Here, Tessé et al. investigated how the complex program of chromosomal events during meiosis (pairing, recombination, and segregation of homologous chromosomes) proceeds in an ordered sequence. Their study now reveals that Mer2 is involved in every step of the DNA-directed meiotic morphogenetic pathway and interhomolog interactions and that Mer2 is highly conserved between fungi, plants, and animals.
Keywords: meiosis, Mer2, recombination, pairing, chromatin compaction SUMOylation, Sordaria
Abstract
Meiosis is the cellular program by which a diploid cell gives rise to haploid gametes for sexual reproduction. Meiotic progression depends on tight physical and functional coupling of recombination steps at the DNA level with specific organizational features of meiotic-prophase chromosomes. The present study reveals that every step of this coupling is mediated by a single molecule: Asy2/Mer2. We show that Mer2, identified so far only in budding and fission yeasts, is in fact evolutionarily conserved from fungi (Mer2/Rec15/Asy2/Bad42) to plants (PRD3/PAIR1) and mammals (IHO1). In yeasts, Mer2 mediates assembly of recombination–initiation complexes and double-strand breaks (DSBs). This role is conserved in the fungus Sordaria. However, functional analysis of 13 mer2 mutants and successive localization of Mer2 to axis, synaptonemal complex (SC), and chromatin revealed, in addition, three further important functions. First, after DSB formation, Mer2 is required for pairing by mediating homolog spatial juxtaposition, with implications for crossover (CO) patterning/interference. Second, Mer2 participates in the transfer/maintenance and release of recombination complexes to/from the SC central region. Third, after completion of recombination, potentially dependent on SUMOylation, Mer2 mediates global chromosome compaction and post-recombination chiasma development. Thus, beyond its role as a recombinosome–axis/SC linker molecule, Mer2 has important functions in relation to basic chromosome structure.
Sexual reproduction is dependent on meiotic recombination in most organisms. DNA recombination not only is important for increasing genetic diversity but also plays critical mechanistic roles in fundamental chromosome dynamics during prophase I of the meiotic program. As a key aspect of these mechanistic roles, every biochemical step of the recombination process, from initiation to completion, occurs in the context of highly organized chromosomes. Meiotic chromosomes are organized into linear arrays of chromatin loops, with conjoined sister linear loop arrays co-oriented and connected by a structural axis that defines the interaction side of the homologous chromosomes during pairing (Moens and Pearlman 1990; for review, see Zickler and Kleckner 1999). Such organization has significance beyond a simple organizational role.
First, association of recombination complexes with chromosome axes is crucial for initiation of recombination through double-strand break (DSB) formation catalyzed by the transesterase Spo11 (e.g., for reviews, see Borde and de Massy 2013; Lam and Keeney 2015). Association of the loop DSB sites with the axes in “tethered loop axis complexes” (TLACs) precedes DSB formation and creates intimate physical contacts between recombination components and axis proteins (Blat et al. 2002; Kumar et al. 2010, Panizza et al. 2011; Miyoshi et al. 2012, 2013; Sommermeyer et al. 2013; Lam and Keeney 2015). Budding and fission yeast Mer2/Rec15 play a critical role in this process (Panizza et al. 2011; Miyoshi et al. 2012, 2013). Mer2 also physically interacts with Spp1, a member of the Set1 COMPASS complex, therefore providing an interesting link with the chromatin state (Acquaviva et al. 2013; Sommermeyer et al. 2013).
Second, in most organisms, interactions of a DSB with an homologous partner promote the spatial juxtaposition of homologous chromosomes (homologs). The resulting DNA/DNA interaction mediates whole-chromosome “pairing” at the structural level, seen as coalignment of homologous axes linked by a series of bridges, which correspond to the positions of DSB-mediated interhomolog interactions (e.g., Albini and Jones 1987; for review, see Zickler and Kleckner 1999, 2015; Oliver-Bonet et al. 2007).
Third, the structural loop/axis association continues to play a central role when a subset of interaxis bridges gives rise to crossovers (COs) with accompanying spatial patterning known as “CO interference.” Concomitantly, these and other bridges nucleate the polymerization of the pairing structure called the synaptonemal complex (SC) (for reviews, see Page and Hawley 2004; Zhang et al. 2014). Moreover, when the SC is assembling, the proteins that mediate recombination become associated with the SC central region (e.g., Anderson and Stack 1988; Moens et al. 2002; de Boer et al. 2006; Oliver-Bonet et al. 2007; Storlazzi et al. 2010; Espagne et al. 2011; Qiao et al. 2012; Reynolds et al. 2013). Finally, chromatin loop/axis relationships continue to be important even after the recombination process has been completed, when the SC disassembles and chromosomes undergo a structural reorganization into a radial loop configuration at diplotene, at which point homologs are linked only by the COs/chiasmata that play a mechanical role for homolog segregation (Rufas et al. 1992; for review, see Hunter 2015).
Here we report the identification of a widely evolutionarily conserved meiotic protein family and, by molecular and cytogenetic studies in the fungus Sordaria macrospora, implicate this molecule as a mediator of virtually all of the above-described events. In previous studies, we identified several mutations (in seven ASY genes) that suppress meiotic defects conferred by a unique allele of cohesin gatekeeper Spo76/Pds5 (spo76-1) (Huynh et al. 1986; van Heemst et al. 1999). Here, molecular identification of ASY2 and ensuing bioinformatics analyses now show that Asy2 corresponds to budding yeast Mer2 and, moreover, reveal the existence of an evolutionarily conserved meiotic protein family with conservation from fungi (fission yeast Rec15 and Coprinus Bad42) to plants (PRD3/PAIR1) and mammals (mouse IHO1). For the latter four molecules, no structural relationship to Mer2 protein has been detected previously (Nonomura et al. 2004; De Muyt et al. 2009; Miao et al. 2013; Stanzione et al. 2016). Our finding that Asy2 is the functional equivalent of Mer2/Rec15 is reinforced by its interaction with Asy3/Rec114, another Spo11 accessory protein in yeast (e.g., Lam and Keeney 2015) and Sordaria (where asy3 is another spo76-1 suppressor).
In the present study, functional analysis of 13 null and nonnull asy2/mer2 mutants plus localization of wild-type and mutant Asy2/Mer2 reveals that the protein localizes all along the chromosomes during meiotic prophase, dependent on axis proteins but not recombination. Moreover, Mer2 acts at multiple steps throughout prophase to mediate the recombination/axis/SC interface. It first mediates formation of pre-DSB complexes and DSB formation (as shown previously for the yeast members of this family) but then also mediates post-DSB homolog recognition/pairing with implications for CO patterning. During synapsis, Mer2 localizes to the SC central region, where it is implicated in the transfer, maintenance, and release of recombination complexes to and from the SC central region. Finally and unexpectedly, after recombination is completed, Asy2/Mer2 plays a global role for timely and regular recompaction of diplotene/prometaphase chromosomes. Moreover, this role is potentially dependent on SUMOylation. Therefore, the Asy2/Mer2 family not only is a conserved axis–recombinosome linker molecule during DSB formation but has additional important functions in relation to basic chromosome structure during the entire meiotic process.
Results
Sordaria Asy2/Mer2 identifies a widely conserved meiotic protein family
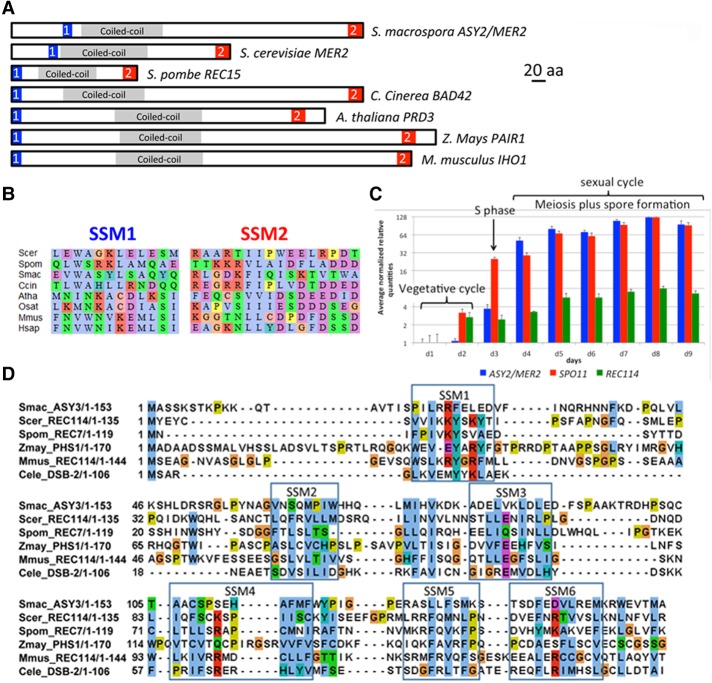
ASY2 (SMAC_02785) was identified by whole-genome sequencing of four asy2 mutant strains by SOLiD technology. No sequence similarity could be detected between Asy2 and proteins in the databases. However, a series of phylogenomics-oriented PSI-BLAST homology searches for Asy2, coupled with multiple sequence alignments of fungal genomes, revealed two weakly conserved short similarity motifs (SSMs)—N-terminal SSM1 and C-terminal SSM2—that flank a centrally located predicted coiled-coil (CC) region (amino acid residues 89–221) (Fig. 1A,B). These motifs are conserved among Sordariomycetes (e.g., Neurospora crassa), Saccharomycetales (Mer2/Rec107), Schizosaccharomycetes (Rec15), and Basidiomycetes (Bad42). Furthermore, we found that CC-containing meiotic proteins also displayed SSM1- and SSM2-related motifs with intraphylum significant conservation in mammals (e.g., mouse IHO1 [Stanzione et al. 2016] and human Q8IYA8), birds (e.g., Gallus gallus XP_004944678), and plants (PAIR1 in rice [Nonomura et al. 2004] and PRD3 in Arabidopsis thaliana [De Muyt et al. 2009]) not previously identified as being Mer2/Rec15 orthologs. These relationships imply functional conservation of the protein among most eukaryotic kingdoms (Fig. 1A,B; Supplemental Figs. S1A,B, S2 [phylogenetic tree]).
