Table 2). This analysis suggests that rs6940 (minor allele [T] frequency=0.22) and rs1057028 (minor allele [T] frequency =0.3) are potentially/probably damaging for the protein function and, therefore, were selected for correlations with periodontal clinical parameters. Sequence alignment of the isoforms shows that the SNPs do not localize in the known functional domains (PYRIN domain, HIN domains and p53 interaction domain) (Supplemental figure 2A–B). However, rs6940 is located between HIN200-1/p53 C-terminus binding and HIN200-2/p53 core domain binding and could be interfering with the 3-D structure and protein function. It is possible that not all 4 isoforms of IFI16 are affected to the same degree based on the different protein sizes (Supplemental figure 2C). Indeed rs1057028 is predicted to probably damage isoform 3 only (Supplemental table 2). Imputation quality was 0.9994 and 0.9999 for rs6940 and rs1057028, respectively. In sum, our analysis identified 2 tight haplotype blocks with several SNPs in the IFI16/AIM2 region that are potentially important in the pathogenesis of periodontal disease. SNPs rs6940 and rs1057028 are predicted to be potentially/probably damaging to the protein function.
Table 1.
SNPs in haplotype blocks associated with the missense SNPs rs6940 and rs1057028 from individuals with Northern and Western European ancestry.
| SNP | Proxy | Distance to lead SNP (bp) | Predicted function |
|---|---|---|---|
| rs7532207 | 244 | intronic | |
| rs74359395 | 2335 | 3′ downstream IFI16 | |
| rs3737522 | 3223 | intronic | |
| rs3018316 | 6710 | intronic | |
| rs2793845 | 7587 | 3′downstream AIM2 | |
| rs2852695 | 8735 | intronic | |
| rs2814770 | 11896 | intronic | |
| rs6940 | rs2814771 | 12430 | intronic |
| rs12098223 | 12974 | intronic | |
| rs3754460 | 17822 | intronic | |
| rs74122232 | 18245 | intronic | |
| rs1633266 | 18691 | intronic | |
| rs1772407 | 18744 | intronic | |
| rs3768519 | 19444 | intronic | |
| rs1616024 | 20370 | intronic | |
| rs3768523 | 20670 | intronic | |
| rs1057027 | 12 | coding | |
| rs861318 | 167 | intronic | |
| rs1633256 | 168 | intronic | |
| rs1772415 | 389 | intronic | |
| rs856057 | 654 | intronic | |
| rs856056 | 680 | intronic | |
| rs856055 | 796 | intronic | |
| rs1057028 | rs856054 | 868 | intronic |
| rs856053 | 885 | intronic | |
| rs1417804 | 1594 | intronic | |
| rs1614182 | 2137 | intronic | |
| rs1633262 | 2499 | intronic | |
| rs1772408 | 3260 | intronic | |
| rs1633265 | 3338 | intronic | |
| rs2570916 | 10257 | intronic | |
| rs855865 | 25989 | 5′ upstream |
Linkage disequilibrium with lead SNP: CEU D′=1, r2=1
IFI16 SNPs are associated with parameters of periodontal disease
Correlation analysis of the clinical parameters showed that the both haplotype blocks rs6940 and rs1057028 had a significant increase in percent sites with extent bleeding on probing (EBOP), while rs1057028 also had significantly higher percentage of extent of probing depth greater or equal to 4mm (EPDGE4) (Table 2). Increased trends were observed for mean PD and percent of extent of gingival score greater or equal to 1 (EGIGE1) (Table 2). This suggests that SNPs in the region of IFI16 and AIM2 affects the biology of the tissues that leads to an increase in the extent of periodontal disease. Further analysis of the microbiological composition of plaque samples shows that several