ABSTRACT
Many microbes exploit host cellular lipid droplets during the host-microbe interaction, but this phenomenon has not been extensively studied for fungal pathogens. In this study, we analyzed the role of lipid droplets during the interaction of Cryptococcus neoformans with macrophages in the presence and the absence of exogenous lipids, in particular, oleate. The addition of oleic acid increased the frequency of lipid droplets in both C. neoformans and macrophages. C. neoformans responded to oleic acid supplementation by faster growth inside and outside macrophages. Fungal cells were able to harvest lipids from macrophage lipid droplets. Supplementation of C. neoformans and macrophages with oleic acid significantly increased the rate of nonlytic exocytosis while having no effect on lytic exocytosis. The process for lipid modulation of nonlytic exocytosis was associated with actin changes in macrophages. In summary, C. neoformans harvests lipids from macrophages, and the C. neoformans-macrophage interaction is modulated by exogenous lipids, providing a new tool for studying nonlytic exocytosis.
KEYWORDS: fatty acids, lipid droplets, Cryptococcus neoformans, nonlytic exocytosis, microscopy, macrophages, host-pathogen interaction, electron microscopy
INTRODUCTION
Cryptococcus neoformans is a ubiquitous, encapsulated basidiomycetous yeast that is an important fungal pathogen worldwide, causing around 180,000 deaths annually, particularly in immunocompromised individuals (1, 2). Infection occurs following the inhalation of spores or desiccated yeasts from contaminated sites in the environment, e.g., from soil, trees, or bird droppings. C. neoformans is usually controlled and eliminated by the immune system in the lung; however, in immunocompromised individuals, the fungi can disseminate to infect the central nervous system, where they cause meningoencephalitis, which is the predominant clinical manifestation of cryptococcosis (2). Recently, the number of cases of cryptococcosis has increased, mainly due to increasing numbers of individuals with impaired immunity due to immunosuppressive therapy (3).
C. neoformans is a free-living organism in the environment, but as a facultative intracellular pathogen, the fungus replicates almost exclusively in alveolar macrophages in the lungs (4). The interplay between the pathogen and the host cell is essential for disease progression because alveolar macrophages comprise a major line of defense between the host and the outside world (5). Various virulence factors have been associated with C. neoformans, including enzymes such as phospholipases, proteases, urease, or laccases, growth at physiological temperature (37°C), melanin synthesis, and the polysaccharide capsule surrounding the cell wall (2, 6, 7). The fungi can modulate some aspects of their intracellular lifestyle, such as inducing phagosomal permeabilization, leading to a higher pH in the phagosome, which also induces host cell apoptosis, in addition to egressing via nonlytic exocytosis (NLE), which may contribute to increased systemic dissemination by a Trojan Horse mechanism (8; C. De Leon-Rodriguez, D. Rossi, M. S. Fu, C. Coelho, I. Guerreri Ros, B. Caballero, S. T. Nolan, and A. Casadevall, unpublished data).
Among contributors to C. neoformans pathogenicity, host lipids or lipids in the environment may play significant roles. Indeed, there is mounting evidence suggesting that C. neoformans uses host cell lipids for growth to influence virulence and to modulate the immune response (9–11). As the phagosomes of macrophages have limited glucose availability (12), C. neoformans therefore must use energy sources other than glucides, for example, lipids. Intracellular C. neoformans upregulates its genes involved in the glyoxylate pathway, which allows the utilization of alternative carbon sources such as fatty acids (FA) from β-oxidation for carbohydrate syntheses and energy production (12, 13). Upon scarcity of glucose, peroxisomal and mitochondrial β-oxidation is activated in C. neoformans when FA are exogenously supplied (10). Interestingly, virulence was attenuated in C. neoformans mutants unable to perform β-oxidation, supporting the hypothesis that the utilization of FA as a carbon source in the nutrient-scarce phagosome is important for the fungus. Additionally, both in animal models and in macrophages in vitro, several cryptococcal lipid metabolism-related gene transcripts, such as FA import and lipid degradation genes, are upregulated (14, 15). These studies highlight the importance of lipid metabolism during cryptococcal infection.
FA synthesis is essential for C. neoformans growth in vitro and in vivo, and its inhibition affects virulence (16). For example, host-derived arachidonic acid can be scavenged by C. neoformans and incorporated into prostaglandins in a phospholipase B1 (PLB1)-dependent manner (9). PLB1 is also thought to interact with host cell membranes and to mediate C. neoformans release from the phagosome (9, 17). Moreover, in stationary-phase C. neoformans, fatty acids such as oleic acid (OA) and palmitic acid were taken up by the fungi, but their fate or use is unknown (10).
Cellular lipid droplets (LD) have gained attention in the field of host-pathogen interactions since many organisms, including viruses, protozoans, or bacteria, can modulate their metabolism by increasing, suppressing, altering, metabolizing, or scavenging their lipids (reviewed in reference 18). LD are ubiquitous structures found in virtually all cell types whose primary functions include the storage of FA for energy via β-oxidation or FA sequestration for lipotoxicity prevention. As such, excess cellular FA are stored in LD following their conversion to the neutral lipid triacylglycerols (TAG) and steryl esters (SE). Approximately 10% of C. neoformans dry weight is composed of lipids, mainly phospholipids (63%) and neutral lipids (30%), with TAG comprising 90% of all cryptococcal neutral lipids (19, 20). In free-growing C. neoformans, TAG within LD accumulate as C. neoformans enters stationary phase (21).
Little is known about the effect of excess FA, host cell LD, and their derived lipids on the C. neoformans-host interaction. In this study, we explored the effects of excess FA on C. neoformans growth axenically and in mouse macrophages and the ability of these fungi to scavenge OA or OA-derived lipids. We show that the fungi stored excess OA in newly formed fungal LD. Excess OA boosted the replication of both extracellular and intracellular C. neoformans and increased the frequency of NLE of C. neoformans from macrophages. This establishes that C. neoformans can scavenge lipids from host cells and responds to them by altering certain aspects of the host-microbe interaction, including NLE.
RESULTS
C. neoformans possesses lipid droplets.
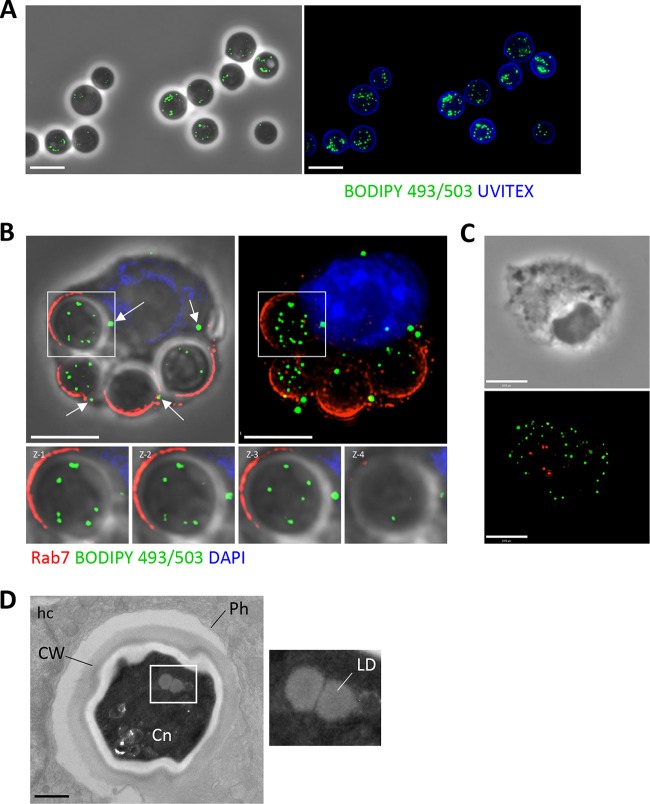
LD are evolutionary conserved organelles found in all cell types and organisms (22). To examine the ability of extracellular C. neoformans to store lipids, the fungi were stained with BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene), which is a specific dye for neutral lipids and therefore stains LD. Fluorescence microscopy revealed the presence of numerous green foci within the fungi (Fig. 1A). The presence of LD was also confirmed within intracellular C. neoformans by infecting bone marrow-derived macrophages (BMDM) with the fungi for 4 h prior to labeling with BODIPY 493/503. LD were detected within C. neoformans, as visualized by sequential z-stack images through the fungi (Fig. 1B). The LD of uninfected BMDM can be seen in Fig. 1C. Ultrastructural studies of intracellular C. neoformans reveal the presence of LD, seen as spherical structures characterized by a homogeneous characteristic, without a lipid bilayer (Fig. 1D). The mean diameter of C. neoformans LD was 200 ± 20 nm.
FIG 1.
Detection of LD in C. neoformans and in infected macrophages. (A) Fluorescence microscopy of Uvitex (cell wall)-labeled extracellular C. neoformans stained with the neutral lipid-specific dye BODIPY 493/503, identifying LD in the fungi. Scale bars, 7 μm. (B) Fluorescence microscopy of BMDM infected with C. neoformans for 2 h, fixed, and immunostained with Rab7 antibody (phagosome), BODIPY 493/503 (LD), and DAPI (nucleus). The presence of LD within the fungi is highlighted, as shown by the sequential z-stack images. White arrows indicate host LD. LD visualized within Rab7 phagosomes are C. neoformans LD. Scale bars, 7 μm. (C) Fluorescence microscopy of uninfected BMDM, stained with BODIPY 493/503 to reveal LD (green) and Rab7 (red). Scale bars, 6 μm. (D) Transmission EM of BMDM infected with C. neoformans (Cn) for 3 h showing LD in the fungi. Scale bars, 500 nm. Ph, phagosome; CW, cell wall; hc, host cell.
