ABSTRACT
Enterococcus faecalis, a member of the human gastrointestinal microbiota, is an opportunistic pathogen associated with hospital-acquired wound, bloodstream, and urinary tract infections. E. faecalis can subvert or evade immune-mediated clearance, although the mechanisms are poorly understood. In this study, we examined E. faecalis-mediated subversion of macrophage activation. We observed that E. faecalis actively prevents NF-κB signaling in mouse RAW264.7 macrophages in the presence of Toll-like receptor agonists and during polymicrobial infection with Escherichia coli. E. faecalis and E. coli coinfection in a mouse model of catheter-associated urinary tract infection (CAUTI) resulted in a suppressed macrophage transcriptional response in the bladder compared to that with E. coli infection alone. Finally, we demonstrated that coinoculation of E. faecalis with a commensal strain of E. coli into catheterized bladders significantly augmented E. coli CAUTI. Taken together, these results support the hypothesis that E. faecalis suppression of NF-κB-driven responses in macrophages promotes polymicrobial CAUTI pathogenesis, especially during coinfection with less virulent or commensal E. coli strains.
KEYWORDS: Enterococcus faecalis, Escherichia coli, catheter-associated UTI, coinfection, immune suppression, macrophage, polymicrobial, urinary tract infection
INTRODUCTION
Enterococcus faecalis is an early colonizer in infants and a ubiquitous member of the human gut microbiome (1). E. faecalis is also associated with up to 70% of wound infections, nearly 10% of bloodstream infections, and up to 30% of catheter-associated urinary tract infections (CAUTI) (2–5). To successfully colonize and persist in the host, pathogens must withstand, modulate, or evade immune-mediated clearance mechanisms. E. faecalis invokes multiple strategies to persist within the host, including the formation of biofilms that prevent phagocytosis by immune cells (6) and the ability to survive within macrophages and neutrophils for extended periods (7–11).
Mammalian cells detect pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs) to trigger nuclear factor kappa B (NF-κB)-dependent host defenses. NF-κB controls the transcription of inflammatory and immune-associated genes, including those for cytokines and chemokines regulating recruitment and activation of immune cells in response to infection (12). E. faecalis infection of macrophages at a low multiplicity of infection (MOI = 10) results in the activation of mitogen-activated protein kinases (MAPKs) and NF-κB, leading to the production of proinflammatory cytokines (13). However, some E. faecalis strains isolated from the gastrointestinal tracts of healthy human infants can suppress MAPK and NF-κB signaling and interleukin-8 (IL-8) expression in intestinal epithelial cells in vitro (14, 15).
Several E. faecalis virulence factors modulate immunity during infection, including aggregation substance (AS), gelatinase, and TcpF (16–19). TcpF is a TIR domain-containing protein and interferes with Toll-like receptor (TLR)–MyD88 interactions, which also depend on MyD88 TIR domain-mediated interactions. As a result, E. faecalis TcpF expression results in decreased NF-κB p65 translocation in RAW macrophages (16, 20). Plasmid-encoded AS promotes phagocytosis and internalization into macrophages via interaction with complement receptor type 3 (21–23). After internalization, AS promotes resistance to superoxide killing, leading to increased survival in macrophages (19). In addition, gelatinase is a secreted, quorum-responsive gene product that facilitates innate immune evasion by cleaving complement components C3, C3a, and C5a to reduce opsonization and decrease neutrophil recruitment (17, 18, 24, 25).
In a mouse urinary tract infection (UTI) model, the cellular response to E. faecalis infection is primarily monocytic and is independent of TLR2 (26). In a CAUTI model, the presence of a urinary catheter alone elicits a strong proinflammatory response in the bladder, composed of neutrophils and monocyte-derived cells (27–29). Infection of catheterized bladders with E. faecalis results in the development of high-titer catheter-associated biofilms and bladder infection despite the presence of a strong inflammatory response induced by catheterization (19). Moreover, in the course of E. faecalis CAUTI, the number of activated bladder-associated macrophages is significantly decreased compared to that in catheterized, uninfected animals (27). Together these observations suggest that E. faecalis can subvert immune-mediated killing to persist within the infected bladder.
During UTI and CAUTI, E. faecalis is often part of a polymicrobial community (30–32). E. faecalis can promote polymicrobial infection by increasing the resistance of coinfecting organisms, such as Pseudomonas aeruginosa and Proteus mirabilis, to clearance by antibiotics (33, 34). Polymicrobial infection by E. faecalis and P. aeruginosa leads to aggravated pyelonephritis more often than monomicrobial infection does (33). E. faecalis and uropathogenic Escherichia coli (UPEC) are also frequently isolated together during CAUTI (35); however, the relationship between these pathogens and its impact on pathogenesis are unknown. Given the frequency with which E. faecalis is found within polymicrobial infections and that E. faecalis can modulate the host immune response within the catheterized bladder, we tested the hypothesis that E. faecalis immune modulation promotes polymicrobial CAUTI. We found that E. faecalis actively subverts E. coli-mediated NF-κB activation and proinflammatory cytokine production in RAW264.7 macrophages in vitro and macrophage-associated proinflammatory expression profiles in catheterized bladders in vivo, culminating in higher-titer E. coli CAUTI.
RESULTS
Live E. faecalis prevents lipopolysaccharide (LPS)- or lipoteichoic acid (LTA)-mediated NF-κB-driven activation in RAW macrophages.
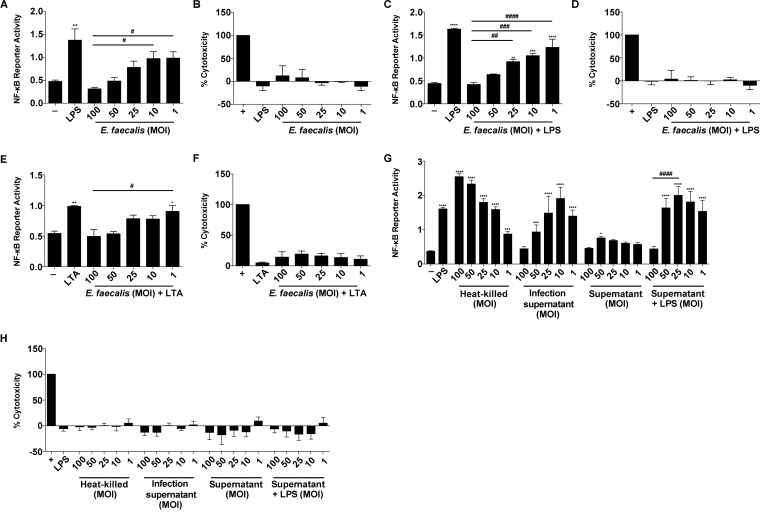
E. faecalis infection during CAUTI induces monocytic infiltration (27). To determine whether E. faecalis immunomodulates monocyte-derived cells, such as macrophages, we assessed NF-κB signaling in mouse RAW264.7 macrophages at 6 h postinfection (hpi). Both E. faecalis strain OG1RF (Fig. 1A) and the multidrug-resistant strain V583 (see Fig. S1A in the supplemental material) activated NF-κB at low multiplicities of infection (MOIs), as previously reported (13). In contrast, neither E. faecalis OG1RF nor V583 activated NF-κB signaling at high MOIs. We simultaneously monitored lactate dehydrogenase (LDH) release into culture supernatants to ensure that the absence of NF-κB activation was not a result of cell death at high MOIs, and we observed no increase in LDH release at any of the MOIs used in this study (Fig. 1B; Fig. S1B).