Figure 1.
Conservation of Mer2 and Rec114 in eukaryotes and expression in Sordaria. (A,B) Localization within the primary structures of fungal, plant, and mouse Mer2 orthologs (A) and alignment (B) of the conserved SSM1 (blue rectangle in A) and SSM2 (red rectangle in A) motifs (see also Supplemental Figs. S1A,B [for details], S2 [for phylogenetic tree]). (C) Comparison of the MER2 (blue), SPO11 (red), and REC114 (green) transcripts during Sordaria vegetative and sexual cycles (RT-qPCR). Histograms reflect the heterogeneity of the meiotic stages in the fruiting bodies. As asci/meiocytes are formed in successive waves (the first ones between days 2 and 3), fruiting bodies from days 4 to 6 contain essentially meiosis I and meiosis II asci but also fewer asci than fruiting bodies from days 7 to 9, which now contain asci at all stages between prophase I and ascospore formation. Relative quantities are scaled to minimal (lowest relative quantity = 1) in 2-log scale. All relative quantities >2 have a P-value of <0.01. Reference genes (PDF2, TIP, UBC, and CIT1) were selected with geNorm as described in the Supplemental Material. The geometric average of the expression of the four reference genes was used for the normalization of the expression of MER2, SPO11, and REC114. (D) Alignment of the N-terminal part of Rec114 orthologs with the six conserved SSMs (framed). (Scer) Saccharomyces cerevisiae; (Spom) Schizosaccharomyces pombe; (Smac) S. macrospora; (Atha) A. thaliana; (Zmay) Zea mays; (Mmus) Mus musculus; (Cele) Caenorhabditis elegans.
Schematic domain organization of the predicted 504-amino-acid Sordaria protein and Saccharomyces cerevisiae Mer2, Schizosaccharomyces pombe Rec15, Coprinopsis cinerea Bad42, A. thaliana PRD3, rice PAIR1, and mouse IHO1 is shown in Figure 1A. These assignments are supported by experimental analyses of function in Sordaria (described below) and previous findings in mice, rice, and Arabidopsis (Discussion). Given the prominence of MER2 in existing literature, we refer to the Sordaria ASY2 gene as ASY2/MER2 or MER2.
Based on RT-qPCR analyses, ASY2/MER2 transcripts are induced during the sexual cycle and the meiotic divisions exactly parallel to the temporal kinetics of transcripts for SPO11, the widely conserved meiotic transesterase that promotes meiotic DSB formation (Fig. 1C; Supplemental Tables S1, S2).
Sordaria Asy3 is Rec114 and identifies another widely conserved meiotic protein family
Mer2/Rec15 interacts physically with Rec114 to promote DSB formation. Bioinformatics analyses identical to those used for ASY2/MER2 (above) revealed that Sordaria ASY3 (discovered in the same genetic screen as ASY2) is Sordaria REC114. The corresponding protein is conserved in plants and mice (Kumar et al. 2010). The N-terminal region of Sordaria Asy3/Rec114 (Fig. 1D) is homologous to the conserved N-terminal domain of S. cerevisiae Rec114, S. pombe Rec7, mouse REC114, and Arabidopsis plus maize PHS1 but also Caenorhabditis elegans DSB-1/2 (Rosu et al. 2013; Stamper et al. 2013), not detected previously. Sordaria REC114 transcripts are meiotically induced but to a lower level than those of SPO11 or MER2 (Fig. 1C; Supplemental Tables S1, S2).
Using budding yeast two-hybrid assays (details in the Supplemental Material), we also found that Asy2/Mer2 (1) makes homodimers via its conserved N-terminal region and (2) interacts with Asy3/Rec114 (Supplemental Tables S3, S4). By deletion analysis, we further determined that the amino acid 1–156 N-terminal region of Sordaria Mer2 (and not the amino acid 287–504 C-terminal region) is required for the interaction with Rec114 (Supplemental Table S4). Since the putative SSM1 motif (amino acid residues 73–86) is included in this N-terminal region, we suggest that it could well be the Rec114-interacting motif.
Domain localization and summary of the phenotypes of the analyzed mer2 mutants
The domain organization of the Sordaria Mer2 protein is shown in Figure 2A. Functional analyses (described below) were performed on 13 different mutants, including the four spo76-1 suppressor alleles (asy2-2, asy2-5, asy2-17, and asy2-19; all now referred to as mer2 alleles); five molecularly generated deletions that eliminate either the entire gene (mer2Δ) or specific domains; a repeat motif at the N terminus (mer2-del1) that includes the S phosphorylation sites (Fig. 2A, in bold)—which, in budding yeast, are necessary for DSB formation (Henderson et al. 2006)—plus the potential CC domains (mer2-del2, mer2-del3, and mer2-del4); and four point mutations in potential SUMOylation sites (mer2-K180R, mer2-186R, mer2-K435R, and mer2-K438R). Their localization along the protein is indicated in Figure 2A.
Figure 2.
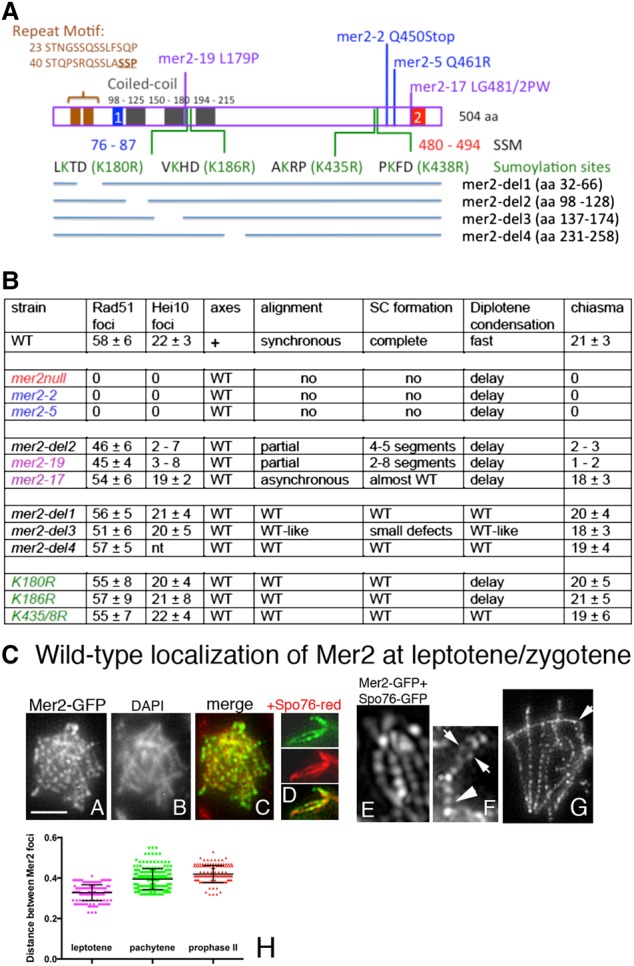
Sordaria Mer2 and phenotypes of the mutants and wild-type Mer2 localization at leptotene/zygotene. (A) Domain organization of the protein. Indicated are the two SSM conserved motifs (in blue and red), the putative CC domains (black rectangles), an N-terminal conserved repeat motif (brown), and the sites of the 12 analyzed mer2 mutants. (B) Prophase I phenotypes of wild type and mutants. (C) Wild-type localization of Mer2 at leptotene and zygotene. (C, panel A) At early leptotene Mer2-GFP foci are regularly spaced along chromosomes. (C, panel B) Corresponding DAPI. (C, panel C) Merge. (C, panel D) Double staining of Mer2-GFP (top) with Spo76-TdTomato (middle) indicates axis colocalization of the two proteins (bottom). (C, panel E) The three-dimensional structured illumination microscopy (SR 3D-SIM) picture double-stained with Mer2-GFP and Spo76-GFP indicates that Mer2 is embedded in the Spo76 axis. (C, panel F) At early zygotene, Mer2 foci remain axis-associated in the unsynapsed regions (arrows) and are single in the synapsed regions (arrowhead). (C, panel G) Late zygotene with remaining Mer2 foci on the axis in the unsynapsed chromosome segment (arrow). (C, panel H) Distances between Mer2 foci at leptotene, pachytene, and prophase II. Mean with standard error of means is marked in black.
Mutants fall into four categories summarized in Figure 2B and described as follows: (1) null alleles defective in recombination, pairing, and progression during the diffuse stage/diplotene transition (mer2Δ, mer2-2, and mer2-5); (2) separation of function alleles nearly normal for recombination initiation but defective in post-DSB pairing/synapsis, with consequences for CO interference but not for SC-related events (mer2-19, mer2-del2, and mer2-17); (3) other separation-of-function alleles that are wild type-like for recombination and pairing but defective for chromosome morphogenesis at the diffuse stage/diplotene transition (mer2-del3, mer2-K180R, and mer2-186R); and (4) mer2-del4, mer2-K435R, and mer2-K438R, which are wild type for all tested processes.
The Asy2/Mer2 protein exhibits dynamic localization to chromosomes throughout meiosis
Mer2 localization to chromosomes in wild type and mutants was examined by single-cell imaging using a fully functional Mer2-GFP C-terminal fusion (expressed under the MER2 promoter control) (details in the Supplemental Material). Mer2 is first visible as a diffuse nuclear signal during S phase (Supplemental Fig. S3A,B). It appears on chromosomes at early leptotene and remains along chromosomes from leptotene through meiotic division II but with marked differences in localization at different stages.
