ABSTRACT
Oral bacteria are the main trigger for the development of periodontitis, and some species are known to modulate neutrophil function. This study aimed to explore the release of neutrophil extracellular traps (NETs), associated antimicrobial proteins, and reactive oxygen species (ROS) in response to periodontal bacteria, as well as the underlying pathways. Isolated peripheral blood neutrophils were stimulated with 19 periodontal bacteria. NET and ROS release, as well as the expression of NET-bound antimicrobial proteins, elastase, myeloperoxidase, and cathepsin G, in response to these species was measured using fluorescence-based assays. NET and ROS release was monitored after the addition of NADP (NADPH) oxidase pathway modulators and inhibitors of Toll-like receptors (TLRs). Moreover, bacterial entrapment by NETs was visualized microscopically, and bacterial killing was assessed by bacterial culture. Certain microorganisms, e.g., Veillonella parvula and Streptococcus gordonii, stimulated higher levels of ROS and NET release than others. NETs were found to entrap, but not kill, all periodontal bacteria tested. NADPH oxidase pathway modulators decreased ROS production but not NET production in response to the bacteria. Interestingly, TLR inhibitors did not impact ROS and NET release. These data suggest that the variability in the neutrophil response toward different bacteria may contribute to the pathogenesis of periodontal diseases by mechanisms such as bacterial avoidance of host responses and activation of neutrophils. Moreover, our results indicate that bacterium-stimulated NET release may arise in part via NADPH oxidase-independent mechanisms. The role of TLR signaling in bacterium-induced ROS and NET release needs to be further elucidated.
KEYWORDS: neutrophils, NETs, reactive oxygen species, periodontitis, oral bacteria
INTRODUCTION
Periodontitis is initiated by the accumulation of microbial biofilms at and below the gingival margin. Indeed, it has been estimated that ∼700 oral bacterial species and ∼1,200 predominant phylotypes exist (1–3). Of these bacterial species, 5 major bacterial complexes (red, orange, yellow, green, and purple) have been identified by Socransky et al. using DNA probes (4) and are referred to here as Socransky complexes. The clustering and ordination analysis allowed them to assign microbial species to a color complex on the basis of the strength of the association with each other and the clinical staging of periodontitis. The biofilms which develop during disease are orchestrated to maximize their adherence, communication, and survival. The accumulation of bacterial species within the biofilm enables its development and perseverance, and certain bacteria, such as Fusobacterium nucleatum, are key orchestrators of biofilm formation and maturation (5).
In susceptible individuals, dysbiosis and an aberrant host-microbe equilibrium can result in the onset of disease (6), where the microbial biofilm thrives by exploiting the host inflammatory response. This process fuels a vicious cycle of bacterial accumulation, inflammation, and subsequent tissue destruction. The acute inflammatory reaction is predominantly mediated by neutrophils via activation of innate neutrophil-derived defense mechanisms and also the activation of the acquired cellular and humoral immune system and is initially protective. In periodontitis, however, the aberrant neutrophil response is reputed to contribute to collateral tissue damage and the formation of disease-associated molecular patterns, which perpetuate the inflammation, leading to chronicity (7). Furthermore, the inflammatory state itself supplies nutrients to pathogenic bacteria, such as Porphyromonas gingivalis, e.g., iron from heme, supporting its survival and proliferation (8).
Exaggerated immune activity is also observed in peripheral blood neutrophils from patients with both chronic and aggressive periodontitis. These neutrophils are reportedly hyperreactive in response to a microbial challenge in terms of their release of reactive oxygen species (ROS), but they are also hyperactive in the absence of an exogenous stimulus (9–11). In addition, excessive neutrophil-driven proteolytic activity and proinflammatory cytokine production have been observed in periodontitis and are associated with pathogenicity (7). One of the mechanisms by which neutrophils combat microorganisms is through the production of neutrophil extracellular traps (NETs), whereby decondensed DNA is released into the extracellular environment to immobilize and potentially kill invading bacteria. NET release is reported to be dependent on the production of ROS, such as hydrogen peroxide (H2O2), via superoxide generation by the enzyme NADPH oxidase (12).
Little is known about the differential interactions between oral bacteria and neutrophils; however, there is evidence that certain species and strains can evoke different neutrophil responses (13–16). This study aimed to elucidate the ability of bacterial species and strains frequently isolated from the oral cavity of healthy and diseased individuals to activate ROS and NET responses in neutrophils. The ability of NETs to entrap and kill bacteria, along with the expression of the antimicrobial and NET-associated proteins neutrophil elastase (NE), myeloperoxidase (MPO), and cathepsin G (CG), was also analyzed.
(This study arose from Phillipa C. White's Ph.D. thesis [17].)
RESULTS
Neutrophil ROS release in response to periodontal bacteria.