FIG 1.
E. faecalis prevents NF-κB-driven macrophage activation. Mouse RAW267.4 macrophages were infected with live E. faecalis OG1RF alone at the specified MOI or treated concurrently with either LPS (100 ng/ml) or LTA (100 ng/ml) for 6 h prior to measurement of NF-κB-driven SEAP reporter activity and cytotoxicity (LDH activity). (A and B) NF-κB-driven SEAP reporter activity (A) and LDH activity (B) of RAW264.7 macrophages infected by E. faecalis alone. (C and D) NF-κB-driven SEAP reporter activity (C) and LDH activity (D) in the presence of E. faecalis and LPS. (E and F) NF-κB-driven SEAP reporter activity (E) and LDH activity (F) in the presence of E. faecalis and LTA. (G and H) NF-κB-driven SEAP reporter activity (G) and LDH activity (H) upon stimulation with heat-killed E. faecalis at the indicated MOI, with infection supernatant, or with bacterial culture supernatant, with or without LPS. Culture supernatants were collected postinfection for SEAP reporter assays and LDH assays. For NF-κB-driven SEAP reporter assays, exposure to medium alone (−) represents background NF-κB reporter activity, and stimulation with LPS or LTA represents a positive control for reporter activity. For LDH assays, Triton X treatment served as a positive control (+) for cell death. Data from 3 independent experiments were combined; mean values were graphed, and error bars represent standard errors of the means (SEM). Statistical analysis was performed using one-way ANOVA followed by Tukey's post hoc test for multiple comparisons. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; #, P < 0.05; ##, P < 0.01; ###, P < 0.001; ####, P < 0.0001. Asterisks are for comparisons to medium-only controls, and number (#) symbols are for comparisons to an MOI of 100.
E. faecalis can attenuate proinflammatory cytokine secretion in intestinal epithelial cells (15). To determine whether E. faecalis actively prevented NF-κB-mediated transcription or simply failed to induce NF-κB-mediated transcription at high MOIs in macrophages, we tested whether E. faecalis could prevent NF-κB-driven activation in the presence of TLR agonists that initiate NF-κB signaling. We exposed macrophages to LPS or LTA simultaneously with E. faecalis for 6 h, quantified both NF-κB-mediated transcription and LDH release, and observed a dose-dependent inhibition of LPS- and LTA-induced NF-κB activation by E. faecalis (Fig. 1C and E; Fig. S1C), in the absence of cytotoxicity (Fig. 1D and F; Fig. S1D).
To determine whether the absence of an NF-κB transcriptional response was due to an E. faecalis secreted factor, we examined the macrophage response to heat-killed E. faecalis or cell-free bacterial supernatants at MOI equivalents ranging from 1 to 100. We observed that heat-killed E. faecalis activated NF-κB at all MOIs, in an inverse manner to that for live intact cells (Fig. 1G), in the absence of cytotoxicity (Fig. 1H). Supernatants from infected macrophages showed NF-κB activation similar to that of live E. faecalis cells (Fig. 1G). To rule out the possibility that NF-κB activation was not due to cytokines secreted by RAW264.7 macrophages during infection, we also exposed macrophages to supernatants from bacterial cultures grown in the absence of macrophages. Supernatants from bacterial cultures weakly activated NF-κB alone and did not suppress LPS-mediated induction of NF-κB activity, except at an MOI of 100 (Fig. 1G). Together, these data suggest that E. faecalis actively prevents NF-κB activation via a process requiring a heat-modifiable factor that is secreted into cell supernatants during coculture with macrophages and that is produced in the absence of macrophages only at very high MOIs.
After determining that the bacterial factor involved in immunomodulation is secreted and heat modifiable, we examined the roles of several factors previously implicated in immune modulation or interactions with phagocytes. The gelatinase encoded by gelE is induced and secreted at high cell densities, akin to the high-cell-density NF-κB activity modulation we observed, and is regulated by the fsr quorum sensing system (18, 25, 36). Multiple peptide resistance factor (MprF) is involved in evading killing by phagocyte-associated antimicrobial peptides in E. faecalis (37, 38) and is implicated in virulence factor secretion in Listeria monocytogenes (39). Therefore, we exposed macrophages to E. faecalis mutants in each factor simultaneously with LPS. However, we observed a dose-dependent suppression of NF-κB for all mutants tested, similar to that of the parental OG1RF strain (Fig. S2). Thus, the immunomodulation phenotype that we observed is likely due to another, uncharacterized secreted bacterial factor(s).
E. faecalis suppresses NF-κB-dependent cytokine and chemokine production in RAW macrophages.
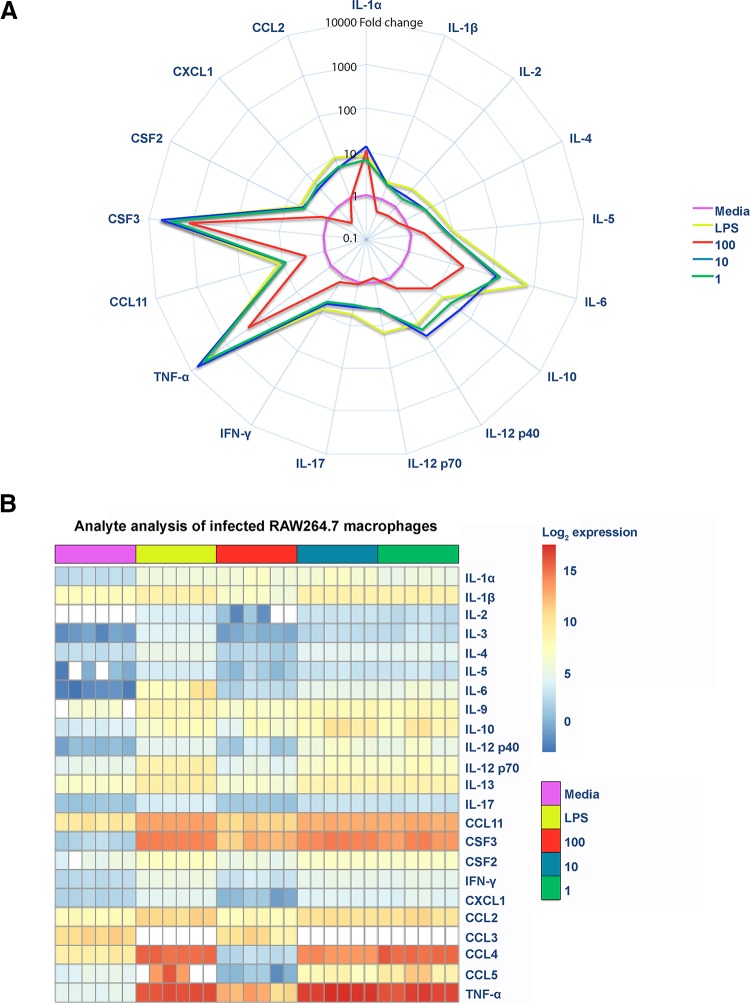
E. faecalis modulates cytokines, such as IL-8, tumor necrosis factor alpha (TNF-α), and IL-1β, in intestinal epithelial cells (13, 14). To investigate whether E. faecalis suppresses cytokine production in infected macrophages, we measured release in the absence of LPS of a variety of cytokines and chemokines whose expression is dependent on NF-κB activation. We observed an overall increase of both pro- and anti-inflammatory cytokines and chemokines at MOIs of 10 and 1, similar to that observed in LPS-treated cells (Fig. 2A and B). Strikingly, at an MOI of 100, we observed a global decrease in cytokine, chemokine, and growth factor expression compared to that with an MOI of 10 or LPS exposure (Fig. 2A). Moreover, at an MOI of 100, we observed that most of the analytes (gamma interferon [IFN-γ], CCL11, CSF2, IL-4, IL-17, IL-12p40, IL-12p70, IL-2, IL-1β, CCL2, CXCL1, and IL-5) were present at levels similar to those for the medium control (Fig. 2A and B; Fig. S3A). Principal component analysis of analytes revealed that the profile for an MOI of 100 overlapped the profile for uninfected macrophages, suggesting that analytes were not expressed despite greater numbers of E. faecalis organisms (Fig. S3B). Therefore, these data suggest that E. faecalis suppression of NF-κB activation at a high MOI led to an overall suppression of cytokine and chemokine expression (Fig. 2).