C. neoformans takes up exogenous lipids.
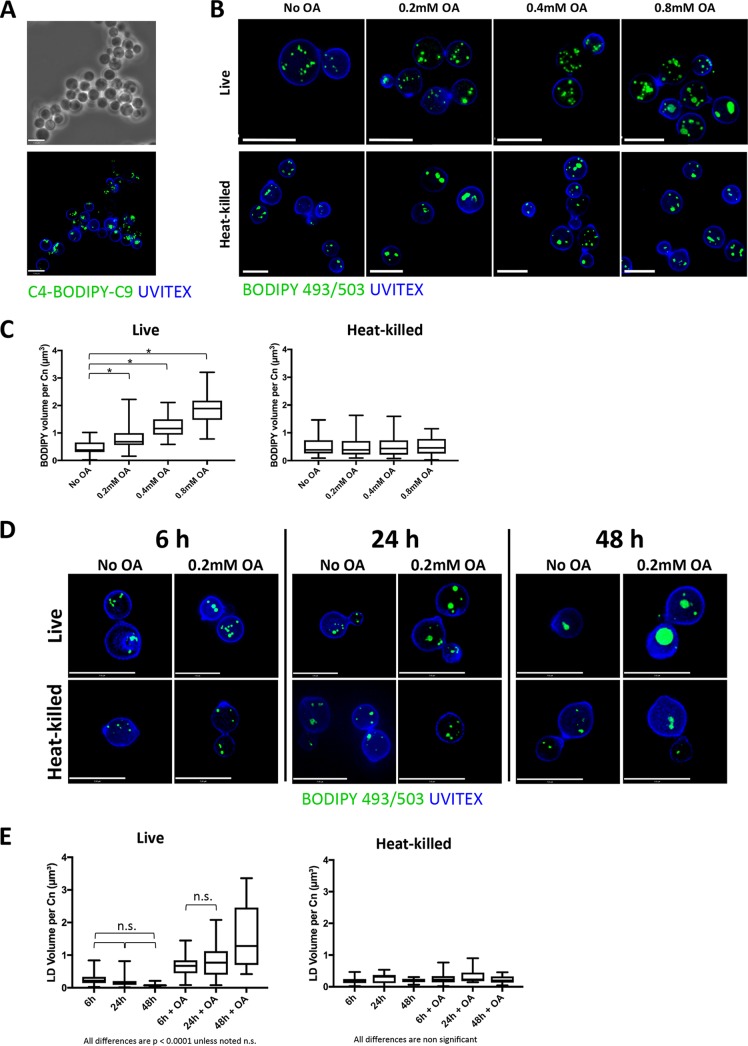
OA is widely used in the lipid field to induce LD formation, as it is rapidly and efficiently converted to neutral lipids which are stored therein (23). We wanted to examine how competent extracellular C. neoformans is at scavenging FA introduced in excess in its environment and what the fate of these lipids could be within the fungus. Extracellular C. neoformans was incubated for 2 h with a 10 mM concentration of the fluorescently labeled FA analog C4-BODIPY-C9 (5-butyl-4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-3-nonanoic acid), which has the fluorophore BODIPY 500/510 linked to the FA chain, to track its internalization into the fungi. This incubation led to intensive fluorescence staining reminiscent of that of LD in C. neoformans, suggesting that C. neoformans can scavenge free FA from the medium and subsequently store them (Fig. 2A). Membranes were not fluorescing, which may be due to very low resolution and staining. To verify the ability of C. neoformans to actively internalize FA-like lipids and subsequently store them, we then incubated extracellular C. neoformans with the OA in excess, which leads to the formation of TAG from diacylglycerol (DAG) and SE for sterols in many organisms, thus stimulating LD biogenesis (23–28). Concentrations of 0.2, 0.4, and 0.8 mM OA were added to the medium, where either live or heat-killed (HK; as negative controls) C. neoformans cells were suspended for 2 h prior to being stained with BODIPY 493/503 (Fig. 2B). As OA concentrations increased, the size of LD in live C. neoformans increased substantially, a phenomenon not detected in HK fungi. The net fluorescence observed represented LD exclusively and not OA specifically. However, following OA uptake, it was converted to nonpolar lipids such as TAG, which then fluoresced within LD. Since the cells were fixed, the BODIPY signal seen was representative of nonpolar lipids and LD exclusively. The total volume of BODIPY 493/503 within individual live or HK fungal cells was measured using Volocity software (Fig. 2C). In live C. neoformans, the total BODIPY volume increased proportionally to OA concentrations, whereas it remained relatively constant in HK C. neoformans, suggesting that FA uptake is an active process in C. neoformans. Interestingly, LD remained intact in HK cells. Despite HK cells being in general smaller than live C. neoformans cells, we did not observe any size differences that would account for the differences seen in LD volumes. For example, Fig. 2B shows live and HK cells that were generally similar in size, and Fig. 2C shows that their LD volumes were similar. To assess whether different growth stages could affect LD volume in C. neoformans, live and heat-killed fungi were grown with or without 0.2 mM OA for 6 h, 24 h, and 48 h, followed by visualization by microscopy (Fig. 2D). The LD volume in live C. neoformans without OA supplementation did not significantly change over time; however, a trend toward decreased LD volume was seen, suggestive of utilization of lipid stores (Fig. 2E). The addition of 0.2 mM OA during growth led to a significant increase in LD volume after 48 h. The volume of LD in heat-killed C. neoformans did not differ over time or with OA incubation, again suggesting that OA uptake and LD formation is an active process by C. neoformans.
FIG 2.
Distribution of FA in extracellular C. neoformans. (A) Fluorescence microscopy of C. neoformans grown for 2 h in the presence of 10 mM C4-BODIPY-C9, fixed, and stained with Uvitex for the fungal cell wall. Scale bars, 7 μm. (B) Fluorescence microscopy of Uvitex-labeled live and HK C. neoformans incubated with 0.2 mM, 0.4 mM, or 0.8 mM OA or with no OA (control) for 2 h and stained with BODIPY 493/503 (LD). Scale bars, 7 μm. (C) Quantification of the LD volume in extracellular C. neoformans (Cn) when grown with excess OA. *, P < 0.0001 by one-way ANOVA and Tukey's multiple-comparison test. (D) Fluorescence microscopy of Uvitex-labeled live and heat-killed C. neoformans incubated with no OA or 0.2 mM OA for 6 h, 24 h, or 48 h and stained with BODIPY 493/503 (LD). Scale bars, 7 μm. (E) Quantification of the LD volume in extracellular C. neoformans when grown with 0.2 mM OA for 6 h, 24 h, and 48 h. Experiments were performed in triplicate, and all comparisons are significant at a P of <0.0001 by one-way ANOVA and Tukey's multiple-comparison test unless noted as nonsignificant (n.s.). In the graph of HK C. neoformans, there are no statistically significant differences by one-way ANOVA.
Fatty acids stimulate C. neoformans replication.
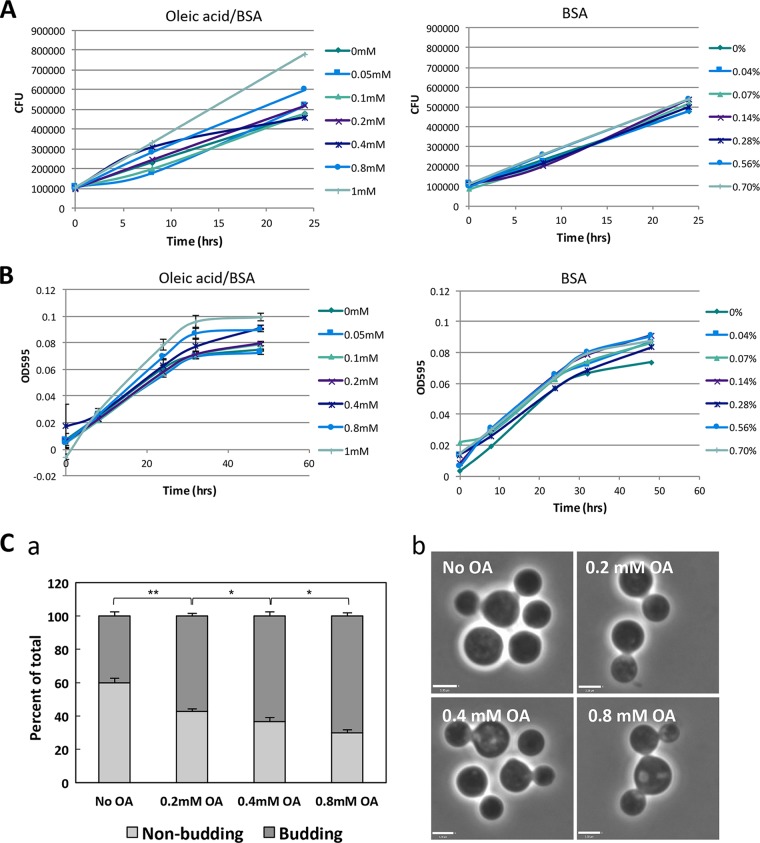
Next, we investigated whether FA uptake may modify the development and growth properties of extracellular C. neoformans. Fungal cells were incubated with concentrations of OA ranging from 0 to 1 mM in Dulbecco's modified Eagle medium (DMEM), and the CFU were counted after 8 h and 24 h (Fig. 3A). Higher CFU counts were observed to be proportional to OA concentrations, whereas no differences were observed upon the addition of bovine serum albumin (BSA). This was confirmed using absorbance, which was measured 8 h, 24 h, 32 h, and 48 h after BSA or OA incubation (Fig. 3B). The process of budding reflects cellular division and growth; therefore, increased numbers of budding events mark increased replication. We further confirmed the increased growth rate with OA by quantifying budding instances by phase microscopy. The percentage of C. neoformans cells on fixed coverslips that were seen in the process of budding was significantly higher in the presence of higher OA concentrations after 2 h of incubation with the fatty acid (Fig. 3C, panel a). Examples of C. neoformans seen budding are shown in Fig. 3C, panel b. Interestingly, in the presence of OA, some fungal cells were seen with multiple buds (up to four), suggesting either accelerated growth rates or defects in proper progeny formation.