periodontal pathogen counts (plaque-adjusted) were also significantly higher in both haplotype blocks, including Pg, Tf and Cr. Loads of Pg were over 274 times higher for rs6940 and 90 times higher for rs1057028 when comparing 2.2 individuals (homozygous for the minor allele) to 1.1 individuals (homozygous for the major allele). Additional organisms were significantly increased in rs1057028, including of Fn and Aa, with similar trends observed for rs1057028 homozygous minor alleles (Table 3). This finding further supports that SNPs in the IFI16 and AIM2 region potentially affects the biological host response of the individual that leads to increased numbers of periodontal pathogens present in plaque samples. Cytokine analysis showed that individuals with haplotype block rs6940 had a 4-fold increased concentration in levels of GCF-IL-1β, with a trend of significance for rs1057028 of 2-fold increase (Table 4). Since IL-1β is a well-known pro-inflammatory cytokine implicated in periodontal disease progression 34, the finding of increased levels of this cytokine in the GCF in the presence of SNPs in the IFI16 and AIM2 regions suggest that potential defects in IFI16/AIM2 can alter the inflammatory response of an individual and further influence disease status. Individuals with both haplotype blocks showed an increased percentage of severe periodontal disease (BGI-P3) when compared to healthy controls (Table 5), reaching statistical significance for haplotype block rs1057028 (p=0.02). The BGI classification accounts for PD and BOP (and not CAL), representing the individual’s current disease activity and inflammation. Because the presence of SNPs in the IFI16 and AIM2 regions lead to an increase in number of individuals with periodontal disease/inflammation, it suggests that the functional defect increased the predisposition of the individual developing the multifactorial condition of periodontal disease. Taken together, the data suggest that the presence of these SNPs alter the normal host response that leads to an increased predisposition to develop periodontal disease, observed by higher numbers of periodontal microorganisms, increased measurements of periodontal disease and higher number of individuals with clinical disease.
Table 2.
Clinical measurements among individuals with SNPs in haplotype blocks, mean (standard deviation).
| SNP | n | EPDGE4 (%) | Mean PD (mm) | EBOP (%) | EGIGE1 (%) | Mean CAL (mm) |
|---|---|---|---|---|---|---|
| rs6940* | ||||||
| 1.1 | 3590 | 6.84 (0.16) | 1.86 (0.01) | 23.1 (0.33) | 22.0 (0.46) | 1.64 (0.01) |
| 1.2 | 1057 | 6.84 (0.30) | 1.84 (0.02) | 23.7 (0.61) | 22.4 (0.84) | 1.65 (0.02) |
| 2.2 | 80 | 6.28 (1.13) | 1.82 (0.06) | 28.3 (2.28)† | 24.9 (3.21) | 1.51 (0.1) |
|
| ||||||
| rs1057028* | ||||||
| 1.1 | 3199 | 6.84 (0.17) | 1.86 (0.01) | 23.1 (0.35) | 22.1 (0.49) | 1.64 (0.01) |
| 1.2 | 1367 | 6.64 (0.27) | 1.84 (0.01) | 23.1 (0.54) | 22.1 (0.74) | 1.63 (0.02) |
| 2.2 | 161 | 8.34 (0.79)† | 1.92 (0.04) | 28.8 (1.60)‡ | 25.1 (2.24) | 1.75 (0.07) |
haplotype blocks for both SNPs include additional SNPs shown on table 1
chi-square p-values using 1.1 as the referent category:
p≤0.05,
p≤0.01.
Data was adjusted for plaque index.
Table 3.
Mean levels of periodontal microorganism (standard errors) and relative (fold) changes for rs6940 and rs1057028 haplotype blocks (adjusted for plaque levels).