FIG 2.
E. faecalis suppresses NF-κB-dependent cytokine and chemokine production in RAW macrophages. Mouse RAW267.4 macrophages were infected with live E. faecalis at the indicated MOIs. (A) Spider plot showing the fold changes of cytokines, chemokines, and growth factors detected in filtered supernatants collected at 6 hpi for the depicted conditions. Data were normalized against the medium control, represented in pink, to obtain fold changes. (B) Heat map depicting the log2 transformation of absolute values (measured in picograms per milliliter) of the indicated cytokines, chemokines, and growth factors.
E. faecalis limits E. coli-mediated immune activation during polymicrobial RAW macrophage infection.
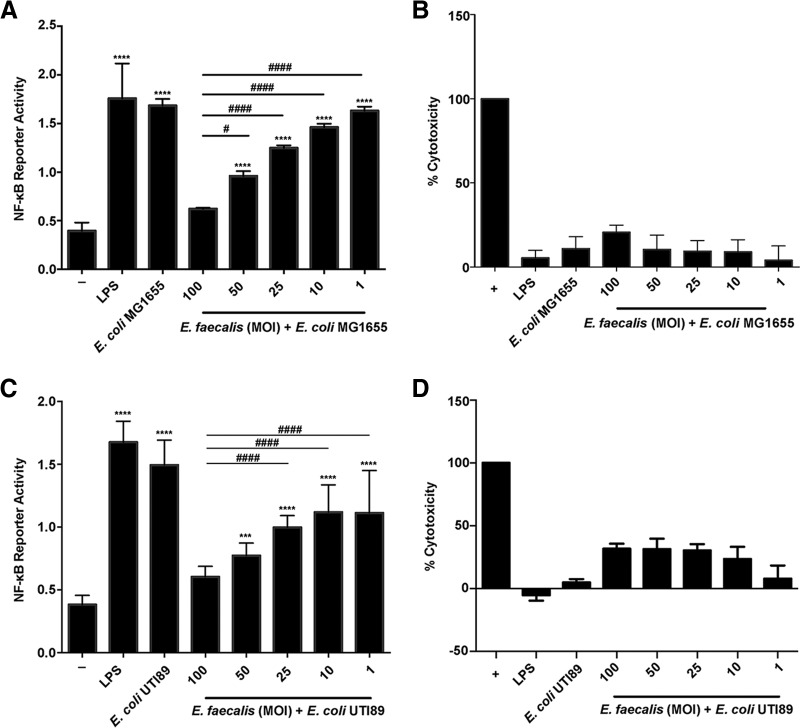
To investigate whether E. faecalis-mediated immune suppression contributed to polymicrobial UTI, we first tested its ability to suppress NF-κB activity in the presence of E. coli in vitro. We determined that RAW macrophages infected with E. coli K-12 strain MG1655 at an MOI of 1 or with E. coli UTI89 at an MOI of 0.125 induced NF-κB activation (Fig. S4A and C) in the absence of cytotoxicity (Fig. S4B and D), whereas higher MOIs were toxic to the mammalian cells (Fig. S4B and D). In addition, we observed minimal NF-κB activity for E. coli UTI89 at a high MOI, consistent with previous reports that the same strain is able to suppress the cytokine response in bladder epithelial cells and suggesting a general immunomodulatory capacity of this strain as well (Fig. S4B and D) (40). We then simultaneously infected macrophages with E. faecalis and E. coli at these predetermined MOIs and observed that while both E. coli strain MG1655 and UTI89 monoinfections induced NF-κB reporter activity equal to that with LPS alone, E. faecalis prevented E. coli-induced NF-κB activity in a dose-dependent manner (Fig. 3). Importantly, E. coli and E. faecalis growth during planktonic coculture in the absence of macrophages was unaffected by the presence of the other organism (data not shown). From this observation, we hypothesized that E. faecalis could similarly suppress the host immune response in vivo.
FIG 3.
E. faecalis suppresses E. coli-induced immune activation in vitro. Mouse RAW267.4 macrophages were stimulated simultaneously with E. faecalis OG1RF and E. coli MG1655 for 6 h prior to measurement of NF-κB-driven SEAP reporter activity (A) and LDH activity (B). Mouse RAW267.4 macrophages were coinfected with E. faecalis OG1RF and E. coli UTI89 before measurement of NF-κB-driven SEAP reporter activity (C) and LDH activity (D). For NF-κB-driven SEAP reporter assays, exposure to medium alone (−) represents background NF-κB reporter activity, and stimulation with LPS represents a positive control for reporter activity. For LDH assays, Triton X treatment served as a positive control (+) for cell death. Data from 3 independent experiments were combined; mean values were graphed, and error bars represent SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey's post hoc test for multiple comparisons. ***, P < 0.001; ****, P < 0.0001; #, P < 0.05; ###, P < 0.001; ####, P < 0.0001. Asterisks are for comparisons to medium-only controls, and number (#) symbols are for comparisons to an MOI of 100.
E. faecalis limits E. coli-mediated immune activation during mixed-species infection.
To investigate whether E. faecalis affects immune-related signaling in vivo, we performed RNA expression profiling on whole bladders at 24 h postcatheterization and -infection. We chose the CAUTI model because we desired an infection model in which E. faecalis would be present at high titers. Whereas E. faecalis does not cause a robust or high-titer infection in the ascending model of UTI (26), it colonizes the catheter and bladder at high CFU in the CAUTI model (27). We compared E. coli UTI89 monospecies infection to E. coli UTI89 and E. faecalis OG1RF coinfection at a 1:1 inoculum ratio. Of the 15,501 detectable genes (adjusted P value [Padj] of <0.05), 2 genes (0.013%) demonstrated increased mRNA levels, while 53 genes (0.34%) demonstrated decreased mRNA levels, between coinfected mice and monoinfected mice. Of these differentially expressed genes, we observed that 31 genes mapped to Gene Ontology (GO) terms, namely, response to external biotic stimulus (GO:0043207), response to other organism (GO:0051707), innate immune response (GO:0045087), response to cytokine (GO:0034097), response to biotic stimulus (GO:0009607), immune effector response (GO:0002252), and regulation of immune response (GO:0050776), that were significantly enriched (Padj < 0.01; Fisher's exact test, corrected for the 218 terms tested [see Materials and Methods]) (Fig. 4A and B; Table S1).