(1) Throughout leptotene and zygotene, Mer2-GFP localizes as rows of regularly spaced foci along all chromosomes (Fig. 2C, panels A–H). There are 60–70 foci per nucleus at early leptotene and 130 ± 8 at late leptotene after axis elongation (n = 50 for each stage), with distances between foci of 0.32 ± 0.03 µm (n = 150 distances) (Fig. 2C, panel H). Colocalization with the axis protein Spo76/Pds5-TdTomato (Fig. 2C, panel D, middle) shows that Mer2 foci are located on the Spo76 signal and thus on the chromosome axis (Fig. 2C, panel D, bottom). A three-dimensional structured illumination microscopy (SR 3D-SIM) picture of colocalization with Spo76-GFP confirmed this axis localization (Fig. 2C, panel E).
(2) Zygotene–pachytene. During zygotene, Mer2 foci move from axes to SC central regions. This change occurs specifically at the same positions where the SC is just forming, implying tight local coupling of Mer2 focus movement and SC installation (Fig. 2C, panels F,G). By pachytene, the 126 ± 6 (n = 70) SC-associated Mer2 foci are dramatically evenly spaced along all seven synapsed homologs (Fig. 3A–D), with distances between foci of 0.39 ± 0.05 µm (n = 200 distances) (Fig. 2C, panel H). 3D-SIM pictures of colocalization with the axis marker Spo76/Pds5-TdTomato show that Mer2 foci are located in the SC central region (Fig. 3D). Concomitantly, per nucleus, five to 10 Mer2 foci are also localized throughout the chromatin rather than associated with the SC/axes (Fig. 3A,D).
Figure 3.
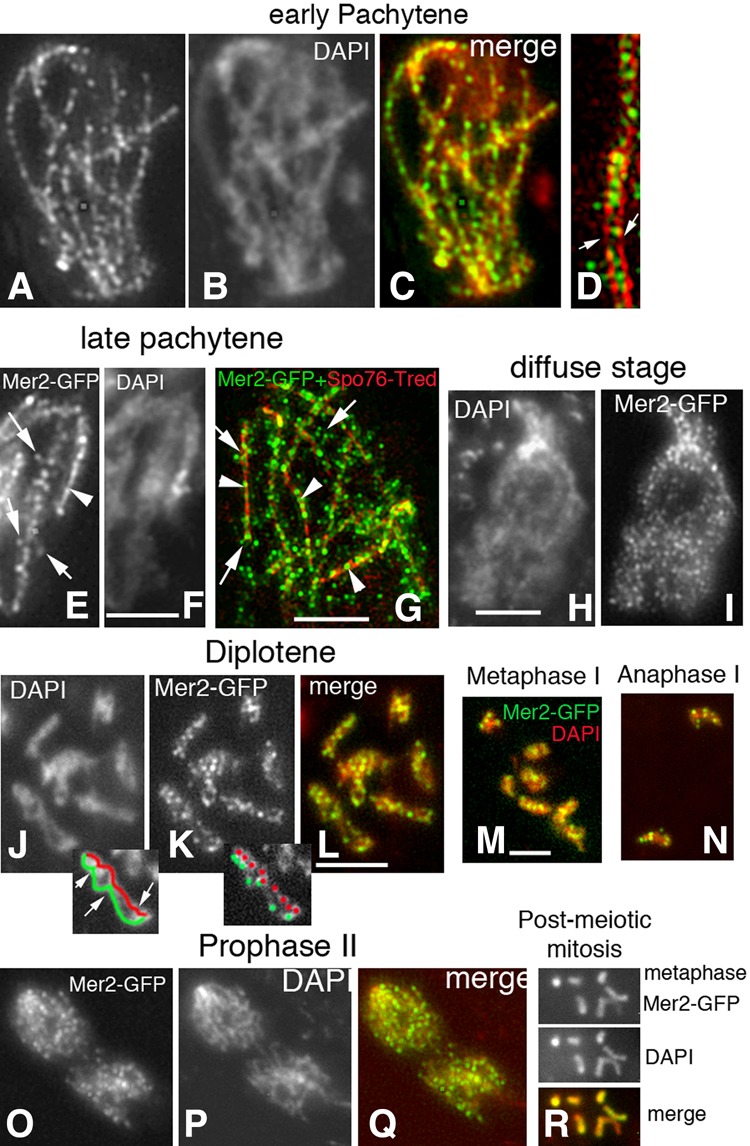
Wild-type Mer2 localization from pachytene to post-meiotic mitosis. (A–C) Pachytene/bouquet nucleus with regularly spaced foci along all bivalents. (B) Corresponding DAPI. (C) Merge. (D) 3D-SIM picture of a bivalent costained with Mer2-GFP and Spo76-TdTomato. Mer2 foci are mostly located between the two homologous red axes (arrows), but a few also decorate the surrounding chromatin. (E,F) During SC disassembly at late pachytene, Mer2 foci show mixed localizations: In some segments, they are still regularly aligned as single foci (arrowhead; right), while, in other segments, they start splitting (arrows; left). (F) Corresponding DAPI. (G) 3D-SIM picture of a late pachytene nucleus (Mer2-GFP and Spo76-TdTomato costaining) comfort observation of E that Mer2 foci form regular arrays only when still SC (red)-associated (arrowheads). (H,I) During the diffuse stage, chromosomes are no longer individualized (H), and Mer2 foci (I) are more numerous than during pachytene (C). (J–L) Spread diplotene nucleus: All bivalents (DAPI in J) show numerous Mer2 foci (K) along their lengths (L; merge). Drawings of the left bivalent (one homolog in red and the other in green) in the two cartoons below J and K show that foci are not specifically localized at chiasma sites (arrows in J). (M,N) Merged pictures of Mer2-GFP and DAPI (red) during metaphase I (M) and anaphase I (N). (O–Q) During prophase II, Mer2 foci (O) are again more numerous and regularly spaced along all chromosomes (Q). (P) Corresponding DAPI. (R) During the post-meiotic mitosis, Mer2-GFP (top) overlaps perfectly with DAPI staining (middle) along all chromosomes (bottom).
(3) Pachytene exit–diffuse stage. At late pachytene, when the SC begins to disassemble via loss of the central region components, Mer2 foci become progressively more irregular in shape and frequently appear as pairs of foci, one on each homolog (Fig. 3E–G, arrows; Supplemental Fig. S3C–E). This pattern suggests that Mer2 foci “split” concomitantly with SC disassembly. Interestingly, split foci seem now to be localized to chromatin loops rather than to the SC/axes (Fig. 3E,G, arrows). Moreover, in such nuclei, foci remain SC-associated in still-synapsed regions (Fig. 3E,G, arrowheads), providing strong evidence for local linkage of these changes to SC disassembly. Pachytene exit leads to the diffuse stage, a prominent universal feature of meiosis (called dictyate stage in mammals) in which individualized chromosomes are no longer discernible (Fig. 3H; more details below). The occurrence of Mer2 splitting is confirmed by the fact that, at the early diffuse stage (Fig. 3H,I), the number of Mer2 foci is nearly double the number seen at mid-pachytene (215 ± 16 [n = 40 nuclei] vs. 126 ± 6 [n = 70]), and foci sizes are smaller (0.17 ± 0.02 µm in diameter compared with the 0.23 ± 0.03-µm diameter of pachytene foci, above).
(4) At diplotene, when individualized chromosomes reappear with closely conjoined sister chromatids linked only at sites of chiasmata, Mer2 forms bright regular foci along each chromosome (Fig. 3J–L), without evidence of specific localization at the chiasma sites (Fig. 3J, arrows). Focus number decreases from early to late diplotene in parallel with increased condensation of the bivalents (from 66 ± 8 to 31 ± 5; n = 50 and n = 30).
(5) Mer2 remains localized on chromosomes through the completion of meiosis and post-meiotic mitoses. Mer2 remains as foci on metaphase I and anaphase I chromosomes (Fig. 3M,N). Their number decreases from 18 ± 3 to 9 ± 3, respectively (n = 20 and n = 10). During prophase of the second meiotic division (Fig. 3O–Q), the number of Mer2 foci increases again (55 ± 5; n = 15), and each chromosome shows rows of regularly spaced foci with distances between foci of 0.42 ± 0.04 µm (n = 130 distances) (Fig. 2CH). Foci remain on metaphase II and anaphase II chromosomes, with numbers reducing with progressive condensation to a final number of five to nine (n = 50) that excludes specific remaining localization to centromeres (seven expected). Thereafter, Mer2 decorates the entire chromosomes during post-meiotic mitosis (Fig. 3R) and disappears upon ascospore germination, which defines return to the vegetative program.
Mer2 localizes to prophase chromosome axes depending on normal cohesin status but is not required for axis development
Further analysis shows that the correct localization of Mer2 is dependent on the normal status of the cohesin complexes. In the absence of the meiotic cohesin Rec8 (rec8Δ), Mer2 foci form (Fig. 4A,B), but their number is reduced (97 ± 10 [n = 30] compared with 130 ± 8 in wild type) (above). This difference in focus number cannot be attributed to defects in mutant axis lengths (53 ± 5 µm compared with 55 ± 4 in wild type; n = 10 and n = 120 nuclei, respectively). In spo76-1, a nonnull mutant of the cohesin gatekeeper Spo76/Pds5, formation of Mer2 foci is even more affected. Their number is strongly reduced (36 ± 11; n = 30), and their localization along chromosomes is highly irregular, with foci showing abnormal shapes (Fig. 4C,D).
Figure 4.