| SNP | N | Pg | Pi | Pn | Tf | Td | Cr | Fn | Aa |
|---|---|---|---|---|---|---|---|---|---|
| rs6940* | |||||||||
| 1.1 | 781 | 2.22 (0.06) | 2.55 (0.07) | 2.68 (0.07) | 2.43 (0.06) | 2.61 (0.07) | 2.66 (0.07) | 2.86 (0.08) | 2.57 (0.06) |
| 1.2 | 255 | 2.27 (0.10) | 2.71 (0.13) | 2.51 (0.13) | 2.49 (0.11) | 2.53 (0.12) | 2.72 (0.11) | 2.96 (0.13) | 2.74 (0.10) |
| 2.2 | 17 | 3.54 (0.43) ‡ | 3.01 (0.54) | 3.47 (0.52) | 3.50 (0.45)† | 3.11 (0.48) | 3.69 (0.48)† | 3.94 (0.55) | 2.99 (0.44) |
| Fold change | 274.34 | 58.41 | 120.34 | 191.54 | 64.87 | 180.11 | 194.47 | 52.20 | |
| rs1057028* | |||||||||
| 1.1 | 711 | 2.24 (0.06) | 2.62 (0.08) | 2.71 (0.08) | 2.42 (0.07) | 2.63 (0.07) | 2.67 (0.07) | 2.86 (0.08) | 2.59 (0.06) |
| 1.2 | 300 | 2.21 (0.10) | 2.48 (0.12) | 2.42 (0.12) | 2.46 (0.10) | 2.47 (0.11) | 2.65 (0.11) | 2.90 (0.12) | 2.58 (0.10) |
| 2.2 | 42 | 2.89 (0.26)† | 3.07 (0.32) | 3.31 (0.31) | 3.13 (0.27)† | 2.92 (0.29) | 3.34 (0.29)† | 3.55 (0.33)† | 3.25 (0.26)† |
| Fold change | 91.55 | 56.83 | 82.21 | 103.40 | 33.64 | 95.42 | 99.37 | 93.48 | |
haplotype blocks for both SNPs include additional SNPs shown on Table 1.
Genotypes, 1.1: homozygous for the major allele, 1.2: heterozygous, 2.2: homozygous for the minor allele.
Chi-square p-values using 1.1 as the referent category:
p≤0.05,
p≤0.01; fold change comparing 2.2 vs. 1.1
Table 4.
Mean log gingival crevicular fluid IL-1β levels (standard errors) and relative (fold) changes contrasting 2.2 vs. 1.1, according to rs6940 and rs1057028 genotypes (adjusted for plaque levels).
| SNP | n | Mean log IL-1β (SE) |
|---|---|---|
| rs6940* | ||
| 1.1 | 3359 | 2.05 (0.01) |
| 1.2 | 976 | 2.04 (0.01) |
| 2.2 | 73 | 2.08 (0.04) |
| Fold change | 3.05 | |
| rs1057028* | ||
| 1.1 | 3006 | 2.05 (0.01) |
| 1.2 | 1255 | 2.04 (0.01) |
| 2.2 | 147 | 2.10 (0.03)† |
| Fold change | 5.13 | |
haplotype blocks for both SNPs include additional SNPs shown on table 1
chi-square p≤0.05 using 1.1 as the referent category; fold change comparing 2.2 vs. 1.1
Table 5.
Distribution of rs6940 and rs1057028 haplotype blocks according to the biofilm-gingival interface (BGI) periodontal disease classification (healthy—n=571 vs. severe periodontitis—n=531).
Descriptive histological distribution of IFI16 and AIM2 in gingival tissues
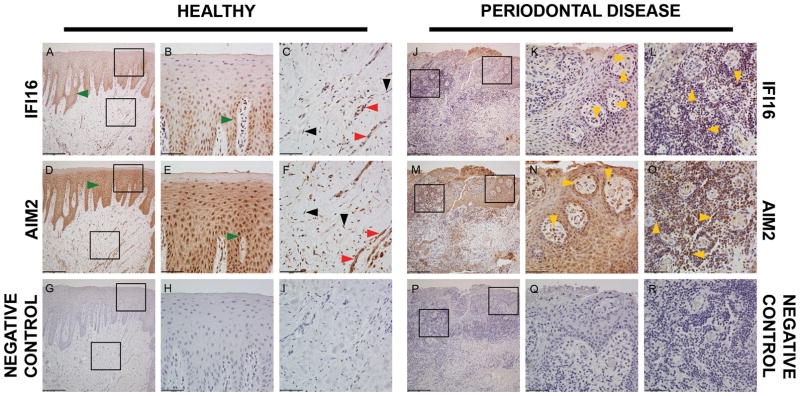
The purpose of this approach was to describe the IFI16 and AIM2 protein expression in cells of the periodontium. IHC analysis was performed in gingival tissue samples of individuals with healthy tissues (n=2) and chronic periodontal disease (n=2). The demographics of this population are shown on Supplementary Table 3. Representative low-resolution (10X) images (Figure 1A, D, G, J, M, P) of healthy and periodontitis tissues show a similar pattern of expression among samples. Both proteins had a homogeneous distribution in the epithelial layer, with minimal-no expression in the keratin layer, among healthy and periodontitis samples (Figure 1B, E, K, N). IFI16 staining was dense in the basal layer (Figure 1A, B). Migrating neutrophils expressing IFI16 and AIM2 were observed in the epithelial layer in a sample derived from individuals with periodontal disease (Figure 1K, N). In the connective tissue (Figure 1C, F, L, O), endothelial cells and cells of the inflammatory infiltrate showed expression of both IFI16 and AIM2. Minor expression of these proteins was observed in fibroblasts in the connective tissue. All samples (n=4) demonstrated a similar pattern of staining for both proteins. This descriptive analysis demonstrated that cells of the periodontal apparatus express IFI16 and AIM2 and, therefore, further supports a potential role of these proteins in the pathogenesis of periodontitis.