FIG 4.
E. faecalis suppresses E. coli-driven inflammation in catheterized mouse bladders. Female C57BL/6NTac mice were implanted with catheters in the bladder and infected with 107 CFU of E. coli UTI89 or 107 CFU each of E. coli and E. faecalis in a 1:1 mixture. After 24 h, bladders were removed and RNA extracted. (A) Binary matrix showing association between differentially expressed genes (rows) and Gene Ontology biological process (GOBP) terms (columns) enriched in the differential expression analysis between E. coli-infected and polymicrobial species-infected animals. Differentially expressed genes that did not map to an enriched GOBP term are not shown. Dark cells indicate genes that are annotated with a GOBP term, and light cells indicate genes that are not. (B) Summary of differential expression in the set of 31 genes shown in panel A. Differential expression of each gene is summarized by the mean (black dot) log2 ratio of expression between E. coli-infected and polymicrobial species-infected animals, where the line indicates the estimated standard error of the mean. (C) To examine whether the observed differential gene expression may be associated with a given ImmGen-defined cell type, we calculated the percentage of the top differentially expressed genes (upregulated in E. coli-infected compared to polymicrobial species-infected animals) that were placed within the top 1% of the distribution of the cell type-specific enrichment scores (73) (purple dots) compared to those for 100 sets of 50 expressed genes drawn at random (gray dots; the violin plot in yellow summarizes the overall distribution, and the vertical bar shows the level of the median). SP, stem and progenitor cells; B, B cells; DC, dendritic cells; MF, macrophages; MO, monocytes; GN, granulocytes; T4, CD4+ cells; T8, CD8+ cells; NKT, natural killer T cells; GDT, γδ T cells; SC, stromal cells; NK, natural killer cells. See Tables S1 and S2 in the supplemental material for related analyses. The experiment was performed twice (n = 3 mice per group per experiment). Representative data from one experiment are shown.
The enrichment of GO terms associated with immune function within downregulated genes during coinfection in the presence of E. faecalis suggested that we might also observe differential gene expression specifically in genes associated with the cell populations responding to CAUTI (29). To test this, we examined the Immunological Genome Project (ImmGen) database, which comprises publicly available data from a collection of immune cell types in C57BL/6J mice (13, 41). We found that within the top 50 differentially regulated genes between the monoinfected and coinfected groups, genes specific for dendritic cells, macrophages, and monocytes were overrepresented and showed decreased mRNA levels in coinfected animals, suggesting a reduced infiltration or activation of these cells in the bladder following coinfection compared to that following monoinfection (Fig. 4C; Table S2).
E. faecalis limits E. coli-mediated immune activation and promotes E. coli virulence during mixed-species CAUTI.
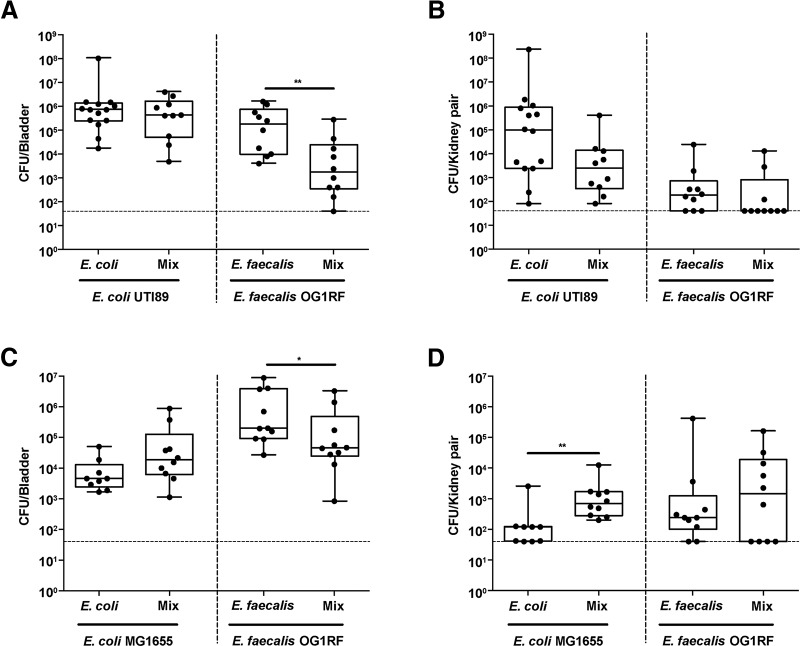
Based on downregulation of transcripts associated with interferon regulation (oas and ifi) and monocytic chemotaxis (CCL12) during E. faecalis-mediated immune suppression in vivo, we hypothesized that suppression allows UPEC to better colonize the bladder in the presence of E. faecalis. To test this in a CAUTI model, we coinfected catheterized mice with 107 CFU of E. faecalis OG1RF and 107 CFU of E. coli UTI89 and observed no significant differences in E. coli titers compared to those for monomicrobial E. coli infection (Fig. 5A and B). In contrast, E. faecalis titers during coinfection were significantly lower in the bladder but not in the kidneys, which could be a result of tissue tropism of E. faecalis for the kidneys or due to enhanced clearance as a result of the E. coli-driven immune activation, as previously described for E. coli-group B streptococcus coinfection in the bladder (Fig. 5A and B) (26, 42). We postulated that the immunomodulatory capability of UPEC strain UTI89 may be sufficient to cause high-titer CAUTI such that E. faecalis cannot further augment infection (40). Therefore, we hypothesized that colonization by a nonpathogenic, commensal-like E. coli strain, such as K-12 strain MG1655, which is deficient for LPS O-antigen expression, may be enhanced by E. faecalis-mediated immune modulation (43). To test this, we infected catheterized mice with 107 CFU of E. coli K-12 strain MG1655 alone or 107 CFU each of E. coli and E. faecalis (equal ratio). Similar to the results for infection with UTI89, the number of E. coli CFU at 24 hpi was not different following coinfection with E. faecalis in the bladder from that for E. coli monospecies infection, and the number of E. faecalis CFU was significantly decreased (Fig. 5C). In contrast, the number of E. coli CFU was significantly increased in the kidneys following coinfection with E. faecalis, while the number of E. faecalis CFU was unchanged (Fig. 5D). Collectively, these infection studies show that the presence of immunomodulatory organisms, such as E. faecalis, in the context of a polymicrobial CAUTI can increase the pathogenicity of otherwise nonvirulent infectious organisms and increase host vulnerability to infection by otherwise commensal organisms.
FIG 5.