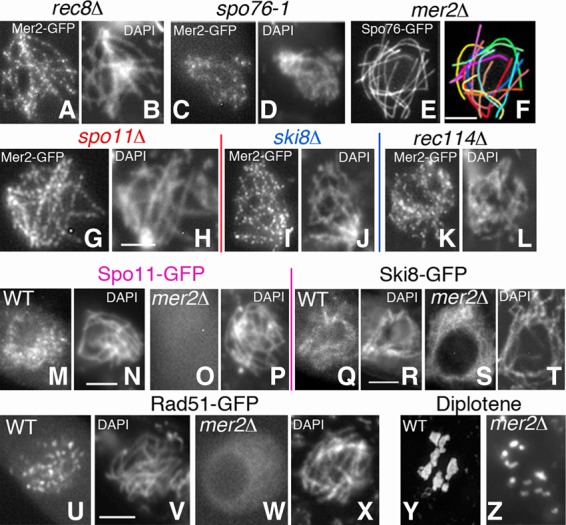
Mer2 localization depends on cohesins, not on DSBs, but is required for DSB formation. (A–D) Mer2-GFP in rec8Δ (A) and spo76-1 (C) leptotene. (B,D) Corresponding DAPI. (C) Mer2 foci are especially abnormal in spo76-1 throughout prophase. (E,F) mer2Δ remains asynaptic until what should be late pachytene by ascus size, but axes are wild type-like by Spo76-GFP staining. (F) Drawing of the 14 univalents (see Supplemental Fig. S3F,G for Rec8-GFP localization). (G–L) Mer2-GFP is and remains axis-located from leptotene to pachytene in spo11Δ, ski8Δ, and rec114Δ. (H,J,L) Corresponding DAPI. (M–P) Spo11-GFP is visible during wild-type leptotene (M) but not in mer2Δ (O). (N,P) Corresponding DAPI. (Q–T) Ski8-GFP localization in wild-type late leptotene (Q) and mer2Δ (S) of same ascus size. (R,T) Corresponding DAPI. (U–X) Leptotene: Rad51-GFP is present in wild type (U) but not in mer2Δ (W). (V,X) Corresponding DAPI (see Supplemental Fig. S4I–W for Rad51-GFP localization in nonnull mer2-19 and mer2-17). (Y,Z) Diplotene: seven chiasmate bivalents in wild type (Y) versus 14 univalents in mer2Δ (Z). Bars, 2 µm.
In contrast, Mer2 is not required for normal axis formation. Axis morphology, as defined by visualization with Spo76/Pds5-GFP (Fig. 4E,F) and cohesin Rec8-GFP (Supplemental Fig. S3F,G), is indistinguishable from wild type in mer2Δ from early leptotene to the end of pachytene. Correspondingly, axis lengths are wild type-like (51 ± 7 µm and 55 ± 4 µm; n = 75 and n = 120, respectively), confirming normal basic chromosome organization in the mutant. These observations imply that the functional interaction between Mer2 and the cohesin axis has functions (below) other than a possible role in basic axis morphogenesis.
Mer2 localization is independent of recombination but is required for the assembly of pre-DSB recombination complexes and DSB formation
Mer2 localization is not dependent on recombination initiation. In spo11Δ (Fig. 4G,H) and ski8Δ mutants (Fig. 4I,J), both of which fail to make DSBs (Tessé et al. 2003), the number of Mer2 foci is essentially the same as in wild type: Unsynapsed axes of these mutants exhibit 124 ± 11 and 131 ± 8 foci, respectively (n = 40 leptotene nuclei by ascus size) as compared with 130 ± 8 in wild-type leptotene (above). Moreover, foci remain axis-associated (with identical numbers) from leptotene to what should be pachytene by ascus size (n = 30 nuclei for each mutant). Mer2 also localizes on chromosomes in the absence of Rec114, but the number of foci is reduced (82 ± 6; n = 20), and foci are (and remain) less regularly spaced than in wild type from leptotene to what should be pachytene by ascus size (Fig. 4K,L).
In contrast, Mer2 localization is essential for the assembly of axis-associated pre-DSB recombination complexes. In wild type, Spo11-GFP appears first as chromatin-associated foci during or just after S phase, and early leptotene nuclei exhibit numerous foci located throughout chromatin (Fig. 4M,N). In contrast, in mer2Δ, Spo11-GFP staining is undetectable at any stage from S phase, when it should first appear, through leptotene (Fig. 4O,P) and pachytene (by ascus size). Interestingly, although reduced in intensity, Ski8-GFP localization along chromosomes is almost wild type-like in mer2Δ (Fig. 4Q–T). This is unexpected given that Ski8 localization is dependent on Spo11 in wild type (Tessé et al. 2003). We infer that Mer2 is required for this dependency (Discussion).
In accord with the above findings, Mer2 is also essential for DSB formation, by three criteria. (1) Rad51 foci, which mark DSB sites in wild type (Fig. 4U,V), are absent from mer2Δ chromosomes (Fig. 4W,X). (2) The spo76-1 mutant (above) and the endonuclease mre11Δ mutant arrest meiotic progression at anaphase I onset—the latter with broken chromosomes (E Espagne, unpubl.). Defects in both mutants are eliminated by mutations that block DSB formation; notably, spo11Δ and ski8Δ. The absence of Mer2 analogously suppresses these defects. (3) The mer2Δ mutant is defective for all of the events of meiotic prophase that are known to be DSB-dependent in Sordaria (Storlazzi et al. 2003; Tessé et al. 2003). (1) Coalignment or synapsis of homologs was never observed in >100 mer2Δ nuclei from leptotene through the stage that should be pachytene by ascus size (Fig. 4E,F, above). (2) Post-DSB recombination markers Mer3, Msh4, and Hei10 foci are absent (data not shown). (3) While wild-type diplotene nuclei exhibit seven bivalents (Fig. 4Y), mer2Δ diplotenes exhibit 14 univalents (Fig. 4Z), implying the complete absence of COs (n = 50 each).
In summary, Mer2 localizes to early prophase chromosome axes specifically in order to mediate the assembly of pre-DSB recombination complexes and thus DSBs without any detectable impact on axis status.
Mer2 is required after DSBs for spatial homologous juxtaposition and pairing
DSBs mediate not only DNA recombination per se but also, concomitantly, chromosome pairing. After DSB formation, one DSB end identifies a partner sequence on the homologous chromosome. The resulting DSB/partner interaction then becomes associated with its underlying partner axis, and the two homologous axes become juxtaposed in space. In wild type, coalignment of homologous axes occurs regularly and synchronously on all chromosomes in each given nucleus even in the presence of an occasional entanglement/interlocking (Fig. 5A). Pairing is followed by SC formation, again with striking per-nucleus synchrony (Fig. 5B). Fifteen percent to 20% of nuclei show entanglements at late leptotene/zygotene (Fig. 5A), but these are resolved such that no interlockings remain by early pachytene (Storlazzi et al. 2010).
Figure 5.
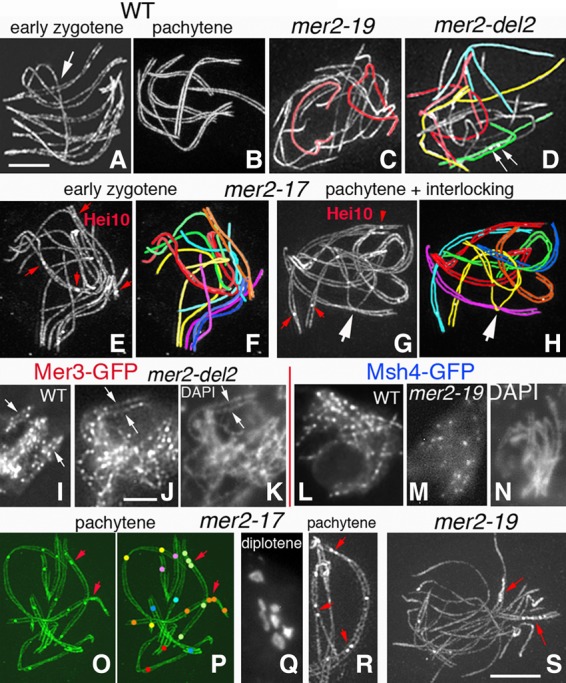
Pairing/synapsis defects in three mer2 nonnull mutants. (A–H,O,R,S) 3D-SIM pictures of Spo76-GFP and Hei10-GFP costaining. The seven bivalents are distinguishable by their lengths and colors (e.g., the two longest are red and yellow, and the smallest is blue). (A,B) Coalignment (A) and synapsis (B) of wild-type homologs are always synchronous, even when two are entangled (arrow in A). (C,D) Pairing/synapsis is either completely defective (C) or partially defective (D) in mer2-19 and mer2-del2. Nevertheless, Hei10 foci form in the few synapsed segments (arrows in D). More nuclei are shown in Supplemental Figure S3, H–L. (E,F) All mer2-17 zygotene nuclei exhibit a mixture of partially aligned (yellow, green, and purple pairs in F) and partially synapsed (red, blue, and orange pairs) homologs, the latter with Hei10 foci (red arrows in E). (G,H) Interlocking in mer2-17: The purple bivalent is entrapped in the yellow pair (arrow). Red arrows point to Hei10 foci. (I–K) Mer3 foci form normally in the three mutants (here mer2-del2) and, like in wild type (I), occur in matching pairs in the segments where coalignment occurs (arrows in J). (K) Corresponding DAPI. (L–N) Note the difference in number and spacing of Msh4 foci in wild type (L) and mer2-19 (M). (N) Corresponding DAPI. (O–R) mer2-17 exhibits complete SC formation with at least one Hei10 focus per bivalent (O,P; red arrows point to close foci) and seven chiasmate bivalents (Q) at diplotene. (R) Example of one bivalent (right) with close Hei10 foci (top and bottom) and another bivalent (left) with close foci adjacent to an open space due to an interlocking (red arrows). (S) At pachytene, mer2-19 remains mostly asynaptic except for few synapsed segments where Hei10 foci accumulate (red arrows). Bars, 2 µm.