Figure 1.
Immunohistochemical detection of IFI16 and AIM2 in human gingival tissues. Representative images of tissue sections from a healthy individual (A–I) and an individual with periodontal disease (J–R) according to the ADA/AAP classification. stained with the indicated antibodies (left and right rows). A, D, G, J, M, P represent 10× magnification (scale bar = 200.00μm); B, C, E, F, H, I, K, L, N, O, Q, R represent 40× magnification (scale bar = 50.00μm) of the square inserts located in the figures with 10× magnification in the epithelial layer and connective tissue layer. Bottom row are negative controls. Green arrows=epithelial cells; black arrows=fibroblasts; yellow arrows=leukocytes; red arrows=endothelial cells.
DISCUSSION
Our study characterized clinical and biological periodontal data among a sizeable group of participants and examined their association with SNPs in the IFI16/AIM2 locus. We found 2 haplotype blocks, one including a missense SNP rs6940 and a variant close to neighboring gene AIM2, and a second block including missense SNP rs1057028 that were significantly associated with periodontal disease parameters, including increased extent PD and BOP, increased GCF-IL-1β levels, higher loads of periodontal pathogens and higher prevalence of severe periodontal disease. Prediction analysis indicated that the function of IFI16 is altered by the presence of rs6940 and rs1057028. Several additional SNPs were in perfect linkage disequilibrium with the index SNPs. These polymorphisms are quite common in the general population with minor allele frequencies of 0.22 for rs6940 (minor allele: T) and 0.3 for rs1057028 (minor allele: T). We identified that IFI16 and AIM2 are expressed in epithelial cells, fibroblasts, endothelial cells and leukocytes of gingival tissues. The presence of these proteins in inflammatory cells of gingival tissues, including the finding of neutrophils in the epithelial layer (Figure 1K, N) suggests a role of IFI16 and AIM2 in the response to periodontal pathogens. Taken together, these findings propose that these proteins and specific polymorphisms may have an important role in periodontal disease pathogenesis.
Previous GWAS analysis by our group have highlighted loci potentially associated with clinically derived-disease definitions, like chronic periodontal disease,10, 12 and high levels of specific periodontal pathogens.35 However, no single marker met the strict genome-wide significance. Only 4 loci met gene-centric statistical significance. 12 This suggested the existence of several distinct conditions with different genetic and biological backgrounds with similar overlapping clinical presentations of periodontal tissue loss. Therefore, a new approach was utilized by defining the disease phenotype as complex traits as previously described.14, 36 With this approach, six PCT were identified by levels of 8 periodontal pathogens, local inflammatory response (GCF-IL-1β) and clinical data.14 PCT1 was defined by a uniformly high periodontal pathogen load and significantly correlated with clinical periodontal disease parameters. Six loci were associated with PCT1, which included IFI16/AIM2. While there was a degree of anticipation that the IFI16/AIM2 haplotype blocks would be correlated with some periodontal microorganisms since the lead SNP was identified from an analysis that used a microbial community structure, the individual microorganisms correlated with the haplotype blocks were not known. In addition, the clinical measurements and disease significance in this population, and tissue distribution of IFI16 and AIM2 were further characterized. Therefore, the present study further refines the findings derived from the PCT1 analysis and defines the clinical and biological characteristics of individuals with SNPs in the IFI16/AIM2 region.