E. faecalis promotes E. coli MG1655 infection during mixed-species CAUTI in vivo. Female C57BL/6NTac mice were implanted with 5-mm silicon catheters in the bladder and infected with 107 CFU of E. coli UTI89 or MG1655 alone, 107 CFU of E. faecalis OG1RF alone, or a 1:1 mixture of 107 CFU of E. coli and 107 CFU of E. faecalis. (A and B) Bladder (A) and kidney (B) titers from E. coli UTI89 and E. faecalis mono- and coinfections. (C and D) Bladder (C) and kidney (D) titers from E. coli MG1655 and E. faecalis mono- and coinfections. After 24 h, bladders and kidneys were removed and CFU enumerated. Data from 2 independent experiments were combined (5 to 7 mice per group). Boxes represent the 25th and 75th percentiles, with the middle line indicating the median. Whiskers represent the minimum and maximum values of the data set. Significance was determined by the nonparametric Mann-Whitney U test. *, P < 0.05; **, P < 0.01. The dashed horizontal line represents the limit of detection (LOD) of the assay. Titers below the LOD were assigned the value of the LOD for visualization on the log scale and a value of 0 for statistical analyses.
DISCUSSION
Bacterial immunomodulatory functions can alter infection sites, leading to increased susceptibility to colonization and persistence (16, 44). E. faecalis can augment the immune response in a variety of cell types, including intestinal epithelial and mouse macrophage cell lines (14–16). Recently, it was shown that E. faecalis strains V583 and E99 suppress NF-κB activation of intestinal epithelial cells and RAW264.7 macrophages at an MOI of 100 (13–16). In contrast to reports of high-MOI immune suppression by V583 and E99, infection of RAW264.7 macrophages and bone marrow-derived macrophages with E. faecalis strain E99 at an MOI of 10 results in NF-κB activation (13–16). These discrepant reports of NF-κB activation and suppression by E. faecalis underscore the need for further investigation into E. faecalis immunomodulatory activities within macrophages. Here we resolve previous conflicting reports and show that both E. faecalis strains V583 and OG1RF prevent NF-κB activity in RAW264.7 macrophages in a dose-dependent manner.
Several E. faecalis virulence factors modulate immunity during infection, including aggregation substance (AS), gelatinase, and TcpF (16–19). The E. faecalis OG1RF strain used in this study lacks the plasmid encoding AS, so AS is not the immunomodulatory factor in this work. In addition, it was shown previously that gelatinase as well as the fsr quorum sensing system that regulates gelatinase expression does not contribute to NF-κB immunomodulation (25, 36). Finally, TcpF is present in E. faecalis V583 and enriched in UTI isolates, but it is absent in OG1RF (16, 20). Since we observed NF-κB modulation by both E. faecalis OG1RF and V583, TcpF is unlikely to be the factor mediating high-level NF-κB suppression in macrophages. Instead, our data suggest that the E. faecalis factor which prevents NF-κB activity is a previously uncharacterized, heat-modifiable molecule. Other Gram-positive pathogens secrete heat-modifiable immunomodulatory molecules. For example, Staphylococcus aureus superantigen-like proteins (SSLs) have immunomodulatory functions, such as inhibiting IgA-mediated immune responses and targeting neutrophils to limit attachment to endothelial cells (45–50). SSL3 can downregulate TLR2-mediated production of IL-8 by binding competitively with PAMP ligands of TLR2 (51). Our work indicates that E. faecalis may possess similar secreted factors that modulate NF-κB activation and prevent bacterial clearance by host immune cells.
A large proportion of E. faecalis infections are polymicrobial, and E. faecalis is frequently coisolated with E. coli from urinary tract and wound infections (35, 44, 52–54). Given the prevalence of E. faecalis in polymicrobial interactions, we performed in vitro and in vivo experiments to study the contribution of E. faecalis to coinfection outcomes. We found that E. faecalis prevented NF-κB activity during coinfection with live E. coli K-12 strain MG1655 and UPEC strain UTI89 in vitro and augmented E. coli K-12 strain MG1655 titers in the kidneys. However, we did not observe E. faecalis-mediated augmentation of CAUTI for UPEC strain UTI89. We propose that this is because E. coli UTI89 is already highly virulent and inflammatory in vivo, so E. faecalis-mediated immune suppression cannot overcome such a strong proinflammatory response. Rather, our data suggest that E. faecalis may help to increase the fitness of less virulent or commensal strains, such as E. coli MG1655. Similar to our findings in this study, the Gram-positive uropathogens Staphylococcus saprophyticus and group B streptococcus induce minimal proinflammatory responses in the urinary tract, and the latter suppresses proinflammatory responses in vitro and limits UPEC pathogenesis in mice (42, 44, 55–57). Taken together, our findings suggest that E. faecalis modulation of the immune response may promote the survival of commensal bacteria, which increases the chance of UTI.
Synergistic polymicrobial infections are increasingly recognized for their contributions to both disease severity and persistence (31, 44). Here we show that E. faecalis modulates the host response and promotes infection by a coinfecting E. coli strain which is otherwise nonvirulent. Importantly, the presence of E. faecalis in the urinary tract, especially when titers are low, has historically been considered a commensal contaminant of questionable pathogenic significance (58). Our findings call into question that supposition and raise the prospect that E. faecalis not only augments some E. coli infections but also may also promote infection by a larger spectrum of less fit or commensal E. coli strains, similar to the effect exerted by group B streptococcus (42). Continued efforts are needed to dissect these polymicrobial molecular interactions to allow for better diagnostics and precision treatment, especially as UTI pathogens are increasingly resistant to antibiotics of last resort (59).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Uropathogenic E. coli (UPEC) strain UTI89 (60, 61) and E. coli K-12 strain MG1655,which is an LPS mutant strain (43, 62), were grown overnight in Luria-Bertani (LB) broth or agar at 37°C under static conditions. E. faecalis strains OG1RF (63) and V583 (64) were grown statically in brain heart infusion (BHI) broth or agar at 37°C overnight. Overnight cultures of bacteria were centrifuged at 6,000 × g for 5 min and resuspended in phosphate-buffered saline (PBS) to an optical density at 600 nm (OD600) of 0.7 (2 × 108 CFU/ml) for E. faecalis and an OD600 of 0.4 (2 × 108 CFU/ml) for E. coli.
Cell culture.
RAW-Blue cells, derived from RAW264.7 macrophages (Invivogen) and containing a plasmid encoding a secreted embryonic alkaline phosphatase (SEAP) reporter under transcriptional control of an NF-κB-inducible promoter, were cultivated in Dulbecco modified Eagle medium containing 4,500 mg/liter glucose (high glucose; 1×) with 4.0 mM l-glutamine, without sodium pyruvate (Gibco) and supplemented with 10% fetal bovine serum (FBS; PAA) supplemented with 200 μg/ml Zeocin at 37°C in 5% CO2.
RAW-Blue macrophage infection.