FIG 3.
Effect of OA on the extracellular growth of C. neoformans. (A and B) Replication of extracellular C. neoformans in the presence of OA (indicated concentrations) or the BSA control was measured by CFU counts (A) or OD595 values (B). Cells were grown in DMEM with or without OA for the indicated times. Experiments were performed in triplicate; data are means ± standard deviations (SD). Comparisons of CFU counts with 0 mM versus 1 mM OA at 24 h were significant at a P of <0.01 by Student's t test. Comparisons of OD595 values with 0 mM versus 1 mM OA at 32 h were significant at a P of <0.01, and those with 0 mM versus 0.05, 0.1, 0.2, 0.4, 0.8, and 1 mM at 48 h were significant at a P of <0.01 by Student's t test. (C) OA stimulates budding of fungal cells. (a) Quantification of the percentage of cells visualized to be budding at various OA concentrations following 2 h of incubation with OA. Data are means ± SD of results of three separate assays, showing significant changes in budding with increasing OA concentrations (two-way ANOVA, Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.0001). (b) Phase microscopy of C. neoformans budding with depicted OA concentration as an example. Scale bars, 3.3 μm.
C. neoformans scavenges neutral lipids from macrophages.
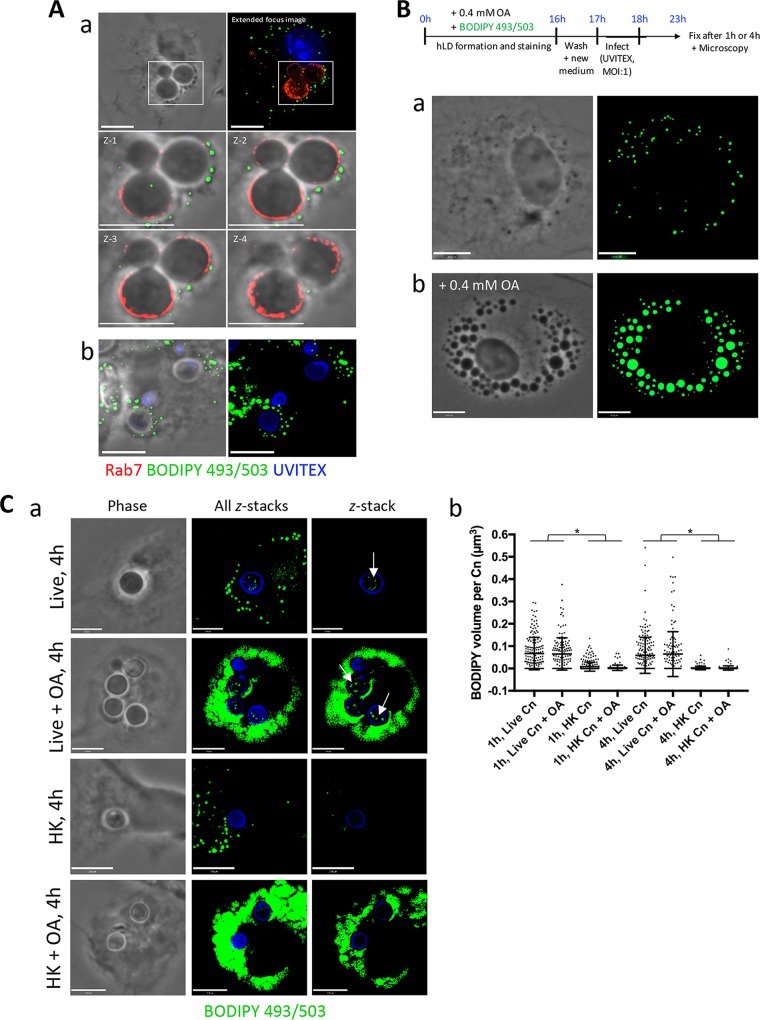
While examining the LD distribution in C. neoformans-infected BMDM using BODIPY 493/503, we noticed that many host LD were located close to the C. neoformans-containing phagosome, both after prolonged infection (4 h) and after a shorter infection time (1 h) with 0.2 mM OA (Fig. 4A, panels a and b, respectively). This led us to hypothesize that C. neoformans may exploit host LD. We therefore wanted to assess whether the fungi could take advantage of these host structures for the purpose of retrieving their lipid content. We first confirmed that BMDM responded to increasing concentrations of OA by inducing the formation of LD. These cells were incubated with no OA, 0.2 or 0.4 mM OA, or control concentrations of BSA (0.14% and 0.28%) for 4 h. BODIPY 493/503 fluorescence revealed an OA concentration-dependent increase in LD volume per BMDM, which was not seen under control conditions (see Fig. S1, panel a, in the supplemental material [graphed in panel b]). Incubation with increasing OA concentrations was not toxic to the BMDM, as an ∼4 to 6% decreased viability was observed with 0.2 and 0.4 mM OA, respectively, and a 10% decrease in viability was noted with 0.8 mM OA via trypan blue staining. We next devised an experimental protocol wherein the macrophage's LD were preloaded with the nonmetabolizable dye BODIPY 493/503 as an LD tracker prior to infection with C. neoformans. BMDM LD formation was stimulated by incubation with 0.4 mM OA and BODIPY 493/503 for 16 h, which resulted in the formation of numerous LD in the cell. We again confirmed the presence of BODIPY-labeled LD in these uninfected cells by fixing and visualizing by fluorescence microscopy the presence of BODIPY-stained LD both under normal conditions and in the presence of 0.4 mM OA (Fig. 4B, panels a and b, respectively). Following extensive washing to remove any remaining dye in the medium, live or HK C. neoformans was allowed to infect these macrophages containing BODIPY-stained LD for either 1 h or 4 h, with or without 0.4 mM OA. Incubation with 0.4 mM OA was performed to attempt to pressure the BMDM to form LD, which may potentially otherwise undergo lipophagy. These infected cells were then fixed and stained with antibody against LAMP1 to pinpoint the C. neoformans-containing phagosome in the macrophage. In addition to the host cell's LD staining positively in the green channel, some green foci were visible within live cryptococcal cells (arrows), suggestive of direct scavenging of BMDM-derived neutral lipids from LD (Fig. 4C, panel a). Following imaging and processing using Volocity software, the volume of BODIPY 493/503, and therefore neutral lipids scavenged from the BMDM, within individual C. neoformans fungi was measured and graphed (Fig. 4C, panel b). The significant difference in BODIPY-positive volume between live and HK fungi indicates that the scavenging of host-derived neutral lipids is an active process. Moreover, the minute presence of green signal in HK samples suggests that host LD were directed to the phagosome and that their contents may have diffused through into C. neoformans. Thus, these data indicate that C. neoformans is capable of dynamically scavenging neutral lipids from the BMDM LD.
FIG 4.
C. neoformans scavenges OA or OA-derived lipids from its host, the macrophage. (A) Fluorescence microscopy of BMDM after a 2 h infection with C. neoformans, fixed and stained with antibodies for Rab7, Uvitex, and BODIPY 493/503 dye. The close apposition of host LD to the C. neoformans-containing phagosome is shown (a) and with 0.2 mM OA added (b). Scale bars, 7 μm. (B) Scavenging of OA or OA-neutral lipids by C. neoformans from the host BMDM. Schema outlining the experimental protocol is at the top. Briefly, BODIPY 493/503 was coadministered with 0.4 mM OA to BMDM overnight to induce the formation of fluorescent LD in the host cell. Following washes to remove BODIPY from the extracellular medium, live or HK C. neoformans infected the BMDM for 1 h or 4 h with (a) or without (b) 0.4 mM OA. In uninfected cells, the accumulation of the BODIPY dye into their LD is shown and increases with OA incubation. Scale bars, 7 μm. (C) Detection of LD in live C. neoformans following the uptake of FA stored in host LD. (a) BODIPY 493/503-positive foci within C. neoformans (arrows) were visible with no OA added and with 0.4 mM OA added during infection. Shown from left to right are phase-contrast images, extended-focus images (all z-stacks), and single z-stacks. No to little fluorescence was detected within HK fungi. (b) The fluorescence detected within individual fungi was quantified and graphed. Data are means ± SD of results of three separate assays, showing a significant increase in the presence of fluorescence within live C. neoformans (Cn) compared to that in HK C. neoformans. Results for all live versus HK samples are significant at a P of <0.0001 (indicated with an asterisk) by two-way ANOVA and Tukey's multiple-comparison test. Scale bars, 6 μm.