Both IFI16 and AIM2 are PYHIN inflammasome proteins and mediators of innate immune responses.17 Inflammasomes are multiprotein oligomers that promote activation of inflammatory cytokines IL-1β and IL-18.37 The PYHIN inflammasome proteins bind microbial DNA and form caspase-1-activating inflammasomes (AIM2) or drive type I IFN gene transcription (IFI16).17 IFI16 is also a mediator of the AIM2 inflammasome-dependent pathway by directly binding to AIM2.38 Therefore, IFI16 has shown anti-inflammatory effects and AIM2 proinflammatory effects. This suggests that defects in the expression or protein function of IFI16 could dampen the anti-inflammatory response and, thereby increase the proinflammatory response. Studies show that AIM2 is increased in wound healing and in several inflammatory conditions, including psoriasis, atopic dermatitis and venous ulcers.18, 19 Additional support for the role of AIM2 and IFI16 in inflammation is a recent demonstration of a strong increase of both proteins in the mucosa of individuals with active inflammatory bowel disease.22 In accordance to our findings, 3 studies have previously demonstrated the presence of AIM2 in gingival tissues.20, 21, 39 AIM2 expression was increased in gingival tissues from individuals with chronic periodontitis compared to healthy controls and generalized aggressive periodontitis. In addition, Pg infection can activate the AIM2 inflammasome in vitro.39 These findings further support an importance of AIM2 in the pathogenesis of periodontal disease. To our knowledge, no other study has evaluated the expression of IFI16 in periodontal tissues. Further analysis and quantification of this protein in periodontal cells and tissues in various disease states is warranted.
The concept that the host genotype can influence the microbiota and lead to disease has been reported previously. Counts of periodontal pathogens from the red and orange complex are reported to be significantly higher at PD>6mm in IL-1 genotype positive individuals compared to genotype negative individuals.6 IL-6 polymorphisms and haplotypes have also been associated with periodontitis, possibly due to the transcription of IL-6 that then alters tissue levels.7, 40 An alteration in the immune response observed in leukocyte adhesion deficiency also leads to significant changes in the subgingival flora and severe periodontitis.8 These results are in support of our data and the concept that changes in the innate immune functions can alter the host response and facilitate the development of diseases. A recent study found that IFI16 rs6940 (also identified in this study) and AIM2 rs855873 (also upstream of AIM2) were associated with increased susceptibility to Behcet disease, a systemic immune-mediated disease characterized by vasculitits and recurrent mucosal ulcerations.41 The study shows rs6940 decreases the expression of anti-inflammatory IFI16 and increases susceptibility to an immune-mediated disease.41 The fact that Behcet is associated with epithelial ulceration and our results show associations of these SNPs with BOP suggests that these variants may impair innate immune responses that maintain epithelial integrity at mucosal surfaces. In fact, individuals with Behcet disease have increased periodontal disease severity compared to healthy controls, suggesting both diseases share pathogenic aspects.42, 43 These findings support the hypothesis of a role of anti-inflammatory IFI16 in periodontal disease pathogenesis and that variants of this gene predispose individuals for an altered immune response to infection that ultimately leads to disease.
Functional analysis was done to help identify SNPs that have a high potential of altering the protein function. Our current analysis indicates that both missense SNPs rs6940 and rs1057028 are not localized in the known PYRIN, HIN-200 or p53-binding domains. However, the predictions suggest that the protein function is potentially/probably altered. Since rs1057028 is localized between both HIN domains it is possible that the variant induces a 3D conformation change to the protein structure. SNP rs6940 is predicted to alter only isoform 3, which is probably related to the different sizes of the isoforms. A previous study suggests that instead of the protein function, SNP IFI16 rs6940 decreases the expression of IFI16.41 It is possible that additional SNPs in high linkage disequilibrium with rs6940 are leading to this effect. However, no functional assays were performed in the study.41 Since the SNP associated with AIM2 is not present in a missense region, predictions for potential alterations could not be performed. Future studies defining the impact of the SNPs present in the 2 haplotype blocks in protein expression and host response should provide additional evidence for the pathogenesis of periodontitis.