Three nonnull mer2 mutants that map to two important domains of the protein, the CC and the C-terminal region (Fig. 2A), exhibit phenotypes diagnostic of a pairing defect. Their analysis is facilitated in Sordaria because stages leading up to metaphase I can be identified independently of chromosomal morphology on the basis of ascus (meiocyte) size, which increases from 20 µm at leptotene to 150 µm at metaphase I.
Two nonnull mutants, mer2-19 and mer2-del2, show similar severe pairing defects: Sixty-five percent of the 100 analyzed nuclei (for each mutant) show no coalignment (Fig. 5C), and 35% show one to five scattered segments of coalignment and one to five patches of the SC, which in every case contain at least one E3 ligase Hei10 foci, which mark the sites of SC-associated CO recombination complexes (Fig. 5D; more examples in Supplemental Fig. S3H–J). In the third nonnull allele, mer2-17, pairing is also defective: Early prophase nuclei (n = 70) show only partial alignment (Supplemental Fig. S3K,L), and 40% show delayed and asynchronous synapsis at zygotene (Fig. 5E,F). Also, in contrast to mer2-19 and mer2-del2, in mer2-17, the SC finally forms all along the chromosomes in 98% of nuclei (Fig. 5G). Also importantly, up to 10% of nuclei exhibit entangled chromosomes or bivalents even through late pachytene (n = 75) (Fig. 5G,H), in contrast to wild type, where interlocks are all resolved by early pachytene (above).
Additional information shows that the pairing defects in these three mutants cannot reflect aberrancies in DSB formation. First, the number of DSBs, as judged by Rad51 foci, is only slightly reduced—by ∼25% in mer2-19 and mer2-del2 and by ∼10% in mer2-17 (n = 50 nuclei for each) (Fig. 2B). Interestingly, mer2-19 and mer2-del2 with 75% of wild-type DSB/Rad51 levels show the same low level of pairing as seen for the ski8 mutants that exhibit only 10% of the wild-type level of Rad51 foci (Tessé et al. 2003). Therefore, a much higher level of pairing should have been observed in the mer2 mutants if the only defect were in DSB formation. Importantly, also in all three mer2 mutants, Rad51 foci appear with wild-type timing without detectable asynchrony, according to similar ascus sizes in wild type and mutants (from cultures started the same day), and with wild-type localizations (Supplemental Fig. S4A–W).
Analysis of Mer3 foci confirms and extends this conclusion. In all three mutants, Mer3 focus numbers are reduced to the same extent as Rad51 foci—by ∼20% in mer2-19 and mer2-del2 (111 ± 9 and 115 ± 6; n = 40 nuclei each) and ∼10% in mer2-17 (121 ± 8; n = 50) as compared with 136 ± 10 in wild type (n = 100). These correspondences suggest that every DSB still gives rise to two Mer3 foci; i.e., one on each homolog axis. Accordingly, all Mer3 foci are axis-associated, and, exactly as in wild type (Fig. 5I), in the limited segments where axis juxtaposition occurs successfully, coaligned axes exhibit matching pairs of Mer3 foci (e.g., mer2-del2 in Fig. 5J,K). By implication, the three pairing-defective mutants are not defective in the formation of axis-associated Mer3 complexes on either DSB end. Also, the fact that the number of Mer3 foci is reduced to the same extent as the number of Rad51 foci argues against a defect in homolog bias as the basis for the pairing defect. If a homolog bias defect were involved, the number of Mer3 foci should be significantly less than the number of Rad51 foci because some DSB ends would not find a partner to set up a Mer3-containing recombination complex, which is not the case.
Interestingly, the severe deficit and asynchrony in SC formation in mer2-19 and mer2-del2 are very similar to the SC defects described previously for a mutant lacking the meiosis-specific MutS homolog Msh4. Since we showed previously that Msh4 acts downstream from Mer3 during the pairing process (Storlazzi et al. 2010), this correspondence further supports a role for Mer2 that is downstream from Mer3 loading. Indeed, Msh4 loading is aberrant in mer2-19 and mer2-del2: (1) The number of foci is reduced by ∼75% at early pachytene (18 ± 5 and 15 ± 9, respectively, as compared with 81 ± 7 in wild type; n = 50, n = 30, and n = 75, respectively), and, (2) contrary to wild type (Fig. 5L), foci are fainter and less regular in spacing and shape (Fig. 5M,N). However, since Msh4 foci normally appear only after coalignment, it is unclear whether these defects reflect a cause or are a consequence of the mutant pairing defect. In mer2-17, the number of Msh4 foci is reduced by only ∼15% (n = 50).
In summary, nonnull mer2 phenotypes identify a Mer2 prophase subfunction specific to the pairing process. DSBs are only slightly reduced in number, Mer3 foci are formed efficiently on both partner axes, and all of these events exhibit normal timing. Also, after coalingment, in mer2-19 and mer2-del2, CO formation occurs in the few short regions of the genome that do successfully complete synapsis, as indicated by the presence of late E3 ligase Hei10 foci (Fig. 5D, arrows; above). SCs and Hei10 foci are analogously well correlated in synapsed regions of mer2-17 (Fig. 5E,G; below). Thus, the three nonnull “pairing” mutants are specifically defective for the homolog coalignment process, not for upstream or downstream processes.
The mer2-17 coaligment defect confers a unique defect in CO patterning
In accord with its modest defect in DSB formation and eventual complete SC formation, the mer2-17 mutant exhibits only an ∼15% reduction in the number of Hei10 foci at mid/late pachytene (19 ± 2 vs. 22 ± 3 in wild type; n = 75 and n = 100, respectively). This decrease concerns all seven bivalents and cannot be attributed to differences in SC lengths: 55.4 μm ± 6.5 μm versus 54.3 μm ± 8.5 μm in wild type (n = 75 and n = 100, respectively). However, analysis of Hei10 focus positions (Fig. 5O,P) further reveals that wild type-like spatial patterning of COs via CO interference is significantly altered. Since CO interference results in a tendency for COs to be evenly spaced along the chromosomes, it is often described by the extent to which interfocus distances are fit by the ν (shape) parameter of a γ distribution. Measurement of 525 bivalents from 75 mer2-17 nuclei and 658 bivalents from 120 wild-type nuclei defined ν-values of 2.52 ± 0.2 and 5.23 ± 0.3, respectively, implying reduced interference in the mutant (Supplemental Fig. S5A). The coefficient of coincidence (CoC) is analogously affected, with significantly higher levels of double COs at shorter interinterval distances in the mutant versus wild type: LCoC = 0.7 µm± 0.18 µm versus 1.3 µm ± 0.1 µm in wild type (Supplemental Fig. S5B). Decreased interference is further confirmed genetically by tetrad analyses made in four adjacent intervals of bivalent 2 in both wild type and mutant (Supplemental Fig. S5C). However, (1) all 525 bivalents exhibit at least one Hei10 focus per bivalent (Fig. 5O,P), and (2) the mutant shows seven chiasmate bivalents at diplotene (n = 60) (Fig. 5Q). Thus, there is no loss of the “obligatory CO,” implying that CO designation is robust.
Reduced interference implies that COs tend to be closer together than normal. However, precise inspection of Hei10 focus localization in mer2-17 further revealed a more specific effect: Forty percent of the 525 analyzed bivalents tend to exhibit very close foci, seen as interfocus distances of 0.1–0.4 µm (Fig. 5P,R, arrows) when the average wild-type focus distance is 2.6 ± 1.1 µm (n = 200 distances). Consequently, the average number of pairs of foci separated by <0.4 µm in mer2-17 is significantly higher than in wild-type nuclei: 1.9 ± 0.13 versus 0.7 ± 0.08 (P < 0.0001; t-test) (Supplemental Fig. S6). Indeed, when the close foci are considered as a single focus, ν-values are now 4.15 ± 0.2 instead of 2.52 ± 0.2, thus still reduced but closer to the wild-type values of 5.23 ± 0.3, indicating that the decreased interference seen in mer2-17 is mostly due to the presence of those close foci. We suspect that closely spaced foci result from the fact that in some chromosome regions, normal coalignment/SC formation was delayed (Fig. 5E), with CO designations therefore channeled into regions of normal SC progression (Discussion). Accordingly, closely spaced foci tend to occur in synapsed regions adjacent to persisting entanglements (Fig. 5R, left).
Analysis of CO interference is not possible in mer2-19 and mer2-del2 mutants because of the small number of SC segments and Hei10 foci per nucleus. However, the occurrence of close foci is even more obvious in the few SC segments that do occur in these mutants versus mer2-17 (Fig. 5S).
Mer2 localization is tightly correlated with the transfer and maintenance of recombination complexes to SC central regions and with SC status
As shown above, during zygotene, Mer2 foci move from axes to SC central regions. This same pattern is also seen for the post-DSB recombination proteins Mer3 and Msh4, implying the existence of a single concerted “transport” step (Storlazzi et al. 2010; Espagne et al. 2011; Discussion). These SC-related focus dynamics of Mer2 are determined by local underlying SC status irrespective of recombination complex status. (1) In null mutants of the SC central region components Zip4 and Sme4, which do not form an SC, Mer2 foci remain on unsynapsed zip4 axes (Fig. 6A) and coaligned sme4 axes (Fig. 6B–D) up to late pachytene, as defined by ascus size. (2) The Mer2 focus shift from axes to SCs still occurs in the absence of Mer3 and Msh4 (thus when both the recombination process and the SC formation are defective) (Storlazzi et al. 2010). Mer2 foci are localized to SC central regions in synapsed regions but remain axis-associated in unsynapsed regions, further highlighting that focus movement is tightly linked to SC formation (Fig. 6E,F). (3) A null hei10 mutant is defective for intermediate and late stages of CO formation but exhibits regular SCs (De Muyt et al. 2014). In this mutant, Mer2 foci occur in wild-type numbers (137 ± 15; n = 40) and move normally to SC central regions (Fig. 6G).