Oleic acid induces LD biogenesis in C. neoformans.
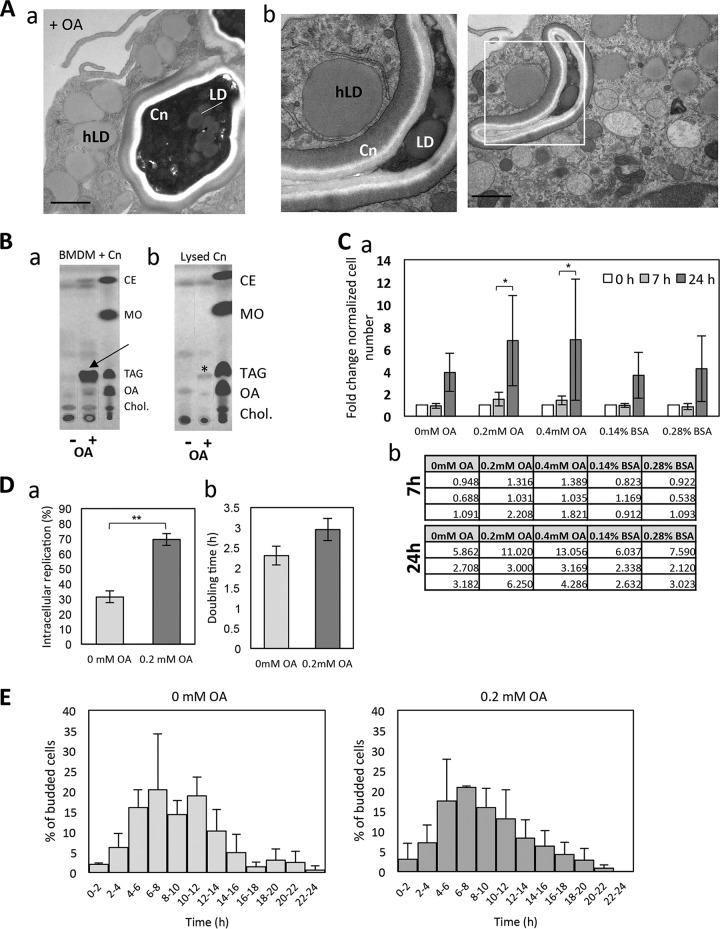
Knowing that OA-derived lipids are scavenged by the fungi, we next examined whether exposure of C. neoformans-infected BMDM to excess OA would have an impact on the intracellular development of the fungi. We first observed by electron microscopy (EM) that upon incubation with 0.2 mM OA, C. neoformans formed many LD (Fig. 5A), in addition to visualizing an accumulation of host LD around the phagosomes, which was more pronounced than without OA added to the medium (Fig. 4A).
FIG 5.
Effect of OA on the intracellular growth of C. neoformans. (A) (a and b) EM of intracellular C. neoformans (Cn) exposed to 0.2 mM OA showing the gathering of host LD (hLD) around the phagosome, in addition to several LD in C. neoformans. Scale bars, 500 nm. (B) TLC analysis of intracellular C. neoformans upon excess OA, showing neutral lipid fractions of cellular extracts of C. neoformans-infected BMDM (a) and C. neoformans isolated from BMDM (b) without (control; −) or with (+) 0.4 mM OA for 4 h. In panel a, OA incubation led to increased TAG and CE formation in BMDM similar to that under uninfected conditions (arrows). In panel b, an increase in TAG was observed in isolated C. neoformans (asterisk). Nonpolar lipid standards are shown in lane 3. MO, methyl oleate; Chol, cholesterol. (C) (a) The survival of C. neoformans in BMDM, which were treated with 0.2 mM and 0.4 mM OA, was determined by CFU counts 7 h and 24 h after phagocytosis. The CFU values at 7 h and 24 h are normalized to those at time zero. Data are the means of triplicates, and error bars are SD. (b) Tables show the absolute numbers per experimental replicate. *, P < 0.01 by two-way ANOVA, multiple-comparison test. (D) BMDM, treated with or without 0.2 mM OA, were infected with C. neoformans. The number of C. neoformans cells which underwent intracellular replication and the time interval between the first and second budding (doubling time) were measured in a 24-h time-lapse movie. The data were obtained from three independent biological experiments. Error bars represent the 95% confidence interval of the mean. **, P < 0.0001 by Fisher's exact test. (E) The start point of C. neoformans replication is not affected by OA. The histogram represents the percentage of cryptococcal cells which underwent first budding at the indicated times after phagocytosis during a 24-h time-lapse movie. Experiments were performed in triplicate. Error bars are SD, with no significant differences observed.
To verify that incubation with OA led to an increase in neutral lipids stored in LD, we performed thin-layer chromatography (TLC) from total lipid extracts isolated from 7 h C. neoformans-infected BMDM, with 0.14% BSA (control; −) or 0.4 mM OA (+) added to the medium (Fig. 5B, panel a). The results were compared to those of purified previously intracellular C. neoformans to identify the neutral lipids produced by the fungi while growing in BMDM (Fig. 5B, panel b). OA incubation led to increased TAG and cholesteryl ester (CE) formation in BMDM. An increase in TAG was observed in isolated C. neoformans grown with OA, whereas no changes in wax esters were seen. This result confirms previously reported findings that C. neoformans stores excess lipids mainly as TAG in its LD and not also as CE as many other organisms do (19, 20).
Oleic acid boosts intracellular C. neoformans replication.
Having determined that C. neoformans scavenges OA-derived lipids from the host cell, we examined whether excess OA would also enhance intracellular cryptococcal replication, as seen with extracellular fungi (Fig. 3). BMDM were infected with C. neoformans in the presence of no added OA or with 0.2 or 0.4 mM OA for 7 h or 24 h. At both concentrations, OA proportionally increased C. neoformans growth (Fig. 5C, panels a and b). This increased growth rate was also evaluated by infecting BMDM with C. neoformans for 24 h, with or without 0.2 mM OA, and quantified by counting cryptococcal cells which had undergone replication (Fig. 5D, panel a). The addition of OA led to ∼70% of observed C. neoformans cells undergoing a replication event, compared to only ∼30% when grown without OA. The average doubling time was also measured by calculating the time interval between the first and second budding (doubling time), which was not statistically different between the two conditions (Fig. 5D, panel b). To determine whether OA may affect the timing of replication, the first budding event of an individual C. neoformans yeast cell was recorded during a 24 h time-lapse movie following phagocytosis by the BMDM (Fig. 5E). No statistically significant differences were observed between the two conditions, indicating that the increased CFU counts in the presence of 0.2 mM OA, and therefore replication rates, were not due to an earlier initial replication of C. neoformans. These data also confirm that OA may be beneficial to the fungi, since replication appears to be stimulated.
Oleic acid increases the frequency of NLE.
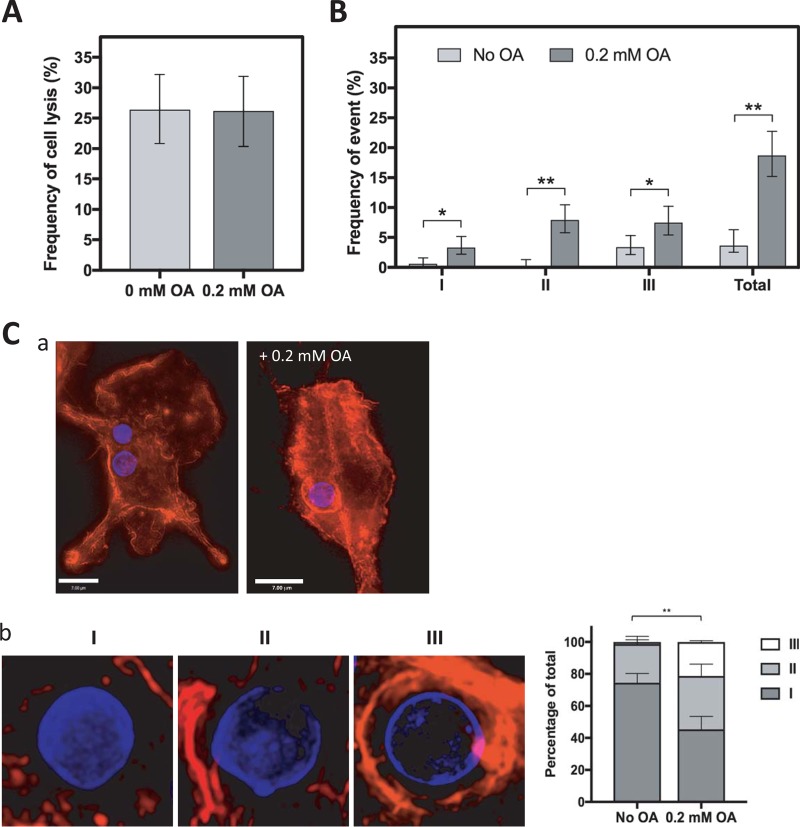
As FA are the building blocks of more complex lipids such as phospholipids and therefore membranes, incubation of cells with OA may lead to changes in membrane lipid composition. Previous studies have shown that in addition to causing changes in lipid metabolism such as the accumulation of neutral lipids, the increase in transcription of diacylglycerol acyltransferases (DGAT) and acyl-coenzyme A cholesterol acyltransferase (ACAT), and increased LD biogenesis, treating cells such as macrophages or epithelial cells with OA raises the baseline levels of this fatty acid in membranes to nearly 40% of all fatty acids (29, 30). Membrane lipid composition may be important for cryptococcal egress from its host cell, which occurs via two distinct mechanisms: lysis and NLE (ranging from partial NLE to cell-to-cell transfer) (31, 32). NLE occurs ∼5% of the time and involves the egress of cryptococcal cells from their host, while leaving the macrophage intact and still viable. The frequency of C. neoformans egress via lysis and NLE was examined and quantified during OA incubation by microscopy from a 24-h live movie of C. neoformans-infected BMDM with or without 0.2 mM OA. No significant difference was observed in the incidence of cryptococcal egress via macrophage lysis (Fig. 6A). In the same 24-h live movie, the frequency of NLE was determined and categorized based on NLE type, which is classified as complete (I) or partial (II) or as cell-to-cell transfer (III) (Fig. 6B). In all three categories, the total frequency of NLE was significantly higher (∼20% versus ∼5%) when the C. neoformans-infected BMDM were grown in the presence of OA.