Although we identified a biologically-relevant region associated with elevated periodontitis parameters, our results have limitations. The validity of the reported SNP associations will need to be further examined and replicated in another new study. However, it is important to note that the identification of these genes was not based on traditional AAP/ADA classification for chronic and aggressive periodontitis, which is utilized in other GWAS studies.10, 44–46 Instead, these genes were first identified in the context of a PCT that utilized a combination of microorganisms and GCF-IL-1β levels.14 In this model, periodontal disease is a group of distinct biological conditions with overlapping clinical presentations. Therefore, future GWAS analysis may consider periodontal complex traits for identifying biologically relevant genes.
CONCLUSION
Together, our results support a role of for IFI16/AIM2 in the pathogenesis of periodontal disease. This association was observed by the correlation of SNP variants with increased measurements of microorganisms in the subgingival plaque, increased levels of GCF-IL-1β, increased periodontal clinical parameters of disease and increased prevalence of severe disease.
Supplementary Material
Plot of the IFI16/AIM2 locus associated with the Socransky trait (periodontal complex trait 1)12.
SNP position
A) Schematic diagram of SNP location relative to protein domains; DAPIN = domain in apoptosis and interferon response (protein-protein interaction domain), NLS = nuclear localization signal, HIN = hematopoietic expression, interferon-inducible nature and nuclear localization (binding to nucleic acids in the cytoplasm and nucleus).
B) Protein isoform sequence alignment indicating domain and SNP positions
C) Representation of different isoforms of IFI16
Acknowledgments
We thank the staff and participants of the ARIC study for their important contributions. The ARIC study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HSN268201100012C) and grants (R01HL087641, R01HL59367, R01HL086694), the National Human Genome Research Institute (contract U01HG004402), the National Institutes of Health (contract HHSN268200625226C), the National Institute of Environmental Health Sciences (grant P30ES010126), and the National Institute of Dental and Craniofacial Research (grants R01DE11551, R01DE021418). Infrastructure was partly supported by a component of the National Institutes of Health and NIH Roadmap for Medical Research (grant UL1RR025005). This study was funded also by the Carolina Postdoctoral Program for Faculty Diversity, University of North Carolina (to J.T.M) and the National Institutes of Health (grant R01DE23836 to S.O., R90DE022527 to S.Z and 5T90DE021986-05 to Y.Z.J). We thank Director of the Histology Research Core Facility at UNC Ms. Jennifer Ashley Ezzell for her assistance with the immunohistochemistry.