RAW-Blue cells were seeded in a 96-well plate at 100,000 cells/well in 200 μl of antibiotic-free cell culture medium. Following overnight incubation, the cells were washed once with PBS, and fresh medium was added. The SEAP reporter assay was established by empirically defining the minimal agonist (LPS or lipoteichoic acid [65]) concentration that induced the maximum SEAP activity in the absence of cell death. Cells were stimulated using EB ultrapure LPS purified from E. coli O111:B4 (100 ng/ml) (Invivogen) or LTA derived/purified from Staphylococcus aureus (100 ng/ml) (Invivogen) as a positive control or medium alone as a negative control. RAW-Blue cells were infected with E. faecalis (at MOIs of 100:1, 50:1, 25:1, 10:1, and 1:1) for 6 h, with or without TLR agonists. Overnight bacterial cultures were centrifuged and resuspended in cell culture medium. For infection experiments, live bacterial cultures were diluted to achieve the desired MOI with macrophages. Alternatively, bacteria were heat killed (80°C for 1 h) prior to addition to macrophage cultures. For coinfection experiments, RAW-Blue cells were simultaneously infected with E. coli K-12 strain MG1655 (MOI of 1:1) or E. coli UTI89 (MOI of 0.125:1) and E. faecalis OG1RF (MOIs of 100:1, 50:1, 25:1, 10:1, and 1:1). Heat-killed bacteria were verified by the absence of viable bacteria on BHI agar.
Collection of bacterial cell-free culture supernatants.
Bacteria were grown in cell culture medium for 6 h, and bacterium-free culture supernatants were collected after centrifugation (6,000 × g) followed by filtration (using a 0.2-μm syringe filter). Alternatively, supernatants were collected after infecting macrophages with bacteria at various MOIs and then filtered by use of a 0.2-μm syringe filter. Sterility of bacterium-free culture supernatants was verified by the absence of viable bacteria on BHI agar.
NF-κB reporter assay.
Postinfection, 20 μl of supernatant was added to 180 μl of Quanti-Blue reagent (Invivogen) and incubated overnight at 37°C. SEAP levels were determined at 640 nm by using a Tecan M200 microplate reader. All experiments were performed in triplicate.
Cell viability assay.
Simultaneously with supernatant collection for SEAP determination, culture supernatants were collected from each well to measure lactate dehydrogenase (LDH) release by using an LDH cytotoxicity assay (Clontech) according to the manufacturer's instructions. Background LDH activity was determined using mock (PBS)-treated RAW-Blue cells. Maximal LDH activity was determined by lysing cells with 1% Triton X. Each condition was carried out in triplicate. The percentage of cytotoxicity was calculated as follows: % cytotoxicity = [(sample absorbance − background absorbance)/(maximal absorbance − background absorbance)] × 100.
Luminex MAP analysis.
Supernatants were collected from RAW-Blue cells at 6 h postinfection and stored at −80°C until assessment by use of a Bio-Plex Pro mouse cytokine 23-plex assay kit (Bio-Rad Laboratories) according to the manufacturer's recommendations (66). All samples were assessed using the same kit lot and at the same time to avoid interassay variability.
Catheterization and bacterial infections.
Catheters were implanted into bladders of mice, followed by bacterial inoculation via a transurethral catheter, as previously described (28, 67). Briefly, 6- to 8-week-old female C57BL/6NTac mice (InVivos Pte Ltd., Singapore) were anesthetized with isoflurane (4%). Inoculum volumes of 50 μl contained bacterial suspensions of either single or polymicrobial species prepared in PBS, as follows: (i) 107 CFU of E. coli K-12 strain MG1655 with 107 CFU of E. faecalis OG1RF and (ii) 107 CFU of E. coli UTI89 with 107 CFU of E. faecalis OG1RF. Single-species control infections (107 CFU of E. coli K-12 strain MG1655, 107 CFU of E. faecalis OG1RF, or 107 CFU of E. coli UTI89) were performed alongside polymicrobial infections. Animals were euthanized by carbon dioxide inhalation and cervical dislocation, and bladders and kidneys were aseptically removed and homogenized in 1 ml PBS for CFU enumeration by serial dilution on MacConkey agar or on BHI agar supplemented with 10 μg/ml colistin and 10 μg/ml nalidixic acid to isolate E. coli or E. faecalis, respectively. To identify bacterial species other than the inoculated E. coli or E. faecalis organisms, serial dilutions were also plated on LB and BHI agar. Animals in which other organisms were found were excluded from our analysis. Data from 2 independent experiments (5 to 7 mice per group) were combined. Animals without catheters at the time of sacrifice were not included in the analyses.
RNA sequencing of RNAs from infected bladders.
Catheterized mice were infected as described above with 107 CFU of E. coli UTI89, alone or mixed at a 1:1 ratio with 107 CFU of E. faecalis OG1RF, in 50 μl PBS. After 24 h, whole bladders were removed and incubated overnight in RNAlater (Qiagen) to allow complete tissue penetration by the protectant prior to storage at −80°C. RNA was extracted as described previously (29). For each sample condition, a total of three sequence libraries were constructed from 50 to 200 ng of rRNA-depleted RNA. Each library (2 nM) was pooled at equal volumes and sequenced using an Illumina Hiseq2500 v.2 sequencer (Illumina), giving 150-bp paired-end sequences.
Analysis of RNA sequencing data.
RNA sequencing results were analyzed as described previously (29). Briefly, reads were quality checked and adapters trimmed with cutadapt-1.4.1, using default parameters. The mm10 mouse genome was used as a reference for tophat-2.0.11.Linux_x86_64 (68), and transcriptional read counts were obtained using HTSeq-0.6.1 (69) with default parameters, using a nonstranded analysis. Unless otherwise stated, all further analyses were performed in the R statistical computing environment (version 3.3.3) (70). Differential analysis of E. coli monospecies-infected animals and E. coli- and E. faecalis OG1RF-coinfected animals was performed using the R/Bioconductor package DESeq2 (version 1.40.1) (71), using default settings from that package. The NCBI file gene2refseq (downloaded 3 March 2016) was used to convert Refseq identifiers to Entrez identifiers for further analysis of Gene Otology annotations and Immunological Genome Project (ImmGen) data for functional analysis.
Functional analysis of the bladder transcriptome.
Processing of ImmGen expression data was performed in R 3.2.2 (70). Briefly, all 681 CEL files (72) were processed using the RMA method with the R/Bioconductor package oligo (version 1.34.0). Annotation was done using the R/Bioconductor package mogene10sttranscriptcluster.db (version 8.4.0), with expression profiles for immune cell types referenced from the work of Jojic et al. (72). An enrichment score was calculated for each immune cell type (73), and we examined differentially expressed genes in the top 1% of the score distribution for each cell type and compared these to equally sized cohorts of randomly selected genes (see Table S2 in the supplemental material). This analysis was conducted separately with up- and downregulated gene sets in the two-group comparison. Ontology analysis was performed using the R/Bioconductor package GO.db (version 3.4.0) and the gene2go file from the NCBI Gene database (downloaded 3 March 2016), using modified code from the R/Bioconductor package ontoTools (version 1.28.0) (74). To test if differentially expressed genes associated with specific gene sets, we constructed 2-by-2 contingency tables and categorized genes based on whether they were differentially expressed or not for each included Gene Ontology biological process (GOBP) term. Random assignment was tested using Fisher's exact test (75) and corrected for the number of terms by use of the Benjamini-Hochberg correction (76). We filtered terms by using their information content (IC) (77), based on a frequency of occurrence between 3 and 4, resulting in a set of 157 included terms that represent an appropriate trade-off between the total number of included terms and the specificity of functional insight. The entire R workflow and input data files are available online (https://github.com/rbhwilliams/Kline-polymicrobial-infection-paper).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism software (version 6.05 for Windows). SEAP assays were analyzed using one-way analysis of variance (ANOVA) with Tukey's post hoc test for multiple comparisons. Cytokine readings for Luminex MAP analysis were analyzed using the Kruskal-Wallis test. CFU titers were compared using the nonparametric Mann-Whitney U test. P values of <0.05 were deemed significant. Cytokine comparisons were performed using the Mann-Whitney U test, and further comparison was done using principal component analysis in R (version 3.3.2), with the packages factoextra (version 1.0.4) and FactoMineR (version 1.34). Heat map data reflect log2 transformation of the raw data and were plotted in R (version 3.3.2) by using the R package pheatmap (version 1.0.8).