FIG 6.
Effect of OA on the mode of C. neoformans egress. (A) The frequency of cell lysis was counted from 24-h time-lapse movies of BMDM treated with or without 0.2 mM OA and infected with C. neoformans. Experiments were performed in triplicate. Error bars represent the 95% confidence interval of the mean. (B) BMDM were treated with or without 0.2 mM OA during a C. neoformans infection. The frequencies of nonlytic exocytosis (NLE) events (type I, complete NLE; type II, partial NLE; type III, cell-to-cell transfer) were counted from 24-h time-lapse movies. Experiments were performed in duplicate. Error bars represent the 95% confidence interval of the mean. *, P < 0.01; **, P < 0.0001 by Fisher's exact test. (C) Fluorescence microscopy of Uvitex-labeled C. neoformans infecting BMDM, 4 h postinfection, stained with Alexa594 –F-actin. (a) A more pronounced actin staining is visible surrounding the C. neoformans-containing phagosome when incubated with 0.2 mM OA. Scale bar, 7 μm. (b) Class distribution of the actin coverage surrounding the C. neoformans-containing phagosome with 0.2 mM OA or no OA (control): little to none (0 to 30%; I), half (30 to 70%; II), and full (70 to 100%; III). **, P < 0.0001 by two-way ANOVA with standard deviations shown, Tukey's multiple-comparison test.
The host cell cytoskeleton, in particular actin, is essential for C. neoformans phagocytosis (33). Previous reports have excluded a role for host actin during the expulsion process of NLE (31, 32); however, a recent study showed that prolonged actin coverage around the phagosome may promote NLE (34). Actin flashes, which are transient and rapid accumulation of filamentous actin around the phagosome, are positively correlated with NLE but do not generate the force required for expulsion. We wanted to examine whether the presence of OA may affect F-actin coating of the phagosome, which may explain the increase in NLE events observed with OA. In C. neoformans-infected BMDM, incubated with 0 or 0.2 mM OA, increased actin accumulation was observed around the phagosome in the presence of OA, as visualized by fluorescence microscopy (Fig. 6C, panel a). To quantify this phenomenon, the percentage of actin coverage was categorized into three classes, i.e., little to no coating (0 to 30%; I), half (30 to 70%; II), and full (70 to 100%; III), and the percentage of actin coverage was graphed based on the proportion of phagosomes exhibiting various actin coverages (Fig. 6C, panel b). We further confirm a potential link between actin coverage and the process of NLE; however, the significance is still unknown, as is the mechanism behind OA-induced host actin accumulation to the phagosome.
Choreography of NLE.
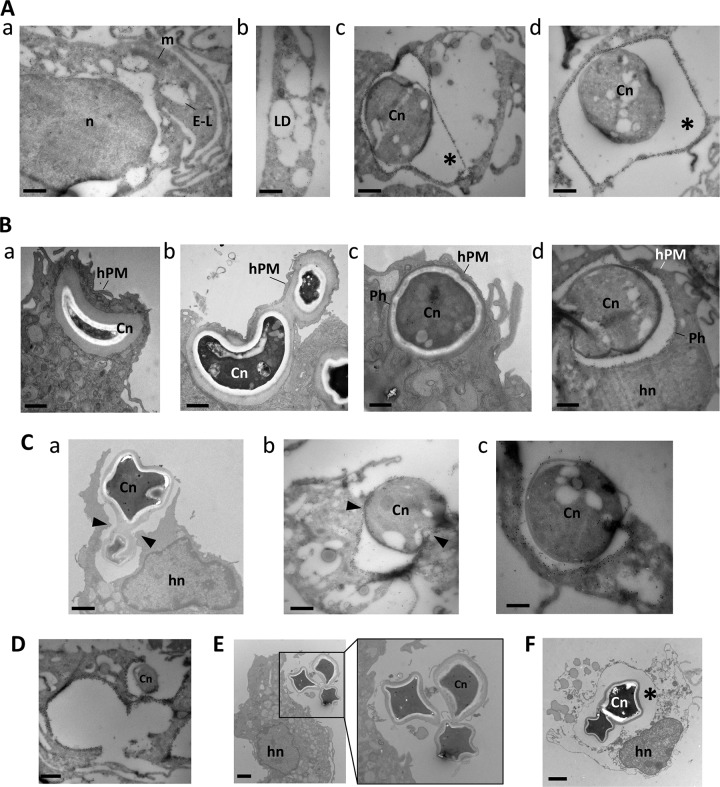
Further investigation into the mechanisms of NLE was pursued at the ultrastructural level. Since the frequency of NLE is significantly increased with OA, we incubated C. neoformans-infected BMDM with 0.2 mM OA for 3 h prior to visualization by EM (Fig. 7). In parallel, we performed immunogold staining for host LAMP1 to track the fate of the phagosomal membrane of C. neoformans. In uninfected cells, the LAMP1 staining was confined to the limiting membranes of electron-lucent organelles, likely corresponding to endolysosomes, particularly abundant in macrophages (Fig. 7A, panel a) and distinct from LD (Fig. 7A, panel b). In infected cells, the LAMP1 was localized to the phagosomes containing the fungi (Fig. 7A, panels c and d). To ensure no cross-reactivity with the capsule of C. neoformans, in particular with the 18b7 antibody, we stained by immunofluorescence both LAMP1 and 18b7 in the red and green channels, respectively (see Fig. S2 in the supplemental material). We saw no LAMP1 staining on extracellular C. neoformans. The red staining observed when the 18b7 antibody was also added was due to the fluorescence being so bright that it bled through to the red channel. An initial event in the escape of C. neoformans from macrophages without host cell lysis was the migration of the phagosome toward the cell periphery, leading to protrusion of the host cell surface (Fig. 7B, panels a to c), reducing the distance between the phagosomal membrane and the host plasma membrane (Fig. 7B, panel d). Subsequently, a process of fusion of the phagosomal membrane with the host plasma membrane occurred, liberating the fungi in the environment; however, a local rupture of the host cell membrane due to the compression of C. neoformans may also ensue, more likely upon egress of two fungi occupying the same phagosome (Fig. 7C). Regardless of the mechanism of egress, the host cell was left with large exocytic cups (Fig. 7D) but remained largely intact, with a resealed plasma membrane (Fig. 7E). This NLE contrasted with lytic egress in which the fungi remained secluded in the phagosome while the host plasma membrane was disrupted, liberating the cell contents (Fig. 7D).
FIG 7.
NLE of Cryptococcus neoformans infecting macrophages. (A) Specificity of LAMP1 staining. (a) Immuno-EM of macrophages with anti-LAMP1 antibodies showing gold particles on the limiting membrane of late endosomes-lysosomes (L-E). n, nucleus; m, mitochondria. (b) Lipid droplet (LD) biogenesis in macrophages incubated with 0.2 mM OA for 24 h. (c and d) Immuno-EM of C. neoformans (Cn)-infected macrophages, immunolabeled for LAMP1, confirming the confinement of the fungus in a LAMP1-containing phagosome. (B) (a to c) Distribution of C. neoformans-containing phagosomes (Ph) in a well-preserved macrophage as observed by EM showing a C. neoformans-containing phagosome close to the host plasma membrane (hPM), protruding from the host cell. hn, host nucleus. (d) Results in panels a to c were confirmed by immuno-EM with anti-LAMP1 antibodies. (C) Egress of C. neoformans from infected macrophages without cell lysis. Panel a illustrates C. neoformans released from a well-preserved macrophage. Cryosections in panels b and c show released fungi from the phagosome positively labeled for LAMP1. (D) Exocytic cups from a multi-infected macrophage. A cryosection stained with anti-LAMP1 antibodies reveals three empty phagosomes following NLE of C. neoformans. (E) Release of C. neoformans in the extracellular medium from an intact host cell. (F) Lysis of macrophages upon C. neoformans infection. EM of macrophages infected with C. neoformans for 3 h, showing two fungi within a phagosomal compartment (asterisk) prior to egress from a necrotic host cell. All bars, 500 nm.