Footnotes
LocusZoom, http://locuszoom.org/
SNPNexus, http://www.snp-nexus.org
UniProt, http://www.uniprot.org/
Santa Cruz, Dallas, TX
abcam, Cambridge, MA
R&D Systems, Inc. Minneapolis, MN
Olympus America Inc. Melville, NY
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1.Michalowicz BS, Diehl SR, Gunsolley JC, et al. Evidence of a substantial genetic basis for risk of adult periodontitis. Journal of periodontology. 2000;71:1699–1707. doi: 10.1902/jop.2000.71.11.1699. [DOI] [PubMed] [Google Scholar]
- 2.Michalowicz BS, Aeppli D, Virag JG, et al. Periodontal findings in adult twins. Journal of periodontology. 1991;62:293–299. doi: 10.1902/jop.1991.62.5.293. [DOI] [PubMed] [Google Scholar]
- 3.Kornman KS. Mapping the pathogenesis of periodontitis: a new look. Journal of periodontology. 2008;79:1560–1568. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- 4.Weir BS. Bioinformatics and approaches to identifying polygenic susceptibility traits. Annals of periodontology/the American Academy of Periodontology. 2002;7:1–7. doi: 10.1902/annals.2002.7.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Mucci LA, Bjorkman L, Douglass CW, Pedersen NL. Environmental and heritable factors in the etiology of oral diseases--a population-based study of Swedish twins. Journal of dental research. 2005;84:800–805. doi: 10.1177/154405910508400904. [DOI] [PubMed] [Google Scholar]
- 6.Socransky SS, Haffajee AD, Smith C, Duff GW. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. Journal of clinical periodontology. 2000;27:810–818. doi: 10.1034/j.1600-051x.2000.027011810.x. [DOI] [PubMed] [Google Scholar]
- 7.Nibali L, D’Aiuto F, Donos N, et al. Association between periodontitis and common variants in the promoter of the interleukin-6 gene. Cytokine. 2009;45:50–54. doi: 10.1016/j.cyto.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Moutsopoulos NM, Chalmers NI, Barb JJ, et al. Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. PLoS Pathog. 2015;11:e1004698. doi: 10.1371/journal.ppat.1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.American Academy of Periodontology Task Force Report on the Update to the 1999 Classification of Periodontal Diseases and Conditions. Journal of periodontology. 2015;86:835–838. doi: 10.1902/jop.2015.157001. [DOI] [PubMed] [Google Scholar]
- 10.Divaris K, Monda KL, North KE, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Human molecular genetics. 2013;22:2312–2324. doi: 10.1093/hmg/ddt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teumer A, Holtfreter B, Volker U, et al. Genome-wide association study of chronic periodontitis in a general German population. Journal of clinical periodontology. 2013;40:977–985. doi: 10.1111/jcpe.12154. [DOI] [PubMed] [Google Scholar]
- 12.Rhodin K, Divaris K, North KE, et al. Chronic periodontitis genome-wide association studies: gene-centric and gene set enrichment analyses. Journal of dental research. 2014;93:882–890. doi: 10.1177/0022034514544506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng P, Wang X, Casado PL, et al. Genome wide association scan for chronic periodontitis implicates novel locus. BMC Oral Health. 2014;14:84. doi: 10.1186/1472-6831-14-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offenbacher S, Divaris K, Barros SP, et al. Genome-wide association study of biologically-informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Human molecular genetics. 2016 doi: 10.1093/hmg/ddw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. Journal of clinical periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 18.Dombrowski Y, Peric M, Koglin S, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Koning HD, Bergboer JG, van den Bogaard EH, et al. Strong induction of AIM2 expression in human epidermis in acute and chronic inflammatory skin conditions. Exp Dermatol. 2012;21:961–964. doi: 10.1111/exd.12037. [DOI] [PubMed] [Google Scholar]
- 20.Sahingur SE, Xia XJ, Voth SC, Yeudall WA, Gunsolley JC. Increased nucleic Acid receptor expression in chronic periodontitis. Journal of periodontology. 2013;84:e48–57. doi: 10.1902/jop.2013.120739. [DOI] [PubMed] [Google Scholar]
- 21.Xue F, Shu R, Xie Y. The expression of NLRP3, NLRP1 and AIM2 in the gingival tissue of periodontitis patients: RT-PCR study and immunohistochemistry. Arch Oral Biol. 2015;60:948–958. doi: 10.1016/j.archoralbio.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Vanhove W, Peeters PM, Staelens D, et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:2673–2682. doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 23.Beck JD, Elter JR, Heiss G, Couper D, Mauriello SM, Offenbacher S. Relationship of periodontal disease to carotid artery intima-media wall thickness: the atherosclerosis risk in communities (ARIC) study. Arterioscler Thromb Vasc Biol. 2001;21:1816–1822. doi: 10.1161/hq1101.097803. [DOI] [PubMed] [Google Scholar]