Figure 6.
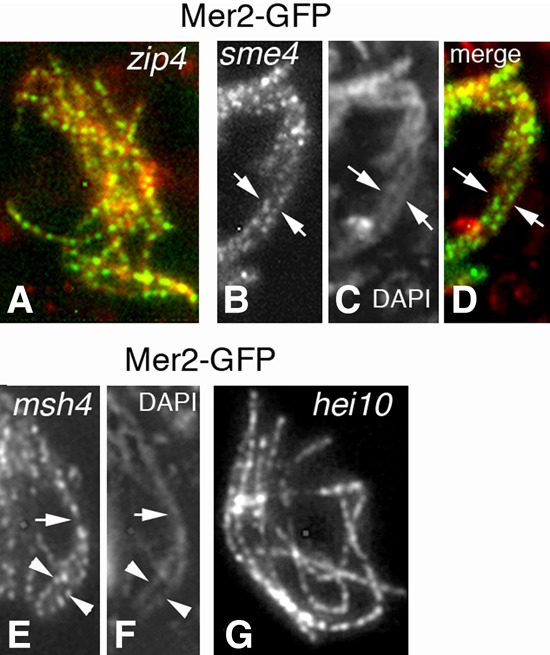
Mer2 localization in the SC plus recombination mutants. (A) In absence of Zip4, Mer2 foci are regularly spaced along all unsynapsed axes like in wild type (distances of 0.42 µm ± 0.06 µm) from leptotene through late pachytene (by ascus size). (B–D) The same timing occurs in the absence of Sme4. As this mutant is not required for homolog pairing, Mer2 foci decorate both coaligned homologs (arrows). (C) Corresponding DAPI. (D) Merge. (E,F) In the absence of Msh4, SCs form only partially: Mer2 remains on axes in unsynapsed regions (arrowheads) but shifts to SC central regions in segments with formed SCs (arrow). (F) Corresponding DAPI. (G) In hei10-null pachytene nuclei, Mer2 localizes along all SCs like in wild type and with similar spacing. Bars, 2 µm.
Interestingly, as described above, SC disassembly triggers Mer2 focus splitting and relocalization on axes/chromatin (Fig. 3E–G). Thus, Mer2 focus dynamics are tightly locally coupled to SC dynamics during both SC installation and disassembly, pointing to a direct interaction between Mer2 and SC central region components. Via this localization, Mer2 appears to be involved in the maintenance of the association of recombination complexes with SC central components as long as the SC is present.
Mer2 mediates global chromosome compaction at the diffuse stage/diplotene transition, potentially dependent on two putative sites for SUMO modification
As described above, wild-type Mer2 foci remain colocalized with Spo76 in still-synapsed regions and with chromatin throughout the diffuse stage (Fig. 7A). Also, at pachytene, most Hei10 foci colocalize with Mer2 foci (Fig. 7B, arrows), but a subset of Hei10 foci (arrowheads in Fig. 7B) does not colocalize with Mer2. During the diffuse stage, all Mer2 foci colocalize with Hei10 foci (Fig. 7C), which implies a persistent correlation between Mer2 and CO recombination complexes during the transition into the chiasmate state.
Figure 7.
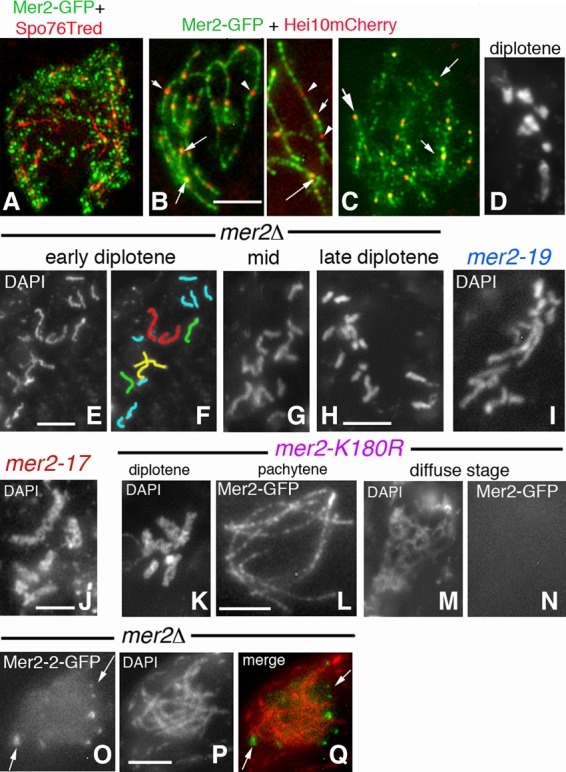
Diffuse stage/diplotene compaction and Mer2-2-GFP localization. (A) 3D-SIM mid-diffuse stage nucleus: Mer2-GFP and Spo76-TdTomato costaining. Mer2 foci are mostly chromatin-associated. (B) At early pachytene, most Hei10 foci colocalize with Mer2 foci (arrows), but a subset of Hei10 foci (arrowheads) does not colocalize with Mer2. (C) At the late diffuse stage, all Hei10 foci colocalize with Mer2 foci (arrows). (D) Wild-type diplotene. (E–H) Chromosome recompaction after the diffuse stage is slow and progressive in mer2Δ. (E) Early diplotene with long kinky chromosomes. (F) Corresponding drawing of the 14 univalents. (G,H) Progressive compaction from mid (G) to late (H) diplotene (see Supplemental Fig. S4X–Z for illustrations of diplotene compaction in spo11Δ, ski8Δ, and msh4Δ). (I–K) Similar diplotene compaction defects and fuzziness in mer2-19 (I), mer2-17 (J), and mer2-K180R (K). (L–N) While Mer2-GFP localization is wild type-like along all pachytene bivalents in mer2-K180R (L), foci are no longer visible from the diffuse stage on (M,N). A similar absence of Mer2 is also seen in the other mer2 mutants (see the text). (O–Q) In contrast to the other mutants, Mer2-2-GFP (O) in a mer2Δ background is not chromosome-located (Q) but forms bright foci around the nucleus from leptotene to pachytene (by ascus size). (P) Corresponding DAPI. Bars, 2 µm.
Both the diffuse stage and diplotene occur in all mer2 mutants. Strikingly, however, with the exception of the molecularly generated mutants with a fully wild-type phenotype (Fig. 2B; above), all other mer2 mutants are defective in the diffuse stage–diplotene transition in two respects. First, contrary to wild-type chromosomes (Figs. 4Y, 7D), mer2Δ diplotene chromosomes are only partially condensed and kinky (Fig. 7E,F) in asci of the same size (chromosome lengths from 38.9 to 7 µm from early to late diplotene compared with 8.9 to 3.1 µm for wild type, respectively; n = 10 nuclei for each). Second, compaction into the diplotene state occurs slowly. In wild type, chromosome recompaction after the diffuse stage occurs rapidly, as suggested by the low frequency of diplotene nuclei seen per fruiting body (5%–10% among the >100 meiocytes in each of the 50 analyzed fruiting bodies; thus, a maximum of 500 in 5000 meiocytes). In contrast, in mer2 mutants, each fruiting body contains >30% diplotene nuclei (thus, >1500 among 5000 meiocytes), which exhibit all intermediates in progression of condensation (Fig. 7E–H), a heterogeneity never seen in wild type. These defects cannot be attributed to the absence of DSBs or other recombination defects because neither is observed in either spo11Δ or ski8Δ (which, like mer2Δ, also form only 14 univalents) (Supplemental Fig. S4X,Y) or mer3Δ or msh4Δ (which show a mixture of univalents and bivalents like mer2-19 and mer2-del2) (Supplemental Fig. S4Z). These findings are especially important because they show that Mer2 plays a general role for global chromosome status and, furthermore, that this role includes an activity that expedites, and is required for, fully global compaction during the early prophase/diplotene transition (Discussion).
It is notable that slow diplotene compaction is observed in the three pairing-defective mutants mer2-19, mer2-del2, and mer2-17 (Fig. 7I,J; above), albeit with a lesser effect in mer2-17 (10%–20% in 10 analyzed fruiting bodies). By implication, the subfunction of Mer2 that mediates homolog–axis juxtaposition (above) also mediates global chromosome compaction at diplotene (Discussion).
Moreover, delayed compaction is seen in two mutants that specifically lack putative sites for SUMO modification, mer2-K180R and mer2-K186R (Fig. 2A), suggesting a role for SUMOylation in the compaction process (Fig. 7K). This role is specific to this stage: The two mutants are fully normal for homolog pairing/synapsis and pachytene Mer2 localization (although as fainter foci as wild-type foci) (Figs. 7L vs. 3A,E) and function by all of the tests described above. Two other putative SUMO sites (K435R and K438R) (Fig. 2A) are not required.
Importantly, also, in the mer2 mutants that exhibit delayed diplotene compaction (Fig. 2B), the corresponding mutant GFP protein, as assayed in a mer2Δ background, is axis- and SC-localized (when formed) up to late pachytene and then disappears prematurely during the diffuse stage as both focus and chromatin staining, (e.g., Fig. 7M,N for mer2-K180R).
In the mer2-2 mutant (null phenotype), the protein is visible from early leptotene to end pachytene as bright foci (five to 20; n = 30) but never on chromosomes (Fig. 7O–Q). While a direct marker for the nuclear envelope is lacking, it is most likely that this mutant Mer2 protein is localized to the nuclear periphery; e.g., via a failure to enter the nucleus. Nonetheless, Mer2-2-GFP staining also disappears at the diffuse stage. The mer2-2 mutation creates a stop codon near the C terminus, and the truncated protein therefore lacks the evolutionarily conserved C-terminal motif. The absence of chromosome localization suggests that this C-terminal domain might also contain a nuclear localization signal.