DISCUSSION
Intracellular pathogens are known to subvert and modulate host LD, either for nutrient supply, replication platforms, or immune system subversion (reviewed in reference 18). Relatively little is known about the exploitation of host LD by pathogenic fungi. Within macrophages as host cells, intracellular C. neoformans actively scavenges neutral lipids originating from host LD and stores them in its own LD, previously characterized (3, 35, 36). In mammalian cells, excess FA must be converted to neutral lipids to avoid lipotoxicity and membrane damage, which is achieved through the generation of SE and TAG from cholesterol and DAG, respectively. The final, rate-limiting step for cholesterol esterification and storage in LD is catalyzed by acyl-CoA:cholesterol acyltransferase 1 (ACAT1) and, for TAG formation, by acyl-CoA:diacylglycerol acyltransferases (DGAT1 and DGAT2) (37–39). The bulk of cryptococcal neutral lipids in LD are TAG, and the genome of C. neoformans contains a DGAT homolog (CnDGAT; accession no. EAL20089.1), which shares 46% identity with Homo sapiens DGAT2 (accession no. NP_079374.2). The CnDGAT hypothetical protein possesses the canonical YFP motif for FA binding and HPHG motif like Saccharomyces cerevisiae ScDGAT2 (40% identity with hypothetical CnDGAT; accession no. XP_571235.1) (40). The ability to synthesize TAG may also serve as a means of avoiding lipotoxicity for C. neoformans, since Schizosaccharomyces pombe mutants impaired in TAG synthesis undergo an apoptosis-like death (41). Moreover, TAG synthesis may occur in response to stresses, as observed for Mycobacterium tuberculosis exposed to nitric oxide or low O2 (42). Future investigation on the function of CnDGAT will inform on the physiological significance of this enzyme both during infection and for C. neoformans growth.
C. neoformans is capable of scavenging neutral lipids directly from macrophage LD, which often accumulate around the C. neoformans-containing phagosome. Under conditions of high concentrations of OA (0.4 mM), which stimulates macrophage LD biogenesis due to neutral lipid accumulation, an increase in FA scavenging was not observed in C. neoformans, suggesting adequate regulation of FA import. It is also potentially a result of host-mediated restriction of lipid import into C. neoformans; however, the process by which C. neoformans accesses lipids no doubt varies when the fungal cell is intracellular versus extracellular. C. neoformans is capable of synthesizing FA, which can be converted to TAG (9), and we show here that C. neoformans is capable of scavenging free FA from its environment and also converting them to TAG, in a concentration- and time-dependent manner. FA transport has not been characterized in C. neoformans, and no proteins homologous to the major mammalian FA transporters, FA binding proteins 1 through 4, CD63, and FATP1 (43) could be retrieved from the genome database. In S. cerevisiae, FA import and activation are mediated by fatty acyl-CoA synthetase (Faa1 and Faa4) and Fat1 for FA use and signaling. Investigation into FA import to the fungi is likely to be a fertile area of investigation, given that FA metabolism and homeostasis are essential for this organism.
The increased availability of FA or neutral lipids can affect microbial replication. A boost in growth is observed when Chlamydia-infected cells are incubated with OA (44). On the other hand, the parasite Toxoplasma gondii relentlessly scavenges OA to the point of enzymatic saturation, leading to an accumulation of lipid deposits in the vacuole (S. J. Nolan, J. D. Romano, and I. Coppens, unpublished data). In extracellular fungal cells, the growth of S. cerevisiae and C. neoformans is boosted upon addition of exogenous OA (10, 45). We confirm this finding for both extracellular and intracellular C. neoformans, which manifest increased fungal replication rates in the presence of OA. This implies that C. neoformans is capable of adequately scavenging, storing, and using FA. The nutritional content of phagosomes is unknown but is thought to be low in glucose (12). Following macrophage internalization and under conditions of limited resources, C. neoformans upregulates extracellular lipases for host FA digestion, carbohydrate transporters, amino acid transporters, β-oxidation enzymes and carriers, and peroxisomal FA transporters (14). C. neoformans contains an essential triacylglycerol lipase gene (CGL1), involved in lipid degradation and FA catabolism, whose expression doubles 2 h after phagocytosis. The growth increase seen in C. neoformans may reflect increased access to FA from the host cell, thus supplementing its FA synthetic machinery.
FA serve many critical roles in the cell, including energy storage, cellular metabolism, and cellular signaling, and are the building blocks of more complex lipids such as phospholipids. Therefore, they influence membrane integrity and dynamics. For C. neoformans to successfully egress from its host cell, membrane integrity must be reconfigured. This is particularly relevant to the phenomenon of NLE observed in ∼5% of C. neoformans infections in macrophages, leaving the host cell intact and in a survival state (31, 32). Little is known of this form of escape, other than it requires viable C. neoformans, a viable host cytoskeleton, and phagosomal permeabilization (34, 46, 47). On this note, it is unknown whether LD may enhance phagosomal permeabilization; however, lipids from LD have been shown to contribute to autophagosomal biogenesis (28). Therefore, any perturbances in FA balances in either the host or fungi may alter membranes and thus affect NLE. In this regard, we observed an increase in NLE upon OA addition. This may be a direct consequence of an increased OA concentration in membranes, thereby affecting their fluidity more propitiously for NLE. However, it may also be due to changes in actin organization around the phagosome. Prolonged host actin coating of the phagosome has been associated with increased NLE, although actin has not been shown to provide the mechanistic force behind C. neoformans discharge, nor does actin accumulate around the phagosome at the moment of expulsion (31, 32, 34). Moreover, cytochalasin D treatment led to a significant increase in NLE events, highlighting the potential contributing role of host actin redistribution to this phenomenon, since prior treatment with C. neoformans had no effect (34). OA can lead to a redistribution of F-actin in umbilical cord blood-derived mesenchymal stem cells, leading to increased mobility of the cells (48). The significance of prolonged host actin redistribution induced by OA to the phagosome in C. neoformans-infected cells is unknown but may lead to clues in deciphering this complex phenomenon.
Our EM studies showed distinctive sequences of events which may further our understanding of NLE with 0.2 mM OA. First, the C. neoformans-containing phagosome appears to be localized in close proximity to the macrophage plasma membrane, to the point of distorting the membrane. Second, the protrusion appears very distinct, forming a small “sack” containing the fungi in the phagosome, still delineated by the host plasma membrane. Third, a potential fusion or scission in the host plasma membrane may occur as C. neoformans egresses from the macrophage while containing host LAMP1-positive debris around the fungal cell. Alternatively, the increase in exocytosis may be a fungus-dictated event to avoid cytotoxicity after C. neoformans senses the presence of excess OA, with its potential for lipotoxicity after enzymatic saturation. We have not looked at whether the survival of C. neoformans outside macrophages after exocytosis is impacted by OA, but given our data, we anticipate that OA would only help fungal survival. Irrespective of mechanism, the finding that OA greatly increases the frequency of NLE suggests that lipid supplementation could be used in future studies of this phenomenon.
In summary, lipid supplementation had profound effects on the C. neoformans-macrophage interaction. The finding that C. neoformans can scavenge host cell lipids during intracellular residence implies a capacity to raid the host cell for nutrients. Ultrastructural analysis suggests that NLE is the result of a carefully orchestrated choreography whereby membranes fuse and are rearranged in a process that transports the fungal cell from the inside a phagocytic cell to the extracellular environment. Under our experimental conditions, it is likely that lipid supplementation affects both the fungal and mammalian cells simultaneously, such that the changes to the outcome of the interaction may reflect emergence phenomena not reducible to either player. Consequently, it is likely that the enormous increase in NLE observed after lipid supplementation probably reflects the effects of lipids on both the host and microbial cells. Our findings suggest that lipid metabolism under conditions of lipid abundance and scarcity is a fertile area of research in the area of fungal cell interactions with macrophages, which may also provide insight into the development of new antifungal drugs.
MATERIALS AND METHODS
Reagents and antibodies.
All chemicals were obtained from Sigma (St. Louis, MO) unless otherwise stated. 5-Butyl-4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-3-nonanoic acid (C4-BODIPY-C9) was purchased from ThermoFisher Scientific (Waltham, MA), and 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503) was acquired from Life Technologies (Carlsbad, CA). The polar and nonpolar lipid mixtures for thin-layer chromatography were obtained from Matreya LLC (State College, PA).
Antibodies used in this study included primary rat monoclonal anti-LAMP1 (1:200) and goat monoclonal anti-Rab7 (1:200) (Santa Cruz Biotechnology, Dallas, TX), rabbit monoclonal anti-LAMP1 (1:100) (Cell Signaling, Danvers, MA), and Alexa Fluor594 phalloidin (ThermoFisher Scientific). Uvitex 2B was bought from Polysciences, Warminster, PA. Secondary antibodies used for immunofluorescence were conjugated to Alexa488 or Alexa594 (Invitrogen, Carlsbad, CA). Monoclonal antibody 18B7 (IgG1), which binds to the cryptococcal glucuronoxylomannan, was used for opsonizing yeast cells.
For oleic acid (OA)-albumin complex preparation, sodium oleate was dissolved in H2O at a concentration of 100 mM and then thoroughly mixed by vortexing for 5 min with 5% fatty acid-free BSA to reach a final stock concentration of 10 mM OA-BSA complexes as previously described (49).
Macrophage and C. neoformans cultivation. (i) Macrophage cells.
Bone marrow-derived macrophages (BMDM) were isolated from the bone marrow of hind leg bones of 5- to 8-week-old C57BL/6 female mice (Jackson Laboratories, Bar Harbor, ME). For differentiation into macrophages, cells were seeded in 100-mm tissue culture (TC)-treated cell culture dishes for 6 to 7 days at 37°C with 9.5% CO2 in Dulbecco's modified Eagle medium (DMEM; Corning, Corning, NY), with the following additions: 20% L-929 cell-conditioned medium, 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA), 2 mM Glutamax (Gibco, Gaithersburg, MD), 1% nonessential amino acids (Cellgro, Manassas, VA), 1% HEPES buffer (Corning), 1% penicillin-streptomycin (Corning), and 0.1% 2-mercaptoethanol (Gibco). On day 3, 3 ml of fresh medium was added, and on day 6, the entire medium was replaced. For experiments, differentiated BMDM were used within 5 days of differentiation.