- 24.Elter JR, Champagne CM, Offenbacher S, Beck JD. Relationship of periodontal disease and tooth loss to prevalence of coronary heart disease. Journal of periodontology. 2004;75:782–790. doi: 10.1902/jop.2004.75.6.782. [DOI] [PubMed] [Google Scholar]
- 25.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayem Ullah AZ, Lemoine NR, Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Brief Bioinform. 2013;14:437–447. doi: 10.1093/bib/bbt004. [DOI] [PubMed] [Google Scholar]
- 28.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Offenbacher S, Barros SP, Singer RE, Moss K, Williams RC, Beck JD. Periodontal disease at the biofilm-gingival interface. Journal of periodontology. 2007;78:1911–1925. doi: 10.1902/jop.2007.060465. [DOI] [PubMed] [Google Scholar]
- 33.Offenbacher S, Lin D, Strauss R, et al. Effects of periodontal therapy during pregnancy on periodontal status, biologic parameters, and pregnancy outcomes: a pilot study. Journal of periodontology. 2006;77:2011–2024. doi: 10.1902/jop.2006.060047. [DOI] [PubMed] [Google Scholar]
- 34.Engebretson SP, Grbic JT, Singer R, Lamster IB. GCF IL-1beta profiles in periodontal disease. Journal of clinical periodontology. 2002;29:48–53. doi: 10.1034/j.1600-051x.2002.290108.x. [DOI] [PubMed] [Google Scholar]
- 35.Divaris K, Monda KL, North KE, et al. Genome-wide association study of periodontal pathogen colonization. Journal of dental research. 2012;91:21S–28S. doi: 10.1177/0022034512447951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suo C, Toulopoulou T, Bramon E, et al. Analysis of multiple phenotypes in genome-wide genetic mapping studies. BMC Bioinformatics. 2013;14:151. doi: 10.1186/1471-2105-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 38.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PloS one. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infection and immunity. 2014;82:112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fife MS, Ogilvie EM, Kelberman D, et al. Novel IL-6 haplotypes and disease association. Genes Immun. 2005;6:367–370. doi: 10.1038/sj.gene.6364186. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz-Fernandez L, Garcia-Lozano JR, Montes-Cano MA, et al. Variants of the IFI16 gene affecting the levels of expression of mRNA are associated with susceptibility to Behcet disease. J Rheumatol. 2015;42:695–701. doi: 10.3899/jrheum.140949. [DOI] [PubMed] [Google Scholar]
- 42.Akman A, Kacaroglu H, Donmez L, Bacanli A, Alpsoy E. Relationship between periodontal findings and Behcet’s disease: a controlled study. Journal of clinical periodontology. 2007;34:485–491. doi: 10.1111/j.1600-051X.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- 43.Arabaci T, Kara C, Cicek Y. Relationship between periodontal parameters and Behcet’s disease and evaluation of different treatments for oral recurrent aphthous stomatitis. J Periodontal Res. 2009;44:718–725. doi: 10.1111/j.1600-0765.2008.01183.x. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu S, Momozawa Y, Takahashi A, et al. A genome-wide association study of periodontitis in a Japanese population. Journal of dental research. 2015;94:555–561. doi: 10.1177/0022034515570315. [DOI] [PubMed] [Google Scholar]
- 45.Freitag-Wolf S, Dommisch H, Graetz C, et al. Genome-wide exploration identifies sex-specific genetic effects of alleles upstream NPY to increase the risk of severe periodontitis in men. Journal of clinical periodontology. 2014;41:1115–1121. doi: 10.1111/jcpe.12317. [DOI] [PubMed] [Google Scholar]
- 46.Shaffer JR, Polk DE, Wang X, et al. Genome-wide association study of periodontal health measured by probing depth in adults ages 18–49 years. G3 (Bethesda) 2014;4:307–314. doi: 10.1534/g3.113.008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plot of the IFI16/AIM2 locus associated with the Socransky trait (periodontal complex trait 1)12.
SNP position
A) Schematic diagram of SNP location relative to protein domains; DAPIN = domain in apoptosis and interferon response (protein-protein interaction domain), NLS = nuclear localization signal, HIN = hematopoietic expression, interferon-inducible nature and nuclear localization (binding to nucleic acids in the cytoplasm and nucleus).
B) Protein isoform sequence alignment indicating domain and SNP positions
C) Representation of different isoforms of IFI16