Ethics statement.
Mouse experiments were performed with ethical approval by the Nanyang Technological University Institutional Animal Care and Use Committee (protocol ARF-SBS/NIE-A0247), which adheres to the National Advisory Committee for Laboratory Animal Research (NACLAR), a national guide which establishes the best practices for the use and care of animals for scientific purposes.
Accession number(s).
RNA sequencing data from this study are available at NCBI's BioProject under accession no. PRJNA335539.
Supplementary Material
ACKNOWLEDGMENTS
We thank Krithika Arumugam for her assistance with sequence data processing. We also thank Gary Dunny and Jennifer Dale for providing the E. faecalis transposon mutants used in this study. This work benefitted from data assembled by the ImmGen Consortium (41).
B.Y.Q.T., H.M.S.G., K.K.L.C., S.B.-T., S.H., R.B.H.W., and K.A.K. were supported by the National Research Foundation and the Ministry of Education Singapore under its Research Centre of Excellence Programme. B.Y.Q.T., H.M.S.G., K.K.L.C., S.B.-T., S.H., and K.A.K. were supported by the National Research Foundation under its Singapore NRF fellowship program (grant NRF-NRFF2011-11) and by the Ministry of Education Singapore under its tier 2 program (grant MOE2014-T2-1-129). K.A.K. and M.A.I. were supported by the Merlion Programme, sponsored by the French Ministry of Foreign Affairs and Nanyang Technological University. F.G. and R.D. were supported by Singapore Immunology Network core funding from the Agency for Science, Technology and Research (A*STAR), Singapore.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00378-17.
REFERENCES
- 1.Gilmore MS, Clewell DB, Ike Y, Shankar N (ed). 2014. Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 2.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 4.Maki DG, Tambyah PA. 2001. Engineering out the risk for infection with urinary catheters. J Emerg Infect Dis 7:342–347. doi: 10.3201/eid0702.010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 6.Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. 2015. How biofilms evade host defenses. Microbiol Spectr 3: MB-0012-2014. doi: 10.1128/microbiolspec.MB-0012-2014. [DOI] [PubMed] [Google Scholar]
- 7.Baldassarri L, Bertuccini L, Creti R, Filippini P, Ammendolia MG, Koch S, Huebner J, Orefici G. 2005. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J Infect Dis 191:1253–1262. doi: 10.1086/428778. [DOI] [PubMed] [Google Scholar]
- 8.Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, Van Winkle WB, Simon SI. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 67:6067–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentry-Weeks CR, Karkhoff-Schweizer R, Pikis A, Estay M, Keith JM. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect Immun 67:2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou J, Shankar N. 2016. The opportunistic pathogen Enterococcus faecalis resists phagosome acidification and autophagy to promote intracellular survival in macrophages. Cell Microbiol 18:831–843. doi: 10.1111/cmi.12556. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, Shankar N. 2014. Enterococcus faecalis infection activates phosphatidylinositol 3-kinase signaling to block apoptotic cell death in macrophages. Infect Immun 82:5132–5142. doi: 10.1128/IAI.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. 2007. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Zou J, Shankar N. 2015. Roles of TLR/MyD88/MAPK/NF-κB signaling pathways in the regulation of phagocytosis and proinflammatory cytokine expression in response to E. faecalis infection. PLoS One 10:e0136947. doi: 10.1371/journal.pone.0136947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Hibberd ML, Pettersson S, Lee YK. 2014. Enterococcus faecalis from healthy infants modulates inflammation through MAPK signaling pathways. PLoS One 9:e97523. doi: 10.1371/journal.pone.0097523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Ng LH, Chow WL, Lee YK. 2008. Infant intestinal Enterococcus faecalis down-regulates inflammatory responses in human intestinal cell lines. World J Gastroenterol 14:1067–1076. doi: 10.3748/wjg.14.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou J, Baghdayan AS, Payne SJ, Shankar N. 2014. A TIR domain protein from E. faecalis attenuates MyD88-mediated signaling and NF-kappaB activation. PLoS One 9:e112010. doi: 10.1371/journal.pone.0112010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. 2007. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun 75:1861–1869. doi: 10.1128/IAI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SY, Shin YP, Kim CH, Park HJ, Seong YS, Kim BS, Seo SJ, Lee IH. 2008. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J Immunol 181:6328–6336. doi: 10.4049/jimmunol.181.9.6328. [DOI] [PubMed] [Google Scholar]
- 19.Sussmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun 68:4900–4906. doi: 10.1128/IAI.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraemer TD, Quintanar Haro OD, Domann E, Chakraborty T, Tchatalbachev S. 2014. The TIR domain containing locus of Enterococcus faecalis is predominant among urinary tract infection isolates and downregulates host inflammatory response. Int J Microbiol 2014:918143. doi: 10.1155/2014/918143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunny GM, Leonard BA, Hedberg PJ. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol 177:871–876. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur J Biochem 222:235–246. [DOI] [PubMed] [Google Scholar]
- 23.Vanek NN, Simon SI, Jacques-Palaz K, Mariscalco MM, Dunny GM, Rakita RM. 1999. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol Med Microbiol 26:49–60. [DOI] [PubMed] [Google Scholar]
- 24.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. 2010. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun 78:4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teixeira N, Santos S, Marujo P, Yokohata R, Iyer VS, Nakayama J, Hancock LE, Serror P, Silva Lopes MF. 2012. The incongruent gelatinase genotype and phenotype in Enterococcus faecalis are due to shutting off the ability to respond to the gelatinase biosynthesis-activating pheromone (GBAP) quorum-sensing signal. Microbiology 158:519–528. doi: 10.1099/mic.0.055574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. 2005. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun 73:2461–2468. doi: 10.1128/IAI.73.4.2461-2468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guiton PS, Hannan TJ, Ford B, Caparon MG, Hultgren SJ. 2013. Enterococcus faecalis overcomes foreign body-mediated inflammation to establish urinary tract infections. Infect Immun 81:329–339. doi: 10.1128/IAI.00856-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. 2010. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect Immun 78:4166–4175. doi: 10.1128/IAI.00711-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau M, Goh HM, Holec S, Albert ML, Williams RB, Ingersoll MA, Kline KA. 2016. Bladder catheterization increases susceptibility to infection that can be prevented by prophylactic antibiotic treatment. JCI Insight 1:e88178. doi: 10.1172/jci.insight.88178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stickler DJ. 2008. Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol 5:598–608. doi: 10.1038/ncpuro1231. [DOI] [PubMed] [Google Scholar]
- 31.Croxall G, Weston V, Joseph S, Manning G, Cheetham P, McNally A. 2011. Increased human pathogenic potential of Escherichia coli from polymicrobial urinary tract infections in comparison to isolates from monomicrobial culture samples. J Med Microbiol 60:102–109. doi: 10.1099/jmm.0.020602-0. [DOI] [PubMed] [Google Scholar]