(ii) Cryptococcus neoformans.
Culturing of C. neoformans serotype A strain H99 yeast cells was done in Sabouraud dextrose broth (Difco, Carlsbad, CA) overnight at 30°C under constant agitation (120 rpm). For infections or growth assays, a 1-ml volume of this culture was centrifuged at 16.3 × g and the pellet was suspended in phosphate-buffered saline (PBS) prior to counting using a hemacytometer (Hausser Scientific, Horsham, PA). Cells were then used at the indicated multiplicity of infection (MOI) or plated as described for each experiment. For experiments using heat-killed C. neoformans, the yeast cells were killed by placing them at 60°C for 1 h.
Intracellular replication assays.
Approximately 1.5 × 105 cells of C. neoformans were added to wells of a 96-well plate with 0 to 1 mM OA or 0 to 0.7% BSA. The plates were incubated at 37°C and 9.5% CO2. For the optical density at 595 (OD595), these plates were incubated for 0, 8, 24, 32, or 48 h and the absorbance at 595 nm was measured using an EMax Plus plate reader (Molecular Devices, Sunnyvale, CA), using as blanks wells containing only medium with different OA or BSA concentrations. For CFU counts, after 0, 8, or 24 h of incubation, the cultures were serially diluted, plated onto Sabouraud agar, and further incubated at 30°C for 48 h for CFU counting.
To measure C. neoformans growth within macrophages, 5 × 104 BMDM were seeded in each well of a 24-well plate in medium containing 0.5 μg/ml lipopolysaccharide (LPS) and 100 U/ml gamma interferon (IFN-γ; Roche). This method of activation was used because of prior experiments in the laboratory, and we wanted our results to be comparable to prior findings. Final concentrations of 0, 0.2, and 0.4 mM OA were added, along with final concentrations of 0.14% BSA and 0.28% BSA as controls for 0.2 mM and 0.4 mM OA additions, respectively, since OA is conjugated to BSA. The plates were incubated at 37°C and 9.5% CO2 overnight. To initiate infection, C. neoformans (1.5 × 105 cells), opsonized with 10 μg/ml 18b7 monoclonal antibody (MAb), was added and encouraged to settle onto a BMDM monolayer culture by using centrifugation at 1,200 rpm for 1 min. The culture was washed twice with medium to remove nonphagocytosed extracellular cryptococcal cells. At various time intervals (0, 7, and 24 h), phagocytized cryptococcal cells were released by lysing the macrophages with sterilized water. The lysates were then serially diluted, plated onto Sabouraud agar, and incubated at 30°C for 48 h for CFU determination.
Quantification of lipid droplet volume and fluorescent dye uptake assays.
BMDM were incubated with 10 μM BODIPY 493/503 for 24 h and 0.4 mM OA for 24 h to induce the formation of fluorescent LD in the host cells. These were then thoroughly washed with PBS and chased for 1 h in control medium. These cells with preloaded BODIPY-stained host LD were then infected with C. neoformans for 1 or 4 h prior to fixation for immunostaining with anti-LAMP1 antibody. In some assays, 0.4 mM OA was added to the medium during C. neoformans infection. For extracellular C. neoformans LD assays, live or heat-killed (HK) C. neoformans cells were grown for the indicated times and then stained with BODIPY 493/503 (1:200) for 30 min and 0.01% Uvitex 2B for 10 min at room temperature and washed twice with PBS prior to visualization by microscopy. Quantification of total host or C. neoformans LD, identified by staining with BODIPY 493/503, was performed using the cropping tool where applicable and the “Find Object” tool in Volocity software for BODIPY measurements, followed by the calculation of the sum of the BODIPY volume by the program, and the results were graphed.
Coverslips were added to the wells of a 24-well plate prior to the addition of 1.5 × 105 cells per well with DMEM containing 0, 0.2, 0.4, and 0.8 mM OA. These plates were then incubated at 37°C, 9.5% CO2, for 2 h prior to fixation with 4% electron microscopy (EM)-grade paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA). The cells were stained with BODIPY 493/503 (1:200) and 0.01% Uvitex 2B prior to visualization by fluorescence microscopy and quantification.
TLC.
BMDM (1 × 107 cells) were grown in medium containing 0.5 μg/ml LPS and 100 U/ml IFN-γ in 100-mm TC-treated plates in addition to a final concentration of either 0.4 mM OA or 0.28% BSA. These cells were incubated at 37°C, 9.5% CO2, overnight. For C. neoformans infections, 1 × 107 cells of C. neoformans were added with 10 μg/ml 18b7 MAb for 7 h, after which the cells were washed three times prior to lipid extractions. For cryptococcal cell release and purification, the infected BMDM were lysed by adding sterilized water and vortexing. Total lipids from the same amount of purified fungi or infected/uninfected BMDM between the two experimental conditions (BSA control or 0.4 mM OA) were extracted with chloroform-methanol-water (10:10:3, vol/vol/vol), dried under liquid N2, and then suspended in chloroform. Lipids were fractionated by thin-layer chromatography (TLC) in hexane-diethyl ether-acetic acid (90:10:1, vol/vol/vol) on TLC silica gel 60 plates (Merck, Gibbsburg, NJ) run with lipid standards and visualized with iodine vapor.
Fluorescence microscopy.
For immunofluorescence assays (IFA) on macrophages or extracellular C. neoformans, coverslips were fixed for 15 min with 4% EM-grade paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS and permeabilized with 0.3% Triton X-100 in PBS for 5 min at room temperature (RT). Cells were then incubated in 3% BSA-PBS to block nonspecific binding for 40 min, followed by incubation with primary antibody for 1 h at RT. After the cells were washed three times in 1× PBS for 5 min, they were incubated with secondary antibody (1:2,000) for 45 min at RT with subsequent DAPI (4′,6-diamidino-2-phenylindole) (1:1,000, 5 min) or Uvitex (1:1,000, 10 min) staining if stated. Alexa594 phalloidin was incubated on coverslips for 30 min. Coverslips were mounted using ProLong diamond antifade mountant (Life Technologies) to minimize bleaching during microscopy. Cells were viewed with a Nikon Eclipse 90i microscope equipped with an oil immersion plan Apo 100×, numerical aperture 1.4 objective and a Hamamatsu GRCA-ER camera (Hamamatsu Photonics, Hamamatsu, Japan). Optical z-sections with 0.2-μm spacing were acquired using Volocity software (PerkinElmer, Waltham, MA), which was also used to adjust brightness levels, cropping, and resizing of the images obtained.
Time-lapse imaging.
BMDM were seeded (5 × 104 cells/well) on poly-d-lysine-coated coverslips at the bottom of MatTek petri dishes with a 14-mm microwell (MatTek Brand Corporation) in medium containing 0.5 μg/ml lipopolysaccharide, 100 U/ml IFN-γ, and in some experiments 0.2 mM OA. The cells were then incubated at 37°C with 9.5% CO2 overnight. On the following day, macrophages were infected with C. neoformans (1.5 × 105 cells/well) in the presence of 10 μg/ml MAb 18B7. After 2 h of incubation to allow phagocytosis to occur, the culture was washed five times with fresh medium to remove extracellular cryptococcal cells. Images were taken every 4 min for 24 h using a Zeiss Axiovert 200M inverted microscope with a 10× phase objective in an enclosed chamber under conditions of 9.5% CO2 and 37°C. Videos were taken using an AxioCam MR camera controlled by AxioVision 4.9 software (Carl Zeiss).
EM.
For transmission EM, BMDM infected with C. neoformans for 3 h (with or without 0.2 mM OA) were fixed in 2.5% glutaraldehyde (EM grade; Electron Microscopy Sciences, Hatfield, PA) in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at RT and processed as described previously (50) before examination with a Philips CM120 electron microscope (Eindhoven, the Netherlands) under 80 kV. For immuno-EM, preparations of BMDM infected with C. neoformans for 3 h were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, PA) in 0.25 M HEPES (pH 7.4) for 1 h at room temperature and then in 8% paraformaldehyde in the same buffer overnight at 4°C. The preparations were infiltrated, frozen, and sectioned as previously described (50). The sections were immunolabeled with rabbit antibodies to LAMP1 (1:20 in PBS–1% fish skin gelatin) and then with anti-mouse IgG, followed directly by 10-nm protein A-gold particles (Department of Cell Biology, Medical School, Utrecht University, the Netherlands) before examination with a Philips CM120 electron microscope (Eindhoven, the Netherlands) under 80 kV.
Statistical methods.
Prism software was used to graph and perform statistical analyses of histograms, box plots, and dot plots (La Jolla, CA) using two-way ANOVA, one-way ANOVA with Tukey's test, and multiple comparisons. Microsoft Excel was used for all other graphs and Student's t tests (Redmond, WA).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Casadevall laboratory for helpful discussions during the course of this work. We thank the excellent technical staff of the Electron Microscopy Core Facility at Yale School of Medicine and the Johns Hopkins University School of Medicine Microscopy Facility.
This study was supported by grant R01HL059842 from the NHLBI. A.C. was supported in part by grants 5R01HL059842, 5R01AI033774, 5R37AI033142, and 5R01AI052733.