One way of integrating these phenotypes is to say that binding of Mer2 to DNA/chromosomes during the diffuse stage, potentially dependent on SUMOylation, is required to protect the protein from premature degradation and is a core function of the protein.
Discussion
Molecular identification of Sordaria Asy2/Mer2 has led to bioinformatics identification of a new evolutionarily conserved meiotic protein family with identified members in almost all eukaryotic kingdoms. Additional information supports the bioinformatics inferences. First, all orthologs whose functions have been analyzed are required for DSB formation and thus pairing. These include budding yeast Mer2/Rec107, fission yeast Rec15, Sordaria Asy2/Mer2, mouse IHO1, and plant PAIR1 and PRD3. However, for the latter three molecules, no relationship to Mer2 has been detected previously (Nonomura et al. 2004; De Muyt et al. 2009; Miao et al. 2013; Stanzione et al. 2016). Second, localization to diplotene chromosomes, demonstrated here for Asy2/Mer2, has also been reported for mouse IHO1 (Stanzione et al. 2016). Third, tangled chromosomes and diplotene defects in chromosome compaction, like those described above for mer2 mutants of Sordaria, were also observed in the rice pair1-1 mutant (Nonomura et al. 2004).
The functional and localization analyses presented above further reveal that Mer2 mediates a series of prophase/prometaphase events that correspond to different Mer2/chromosome association patterns. Axis-associated Mer2 first mediates recombination initiation. Next, after DSB formation, it is required for recombination-mediated juxtaposition of homologous chromosomes. Interestingly, Mer3 focus patterns indicate that Mer2 appears to be required after axis-associated recombination complexes appear on both the donor and partner axes. We thus infer that Mer2 plays a role in the process of spatial juxtaposition per se, the nature of which remains currently unknown. Defective pairing is accompanied by aberrant CO patterns, which we attribute to the failure of regular coalignment. During synapsis, Mer2 relocates into the SC central region, where it mediates recombination complex/SC central component associations, with a possible “transportation role” of the recombination proteins into the SC. Finally, Mer2 mediates global chromosome compaction at the diffuse stage/diplotene transition, with concomitant localization to chromatin in their diffuse state and then along reformed axes at diplotene.
These identified roles of Sordaria Mer2 include, but also extend well beyond, the previously described roles of Mer2/Rec107, Rec15, IHO1, PAIR1, and PRD3 in the formation of DSBs and/or pre-DSB recombination complexes (references above; for review, see Lam and Keeney 2015).
At early leptotene, Mer2 mediates the assembly of pre-DSB complexes and DSBs
The role of Sordaria Mer2 in DSBs is defined by the same criteria as those described for yeasts (see above).
(1) Its localization is independent of both DSBs and the presence of Spo11, Ski8, or Rec114 (Li et al. 2006; Panizza et al. 2011; above). Therefore, Mer2 axis association in spo11Δ and ski8Δ is not a consequence of DSB formation, and the lack of DSB formation in those mutants is not due to the absence of Mer2 per se.
(2) Spo11 and Rad51 are not loaded on chromosomes in the absence of Mer2. Interestingly, Ski8, another pre-DSB component, shows a wild type-like localization in mer2Δ. This is surprising because in wild-type Sordaria, like in budding yeast, Ski8 loading and Spo11 loading are interdependent (Tessé et al. 2003; Arora et al. 2004). Thus, the absence of Spo11 localization in mer2Δ would have been expected to concomitantly abrogate Ski8 localization. Instead, the current findings imply that the interdependency of Spo11/Ski8 loading requires Mer2. One interpretation could be that meiotic pre-DSB complexes comprise a set of competing and counterbalancing interactions such that the absence of Mer2 permits promiscuous loading of Ski8.
(3) Localization of yeast Mer2/Rec15 and mouse IHO1 is strongly dependent on axis components Hop1/Red1/Hormad1 and partially dependent on Rec8 (Panizza et al. 2011; Miyoshi et al. 2012; Stanzione et al. 2016). In Sordaria, no orthologs of Hop1/Red1 have yet been found in the genome; however, the axis localization of Mer2 is strongly dependent on the cohesin gatekeeper Spo76/Pds5 as defined by the spo76-1 allele, which is required for chromosome axis morphogenesis (van Heemst et al. 1999) and again is partially dependent on Rec8. We showed previously that DSB formation in the spo76-1 mutant background triggers axis destabilization, dependent on Spo11 (Storlazzi et al. 2003) and, as shown in the present study, Mer2. Thus, Mer2 might be molecularly “embedded” within leptotene chromosome axes.
At mid–late leptotene, Mer2 mediates homolog axis juxtaposition and normal CO patterning
Analysis of mer2 nonnull alleles has allowed the discovery of a new type of role for Mer2 in the process of homologous pairing. Three separation-of-function mutants (mer2-19, mer2-del2, and mer2-17) are specifically defective in the role of axis-associated recombination complexes to effectively mediate the correct juxtaposition of the two homologs in space after DSB formation. We showed previously that regular homolog coalignment requires tight temporal coupling between the formation of nascent DSB/partner contacts and spatial homolog axis juxtaposition at the corresponding positions (Storlazzi et al. 2010). We can now implicate Mer2 as another player in this process. Since both preceding/prerequisite and ensuing events of prophase occur normally in the three pairing-defective mutants, their phenotype identifies a unique specific Mer2 subfunction.
By mediating axis juxtaposition, Mer2 also mediates normal CO patterning and thus interference. The presence of very close Hei10 foci is likely a secondary consequence of the reduced extent of coalignment (plus, in mer2-17, the presence of unsolved interlockings), which in turn implies a reduced number of interaxis “bridges” available as substrates for CO designation. Given this situation, if there were a driving force to provide a particular total number of COs, those COs would be “forced” to occur in a smaller extent of chromosome length and thus be aberrantly close together in the regions that are still available. An analogous situation has been reported in two Arabidopsis mutants: E1 enzyme axr1 of the neddylation complex and kinesin pps1 (Duroc et al. 2014; Jahns et al. 2014).
At zygotene/pachytene, Mer2 mediates the association of recombination complexes with the SC central region
We showed previously that the SC central component Sme4 (Zip1-like) is required for the relocalization of the recombination proteins Rad51, Mer3, and Msh4 from on-axis to within the SC central region in temporal synchrony and local coordination with SC nucleation (Espagne et al. 2011). Mer2 foci move from axes to SC central regions in the same pattern as that described previously for Mer3 and Msh4 foci. In all three cases, this shift is tightly coupled to the formation of SC both functionally and spatially, implying the existence of a single concerted “transport” step, during which Mer2 is part of the “transportation complex” for recombination ensembles. Thereafter, both Mer2 and recombination complexes remain SC-associated throughout pachytene. In the absence of SC central components Sme4 or Zip4, Mer2 remains on axes (like Mer3 and Msh4) until the end of pachytene. Moreover, relocalization of Mer2 foci to SCs is seen in mer3Δ and msh4Δ, which are severely defective in recombination (Storlazzi et al. 2010). Also, Mer2 foci remain on Hei10-containing SC patches seen during the diffuse stage. Thus, Mer2 focus movement is not coupled to the progressing recombination process or the Mer3/Msh4 cargo but to the local underlying axis/SC status.
Taken together, these results strongly suggest that Mer2 participates functionally in recombination/SC interplay by mediating (1) release of recombination complexes from axes, thereby enabling their transportation to SCs; (2) stable association of those complexes with SCs throughout pachytene, as required for proper completion of the recombination process; (3) release of recombination complexes from the SC central region at the end of pachytene (as suggested by the “return” of Mer2 foci on axes/chromatin); and (4) progression of late stage recombination complexes (e.g., Hei10) to chiasmata sites during the pachytene/diplotene transition. These Mer2 roles for recombination complexes would be analogous to its role for development of pre-DSB recombination complexes, except that the protein now mediates the relationship of recombination complexes to underlying SC central regions/components rather than to underlying axes.
At the diffuse stage–diplotene transition, Mer2 mediates global chromosome reorganization and compaction, potentially dependent on SUMOylation
A particularly unexpected finding of the present study is that Sordaria Mer2 is required for timely and regular recompaction of chromosomes as they emerge from the diffuse stage to diplotene. We note that during diplotene/diakinesis, IHO1 also localizes to chromosomes in mouse spermatocytes (but functional analysis is precluded by apoptosis in the mutant) (Stanzione et al. 2016). Moreover, pair1-1 is defective in chromosome compaction, like the Sordaria mer2 mutants (Nonomura et al. 2004). Thus, this later role of Mer2 family proteins may be quite general.
Interestingly, however, even though mer2 mutant chromosomes are delayed and defective in achieving the diplotene state, they subsequently show an almost wild-type condensation at metaphase I. Moreover, Mer2 chromosome association is required during the diffuse stage/diplotene transition because all mutant Mer2-GFPs are lost at this stage. Finally, this role of Mer2 may specifically require SUMOylation: Mutations in two predicted SUMOylation sites confer defects specifically at this stage.
Is homolog coalignment mediated by chromatin/chromosome compaction?
The fact that Mer2 mediates global chromosome reorganization at the diffuse stage/diplotene transition is a striking and unexpected finding. It is also particularly intriguing that the three mer2 alleles that are specifically defective in homolog coalignment are then also defective at the diffuse stage/diplotene transition, which is notably characterized by chromosome compaction. Accordingly, pairing could also involve chromosome compaction.