- 32.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchimori N, Hayashi R, Shino A, Yamazaki T, Okonogi K. 1994. Enterococcus faecalis aggravates pyelonephritis caused by Pseudomonas aeruginosa in experimental ascending mixed urinary tract infection in mice. Infect Immun 62:4534–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamasaki H, Arakawa S, Kamidono S. 1991. Basic studies on the pathogenicity of Enterococcus faecalis: polymicrobial infection with Enterococcus faecalis and Proteus mirabilis in experimental ascending pyelonephritis models. J Infect Chemother 39:651–662. [Google Scholar]
- 35.Ronald A. 2003. The etiology of urinary tract infection: traditional and emerging pathogens. Dis Mon 49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 36.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol 188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, Hartke A, Huebner J. 2012. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 7:e38458. doi: 10.1371/journal.pone.0038458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, Schröder JM, Caparon MG, Normark S, Henriques-Normark B, Hultgren SJ, Kline KA. 2013. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A 110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol 191:3950–3964. doi: 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunstad DA, Justice SS, Hung CS, Lauer SR, Hultgren SJ. 2005. Suppression of bladder epithelial cytokine responses by uropathogenic Escherichia coli. Infect Immun 73:3999–4006. doi: 10.1128/IAI.73.7.3999-4006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng TS, Painter MW. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 42.Kline KA, Schwartz DJ, Gilbert NM, Hultgren SJ, Lewis AL. 2012. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection by uropathogenic Escherichia coli. Infect Immun 80:4186–4194. doi: 10.1128/IAI.00684-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning DF, Wells TJ, Franca FL, Morris FC, Sevastsyanovich YR, Bryant JA, Johnson MD, Lund PA, Cunningham AF, Hobman JL, May RC, Webber MA, Henderson IR. 2013. Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol Microbiol 87:939–950. doi: 10.1111/mmi.12144. [DOI] [PubMed] [Google Scholar]
- 44.Tay WH, Chong KK, Kline KA. 2016. Polymicrobial-host interactions during infection. J Mol Biol 428:3355–3371. doi: 10.1016/j.jmb.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Fraser JD, Proft T. 2008. The bacterial superantigen and superantigen-like proteins. Immunol Rev 225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 46.Langley R, Wines B, Willoughby N, Basu I, Proft T, Fraser JD. 2005. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J Immunol 174:2926–2933. doi: 10.4049/jimmunol.174.5.2926. [DOI] [PubMed] [Google Scholar]
- 47.Wines BD, Willoughby N, Fraser JD, Hogarth PM. 2006. A competitive mechanism for staphylococcal toxin SSL7 inhibiting the leukocyte IgA receptor, Fc alphaRI, is revealed by SSL7 binding at the C alpha2/C alpha3 interface of IgA. J Biol Chem 281:1389–1393. doi: 10.1074/jbc.M509334200. [DOI] [PubMed] [Google Scholar]
- 48.Ramsland PA, Willoughby N, Trist HM, Farrugia W, Hogarth PM, Fraser JD, Wines BD. 2007. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc Natl Acad Sci U S A 104:15051–15056. doi: 10.1073/pnas.0706028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung MC, Wines BD, Baker H, Langley RJ, Baker EN, Fraser JD. 2007. The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Mol Microbiol 66:1342–1355. doi: 10.1111/j.1365-2958.2007.05989.x. [DOI] [PubMed] [Google Scholar]
- 50.Bestebroer J, Poppelier MJ, Ulfman LH, Lenting PJ, Denis CV, van Kessel KP, van Strijp JA, de Haas CJ. 2007. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 109:2936–2943. [DOI] [PubMed] [Google Scholar]
- 51.Bardoel BW, Vos R, Bouman T, Aerts PC, Bestebroer J, Huizinga EG, Brondijk TH, van Strijp JA, de Haas CJ. 2012. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. J Mol Med (Berl) 90:1109–1120. doi: 10.1007/s00109-012-0926-8. [DOI] [PubMed] [Google Scholar]
- 52.Siegman-Igra Y, Kulka T, Schwartz D, Konforti N. 1994. Polymicrobial and monomicrobial bacteraemic urinary tract infection. J Hosp Infect 28:49–56. [DOI] [PubMed] [Google Scholar]
- 53.Cooper RA. 2013. Surgical site infections: epidemiology and microbiological aspects in trauma and orthopaedic surgery. Int Wound J 10(Suppl 1):S3–S8. doi: 10.1111/iwj.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kline KA, Schwartz DJ, Lewis WG, Hultgren SJ, Lewis AL. 2011. Immune activation and suppression by group B Streptococcus in a murine model of urinary tract infection. Infect Immun 79:3588–3595. doi: 10.1128/IAI.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlin AF, Lewis AL, Varki A, Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol 189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kline KA, Ingersoll MA, Nielsen HV, Sakinc T, Henriques-Normark B, Gatermann S, Caparon MG, Hultgren SJ. 2010. Characterization of a novel murine model of Staphylococcus saprophyticus urinary tract infection reveals roles for Ssp and SdrI in virulence. Infect Immun 78:1943–1951. doi: 10.1128/IAI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kline KA, Lewis AL. 2016. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr 4: UTI-0012-2012. doi: 10.1128/microbiolspec.UTI-0012-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Sahm DF. 2002. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob Agents Chemother 46:2540–2545. doi: 10.1128/AAC.46.8.2540-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 63.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 65.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJWM, Geelen E, Sahin Ö Sieuwerts M, Brakenhoff JPJ, Vogels R, Li OTW, Poon LLM, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RHE. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ingersoll MA, Kline KA, Nielsen HV, Hultgren SJ. 2008. G-CSF induction early in uropathogenic Escherichia coli infection of the urinary tract modulates host immunity. Cell Microbiol 10:2568–2578. doi: 10.1111/j.1462-5822.2008.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hung CS, Dodson KW, Hultgren SJ. 2009. A murine model of urinary tract infection. Nat Protoc 4:1230–1243. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Development Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 71.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Best AJ, Knell J, Goldrath A, Joic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemarte-Pelletier A, Malhotra D, Turley S. 2013. Identification of transcriptional regulators in the mouse immune system. Nat Immunol 14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, Xavier RJ. 2010. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115:5376–5384. doi: 10.1182/blood-2010-01-263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carey VJ. 2004. Ontology concepts and tools for statistical genomics. J Multivar Anal 90:213–228. doi: 10.1016/j.jmva.2004.02.001. [DOI] [Google Scholar]
- 75.Wall JV, Jenkins CR. 2003. Practical statistics for astronomers. Cambridge University Press, New York, NY. [Google Scholar]
- 76.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 77.Alterovitz G, Xiang M, Mohan M, Ramoni MF. 2007. GO PaD: the Gene Ontology Partition Database. Nucleic Acids Res 35:D322–D327. doi: 10.1093/nar/gkl799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.