We declare no competing financial interests.
S.J.N., M.S.F., I.C., and A.C. performed conceptualization, data curation, supervision, investigation, funding acquisition, project administration, visualization, and writing. S.J.N. and M.S.F. performed the formal analysis. S.J.N., M.S.F., and I.C. performed the methodology. S.J.N., M.S.F., I.C., and A.C. performed the validation. S.J.N., I.C., and A.C. performed the editing.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00564-17.
REFERENCES
- 1.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kronstad JW, Attarian R, Cadieux B, Choi J, D'Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, Saikia S, Wang J. 2011. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol 9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. 2006. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell 5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voelz K, May RC. 2010. Cryptococcal interactions with the host immune system. Eukaryot Cell 9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vecchiarelli A, Pericolini E, Gabrielli E, Kenno S, Perito S, Cenci E, Monari C. 2013. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol 8:1107–1116. doi: 10.2217/fmb.13.84. [DOI] [PubMed] [Google Scholar]
- 7.Almeida F, Wolf JM, Casadevall A. 2015. Virulence-associated enzymes of Cryptococcus neoformans. Eukaryot Cell 14:1173–1185. doi: 10.1128/EC.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santiago-Tirado FH, Onken MD, Cooper JA, Klein RS, Doering TL. 2017. Trojan horse transit contributes to blood-brain barrier crossing of a eukaryotic pathogen. mBio 8:e02183-16. doi: 10.1128/mBio.02183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright LC, Santangelo RM, Ganendren R, Payne J, Djordjevic JT, Sorrell TC. 2007. Cryptococcal lipid metabolism: phospholipase B1 is implicated in transcellular metabolism of macrophage-derived lipids. Eukaryot Cell 6:37–47. doi: 10.1128/EC.00262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretschmer M, Wang J, Kronstad JW. 2012. Peroxisomal and mitochondrial β-oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 11:1042–1054. doi: 10.1128/EC.00128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erb-Downward JR, Huffnagle GB. 2007. Cryptococcus neoformans produces authentic prostaglandin E2 without a cyclooxygenase. Eukaryot Cell 6:346–350. doi: 10.1128/EC.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 13.Rude TH, Toffaletti DL, Cox GM, Perfect JR. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun 70:5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan W, Kraus PR, Boily M-J, Heitman J. 2005. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 4:1420–1433. doi: 10.1128/EC.4.8.1420-1433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu G, Cheng P-Y, Sham A, Perfect JR, Kronstad JW. 2008. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol 69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chayakulkeeree M, Rude TH, Toffaletti DL, Perfect JR. 2007. Fatty acid synthesis is essential for survival of Cryptococcus neoformans and a potential fungicidal target. Antimicrob Agents Chemother 51:3537–3545. doi: 10.1128/AAC.00442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noverr MC, Cox GM, Perfect JR, Huffnagle GB. 2003. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect Immun 71:1538–1547. doi: 10.1128/IAI.71.3.1538-1547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barisch C, Soldati T. 2017. Breaking fat! How mycobacteria and other intracellular pathogens manipulate host lipid droplets. Biochimie 141:56–61. doi: 10.1016/j.biochi.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Itoh T, Kaneko H. 1977. The in vivo incorporation of [32P]-labeled orthophosphate into pyrophosphatidic acid and other phospholipids of Cryptococcus neoformans through cell growth. Lipids 12:809–813. doi: 10.1007/BF02533269. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko H, Hosohara M, Tanaka M, Itoh T. 1976. Lipid composition of 30 species of yeast. Lipids 11:837–844. doi: 10.1007/BF02532989. [DOI] [PubMed] [Google Scholar]
- 21.Itoh T, Waki H, Kaneko H. 1975. Changes of lipid composition with growth phase of Cryptococcus neoformans. Agric Biol Chem 39:2365–2371. doi: 10.1271/bbb1961.39.2365. [DOI] [Google Scholar]
- 22.Walther TC, Farese RV. 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A 100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto Y, Onoduka J, Homma KJ, Yamaguchi S, Mori M, Higashi Y, Makita M, Kinoshita T, Noda J-I, Itake H, Takanoa T. 2006. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of acyl-CoA synthetase. Biol Pharm Bull 29:2174–2180. doi: 10.1248/bpb.29.2174. [DOI] [PubMed] [Google Scholar]
- 25.Mei S, Ni H-M, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding W-X. 2011. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther 339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thörn K, Bergsten P. 2010. Fatty acid-induced oxidation and triglyceride formation is higher in insulin-producing MIN6 cells exposed to oleate compared to palmitate. J Cell Biochem 111:497–507. doi: 10.1002/jcb.22734. [DOI] [PubMed] [Google Scholar]
- 27.Ahn JH, Kim MH, Kwon HJ, Choi SY, Kwon HY. 2013. Protective effects of oleic acid against palmitic acid-induced apoptosis in pancreatic AR42J cells and its mechanisms. Korean J Physiol Pharmacol 17:43–50. doi: 10.4196/kjpp.2013.17.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. 2014. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol 24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak M, Tardivel S, Sayegrih K, Robert V, Abreu S, Chaminade P, Vicca S, Grynberg A, Lacour B. 2011. Impact of polyunsaturated fatty acids on oxidized low density lipoprotein-induced U937 cell apoptosis. J Atheroscler Thromb 18:494–503. doi: 10.5551/jat.7062. [DOI] [PubMed] [Google Scholar]
- 30.Jackson KG, Bateman PA, Yaqoob P, Williams CM. 2009. Impact of saturated, polyunsaturated and monounsaturated fatty acid-rich micelles on lipoprotein synthesis and secretion in Caco-2 cells. Lipids 44:1081–1089. doi: 10.1007/s11745-009-3366-7. [DOI] [PubMed] [Google Scholar]
- 31.Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 32.Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr Biol 16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Guerra CR, Seabra SH, de Souza W, Rozental S. 2014. Cryptococcus neoformans is internalized by receptor-mediated or “triggered” phagocytosis, dependent on actin recruitment. PLoS One 9:e89250. doi: 10.1371/journal.pone.0089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston SA, May RC. 2010. The human fungal pathogen Cryptococcus neoformans escapes macrophages by a phagosome emptying mechanism that is inhibited by Arp2/3 complex-mediated actin polymerisation. PLoS Pathog 6:e1001041. doi: 10.1371/journal.ppat.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross NT, Hultenby K, Mengarelli Camner P, Jarstrand C. 2000. Lipid peroxidation by alveolar macrophages challenged with Cryptococcus neoformans, Candida albicans or Aspergillus fumigatus. Med Mycol 38:443–449. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi M, Biswas SK, Kita S, Aikawa E, Takeo K. 2002. Electron microscopy of pathogenic yeasts Cryptococcus neoformans and Exophiala dermatitidis by high-pressure freezing. J Electron Microsc (Tokyo) 51:21–27. doi: 10.1093/jmicro/51.1.21. [DOI] [PubMed] [Google Scholar]
- 37.Goodman DS, Deykin D, Shiratori T. 1964. The formation of cholesterol esters with rat liver enzymes. J Biol Chem 239:1335–1345. [PubMed] [Google Scholar]
- 38.Chang TY, Chang CC, Lin S, Yu C, Li BL, Miyazaki A. 2001. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and -2. Curr Opin Lipidol 12:289–296. doi: 10.1097/00041433-200106000-00008. [DOI] [PubMed] [Google Scholar]
- 39.Buhman KF, Accad M, Farese RV. 2000. Mammalian acyl-CoA:cholesterol acyltransferases. Biochim Biophys Acta 1529:142–154. doi: 10.1016/S1388-1981(00)00144-X. [DOI] [PubMed] [Google Scholar]
- 40.Liu Q, Siloto RMP, Snyder CL, Weselake RJ. 2011. Functional and topological analysis of yeast acyl-CoA:diacylglycerol acyltransferase 2, an endoplasmic reticulum enzyme essential for triacylglycerol biosynthesis. J Biol Chem 286:13115–13126. doi: 10.1074/jbc.M110.204412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Chieu HK, Low CP, Zhang S, Heng CK, Yang H. 2003. Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J Biol Chem 278:47145–47155. doi: 10.1074/jbc.M306998200. [DOI] [PubMed] [Google Scholar]
- 42.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glatz JFC, Luiken JJFP, Bonen A. 2010. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev 90:367–417. doi: 10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- 44.Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A 105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walenga RW, Lands W. 1975. Effectiveness of various unsaturated fatty acids in supporting growth and respiration in Saccharomyces cerevisiae. J Biol Chem 250:9121–9129. [PubMed] [Google Scholar]
- 46.Nicola AM, Robertson EJ, Albuquerque P, Derengowski L da S, Casadevall A. 2011. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. mBio 2:e00167-11. doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH. 2011. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol 193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung YH, Lee S-J, Oh SY, Lee HJ, Ryu JM, Han HJ. 2015. Oleic acid enhances the motility of umbilical cord blood derived mesenchymal stem cells through EphB2-dependent F-actin formation. Biochim Biophys Acta 1853:1905–1917. doi: 10.1016/j.bbamcr.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Nolan SJ, Romano JD, Coppens I. 2017. Host lipid droplets: an important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog 13:e1006362. doi: 10.1371/journal.ppat.1006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fölsch H, Pypaert M, Schu P, Mellman I. 2001. Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J Cell Biol 152:595–606. doi: 10.1083/jcb.152.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.