Taken together, our findings imply that Mer2 is not simply a recombinosome/axis linker molecule but instead has broader roles, which include the capacity for interaction with, and direct functional impact on, basic chromosome structure. The specific molecular targets of this activity remain to be defined. However, cohesin complexes are particularly attractive targets. The Sordaria ASY2/MER2 and ASY3/REC114 genes were identified as suppressors of spo76-1, a nonnull allele of the cohesin gatekeeper Spo76/Pds5. Moreover, the spo76-1 mutant exhibits delayed and defective diffuse stage/diplotene compaction plus local chromosome destabilization at CO sites (van Heemst et al. 1999). Furthermore, the spo76-1 and mer2 diplotene defect is also seen in a rec8-null mutant (Storlazzi et al. 2003). Those phenocopies raise the possibility that Mer2 could be a meiotic modulator of Spo76/Pds5 and/or the cohesin/axis status. Therefore, we anticipate that elucidation of the diffuse stage/diplotene role of Mer2 will reveal important new aspects of its molecular activities.
Materials and methods
Identification of Sordaria ASY2/MER2
ASY2 was identified by whole-genome sequencing with SOLiD technology (performed by the Next-Generation Sequencing Platform of the Institut Curie, Paris) of the wild-type reference strain and the four asy2 mutant strains (Huynh et al. 1986). All analyses were carried out using the Galaxy/Analysis and Bioinformatics for Marine Science (ABiMS) site (http://galaxy.sb-roscoff.fr). Mapping of SOLiD reads was achieved by using the “Map with BWA for SOLiD” tool with default parameters and variant calling using GATK2 tools. Mutations were identified by screening of the sequence data of each strain in order to identify single-nucleotide polymorphisms and indels (insertions/deletions) <4 bases that had a read coverage of at least 40% of the average coverage for the specific strain and occurred in the same CDS in all four mutants and not in the wild-type strain. These analyses allowed the identification of a single CDS: SMAC_02785. The mutations identified by next-generation sequencing were confirmed by sequencing of this CDS in both mutant and wild type. To confirm that the found mutations were in the ASY2 gene, we introduced a wild-type ASY2 allele in the ectopic position in each mutant strain: All four mutants displayed a wild-type phenotype.
Bioinformatics
Identification of Mer2 and Rec114 homologs was performed with a series of PSI-BLAST, CS-BLAST, or HHsenser analyses at Max Planck Institute (https://toolkit.tuebingen.mpg.de), using as inputs multiple sequence alignments (MSAs) generated by MAFFT 7.0 (http://mafft.cbrc.jp/alignment/server). MSA inputs included only previously validated homologs, and alternative query sequences were systematically used as the MSA header sequence to improve detection of remote homologs. Ultimately, candidate proteins were validated through PSI-PRED (http://bioinf.cs.ucl.ac.uk/psipred) or Ali2D (https://toolkit.tuebingen.mpg.de) secondary structure predictions. MSAs were colored with ESPript 3.0 (http://espript.ibcp.fr) using the Risler homology matrix (Supplemental Fig. S1A,B) or with the BioJS MSA viewer (http://msa.biojs.net/app) using the Clustal2 color scheme (Fig. 1). The phylogenetic relationship among Mer2 family members was analyzed with PhyML 3.0 (https://toolkit.tuebingen.mpg.de/#/tools/phyml) using the JTT model of amino acid replacement and 200 replicates (Supplemental Fig. S2).
Mutants and GFP tags
Null mer2 and rec114 mutants were generated by single-step gene replacement: A hygromycin resistance cassette replaced the entire ORF. Transformants were selected for hygromycin resistance, and the presence of the deleted allele was confirmed by sequencing and introduction of an ectopic wild-type MER2 or REC114 gene in the corresponding null mutants. The GFP (p-EGFP-1; Clontech) coding sequence was fused to the C terminus of MER2 under the control of the MER2 promoter. After validation by sequencing, the GFP allele was ectopically integrated into a wild-type strain. Mer2-GFP in wild type or in mer2Δ is wild type by all criteria and thus fully functional. Point mutants were created by PCR-based mutagenesis and introduced in wild type by cotransformation with a plasmid encoding the hygromycin resistance cassette. Additional information on plasmid construction, mutants, and GFP tags is in the Supplemental Material.
Cytology
For cytological analysis, GFP, mCherry, TdTomato, and DAPI (0.5µg/mL) signals were observed either on living material or after fixation in 4% paraformaldehyde, with a Zeiss Axioplan microscope with a CCD Princeton camera and with a DeltaVision OMXTM platform (3D-SIM; Applied Precision).
CO interference analyses
For γ and CoC analyses of interference, Hei10 interfocus distances were measured from the center of the first focus to the center of the next focus using ImageJ software (http://rsb.info.nih.gov/ij). Hei10 focus patterns were defined in early–mid-pachytene nuclei (by ascus size) and along 525 mer2-17 and 658 wild-type bivalents. The ν parameter of the γ distribution was performed by the free Wessa γ distribution calculator (http://www.wessa.net/rwasp_fitdistrgamma.wasp). The interinterval distance at CoC = 0.5 was used as a reliable and convenient indicator for CO interference strength (details in Supplemental legend of Fig. S5).
Genetic analysis of interference was performed in 423 wild-type and 230 mer2-17 tetrads issued from crosses with five linked markers of bivalent 2 (Supplemental Fig. S5).
RT-qPCR experiments
Cultures for RNA preparations were performed from day 1 to day 9 at 25°C. The sexual cycle started at day 3. RNAs were extracted from fruiting bodies, quantified, and checked for integrity on a gel. Total RNAs were reverse-transcribed with SuperScript III (Lifes Technologies). See the Supplemental Material for primer designation, cDNA detection, biological replicates, reference genes, and average Cq for each gene. A bilateral Student's test for heteroscedastic samples was used to compare the four biological replicates obtained at day 1 with those of the other days (Supplemental Table S2). The cutoff for significant P-value was set at <0.01. RT-qPCR experiments were MIQE (minimum information for publication of quantitative real-time PCR experiments)-compliant.
Two-hybrid experiments
cDNA sequences were amplified from RT–PCR at day 2 or day 4 (above) with the appropriate primers (Supplemental Material; Supplemental Table S3) and Phusion (ThermoFisher Scientific) or Pfu (Promega). The detailed methods for both colony assay and dosage of β-galactosidase activity are in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank A. Storlazzi for kindly providing Spo76-TdTomato. We are extremely grateful to Edith Heard for providing access to her 3D-SIM microscope. This work, S.T., R.D., E.E., K.B., E.D., R.A., and D.Z. were supported by grants from the Centre National de la Recherche Scientifique (CNRS; Unité Mixte de Recherche 8621 and Institute for Integrative Biology of the Cell) and by a subcontract collaboration with N. Kleckner supported by the National Institutes of Health (NIH; RO1 GM 044794). H.-M.B. was supported by grants from the CNRS (Unité Mixte de Recherche 5547, Centre de Biologie du Développement), and Z.L. was supported by National Natural Science Foundation of China (project 31671293) and Shandong Provincial Natural Science Foundation, China (project JQ201605).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.304543.117.
References
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V. 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Albini SM, Jones GH. 1987. Synaptonemal complex spreading in Allium cepa and A. fistulosum. I. The initiation and sequence of pairing. Chromosoma 95: 324–338. [Google Scholar]
- Anderson LK, Stack SM. 1988. Nodules associated with axial cores and synaptonemal complexes during zygotene in Psilotum nudum. Chromosoma 97: 96–100. [Google Scholar]
- Arora C, Kee K, Maleki S, Keeney S. 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell 13: 549–559. [DOI] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Borde V, de Massy B. 2013. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev 23: 147–155. [DOI] [PubMed] [Google Scholar]
- de Boer E, Stam P, Dietrich AJJ, Pastink A, Heyting C. 2006. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci 103: 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, Lainé-Choinard S, Pelletier G, Mercier R, Nogué F, et al. 2009. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. 2014. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 28: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Lemhemdi A, Larchevêque C, Hurel A, Cuacos M, Cromer L, Horlow C, Armstrong SJ, Chelysheva L, Mercier R. 2014. The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genet 10: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E, Vasnier C, Storlazzi A, Kleckner NE, Silar P, Zickler D, Malagnac F. 2011. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci 108: 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. 2006. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh AD, Leblon G, Zickler D. 1986. Indirect intergenic suppression of a radiosensitive mutant of Sordaria macrospora defective in sister-chromatid cohesiveness. Curr Genet 10: 545–555. [DOI] [PubMed] [Google Scholar]
- Jahns MT, Vezon D, Chambon A, Pereira L, Falque M, Martin OC, Chelysheva L, Grelon M. 2014. Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biol 12: e1001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bourbon H-M, de Massy B. 2010. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hooker GW, Roeder GS. 2006. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z. 2013. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K. 2012. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell 47: 722–733. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Ohta K. 2013. Spatiotemporal regulation of meiotic recombination by Liaisonin. Bioarchitecture 3: 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Pearlman RE. 1990. Telomere and centromere DNA are associated with the cores of meiotic prophase chromosomes. Chromosoma 100: 8–14. [DOI] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci 115: 1611–1622. [DOI] [PubMed] [Google Scholar]
- Nonomura K-I, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2004. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH. 2007. Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod 13: 837–844. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2004. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, Hunter N. 2012. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufas JS, Santos JL, Diez M, Suja JA. 1992. Meiotic chromosome structure: relationship between the synaptonemal complex and the chromatid cores. Genome 35: 1054–1061. [Google Scholar]
- Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. 2013. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanzione M, Baumann M, Papanikos F, Dereli I, Lange J, Ramlal A, Tränkner D, Shibuya H, de Massy B, Watanabe Y, et al. 2016. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat Cell Biol 18: 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98: 261–271. [DOI] [PubMed] [Google Scholar]
- Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. 2014. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci 111: E5059–E5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.