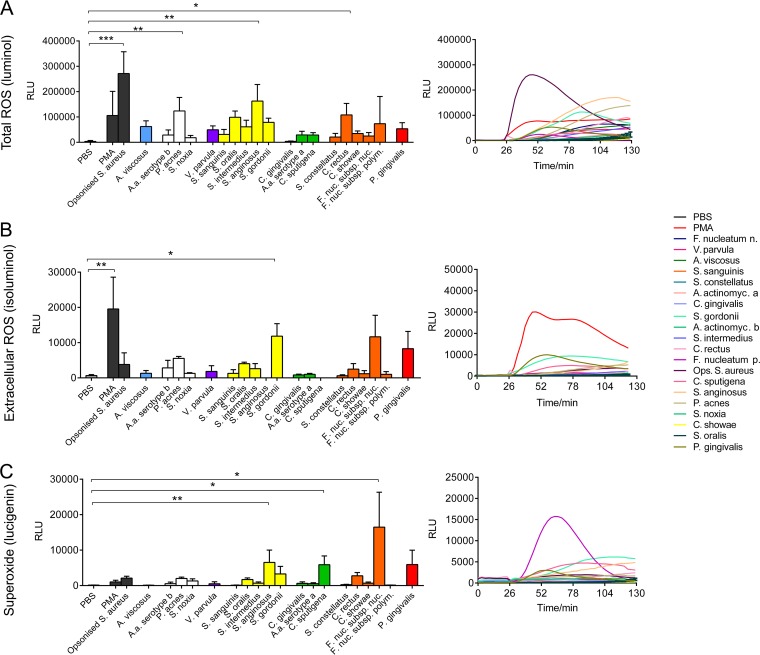
The production of total ROS, extracellular ROS, and superoxide in response to 19 periodontal bacteria and Staphylococcus aureus (Table 1) was determined. Certain bacteria elicited higher total ROS production in neutrophils, which was measured by determination of the amount of luminol chemiluminescence. This was statistically significant for Propionibacterium acnes, Streptococcus anginosus, and Campylobacter rectus as well as the positive control, opsonized S. aureus (Fig. 1A). Consistent with the data expressed as the total peak amount of ROS production, the time course of ROS production, expressed as the area under the curve, demonstrated that ROS production was the highest in response to opsonized S. aureus, followed by S. anginosus. Notably, the increase in the amount of total ROS in response to opsonized S. aureus was more rapid than that following direct stimulation with periodontal bacteria, as illustrated by the sharp elevation of the curve immediately following stimulation. Neutrophil extracellular ROS production was subsequently analyzed by determination of the amount of isoluminol chemiluminescence. Phorbol 12-myristate 13-acetate (PMA; positive control) and Streptococcus gordonii induced significantly higher levels of extracellular ROS than phosphate-buffered saline (PBS) treatment (negative control) (Fig. 1B). The steep time course curve in response to PMA indicates a rapid neutrophil response. Neutrophil extracellular superoxide production was measured using lucigenin. PMA and opsonized S. aureus did not induce significantly higher levels of superoxide production than the PBS control. However, some periodontal bacteria increased extracellular superoxide production in neutrophils, which was statistically significant for S. anginosus, Capnocytophaga sputigena, and Fusobacterium nucleatum subsp. nucleatum (Fig. 1C).
TABLE 1.
Bacteria used, their assignment to Socransky complexes, and growth conditions
| Bacterial species | Strain | Socransky complex | Growth condition |
|---|---|---|---|
| Actinomyces viscosus (A. naeslundii genospecies 2) | ATCC 43146 | Blue | Anaerobic |
| Aggregatibacter actinomycetemcomitans serotype a | ATCC 29523 | Green | Anaerobic |
| Aggregatibacter actinomycetemcomitans serotype b | ATCC 43718 | White | Anaerobic |
| Campylobacter rectus | ATCC 33238 | Orange | Anaerobic |
| Campylobacter showae | ATCC 51146 | Orange | Anaerobic |
| Capnocytophaga gingivalis | ATCC 33624 | Green | Anaerobic |
| Capnocytophaga sputigena | ATCC 33612 | Green | Anaerobic |
| Fusobacterium nucleatum subsp. nucleatum | ATCC 25586 | Orange | Anaerobic |
| Fusobacterium nucleatum subsp. polymorphum | ATCC 10953 | Orange | Anaerobic |
| Porphyromonas gingivalis | ATCC W83 | Red | Anaerobic |
| Propionibacterium acnes | ATCC 11827 | White | Anaerobic |
| Selenomonas noxia | ATCC 43541 | White | Anaerobic |
| Staphylococcus aureus (opsonized) | ATCC 9144 | NAa | Aerobic |
| Streptococcus anginosus | ATCC 33397 | Yellow | 5% CO2 |
| Streptococcus constellatus | ATCC 27823 | Orange | 5% CO2 |
| Streptococcus gordonii | ATCC 10558 | Yellow | 5% CO2 |
| Streptococcus intermedius | ATCC 27335 | Yellow | 5% CO2 |
| Streptococcus oralis | ATCC 35037 | Yellow | 5% CO2 |
| Streptococcus sanguinis | ATCC 10556 | Yellow | 5% CO2 |
| Veillonella parvula | ATCC 10790 | Purple | Anaerobic |
NA, not applicable.
FIG 1.
Neutrophil reactive oxygen species (ROS) production in response to periodontal bacteria. Neutrophil total ROS (A), extracellular ROS (B), and superoxide (C) production levels were quantified (right), and their levels of production in response to periodontal bacteria were assayed using luminol, isoluminol, and lucigenin enhanced chemiluminescence, respectively, over a time course of 130 min (left). ROS release in response to PBS (unstimulated negative control), phorbol 12-myristate 13-acetate (PMA; 25 nM; positive control), and opsonized S. aureus (positive control) was also quantified. Data are presented as relative light units (RLU) and represent the results for neutrophils from five different donors assessed in triplicate wells. *, P < 0.05; **, P < 0.01; ***, P < 0.001. A.a., A. actinomycetemcomitans; A. actinomyc a, A. actinomycetemcomitans serotype a; A. actinomyc b, A. actinomycetemcomitans serotype b; F. nuc. subsp. nuc. and F. nucleatum n, Fusobacterium nucleatum subsp. nucleatum; F. nuc. subsp. polym. and F. nucleatum p, Fusobacterium nucleatum subsp. polymorphum; Ops., opsonized.
Quantification of NET production in response to periodontal bacteria.
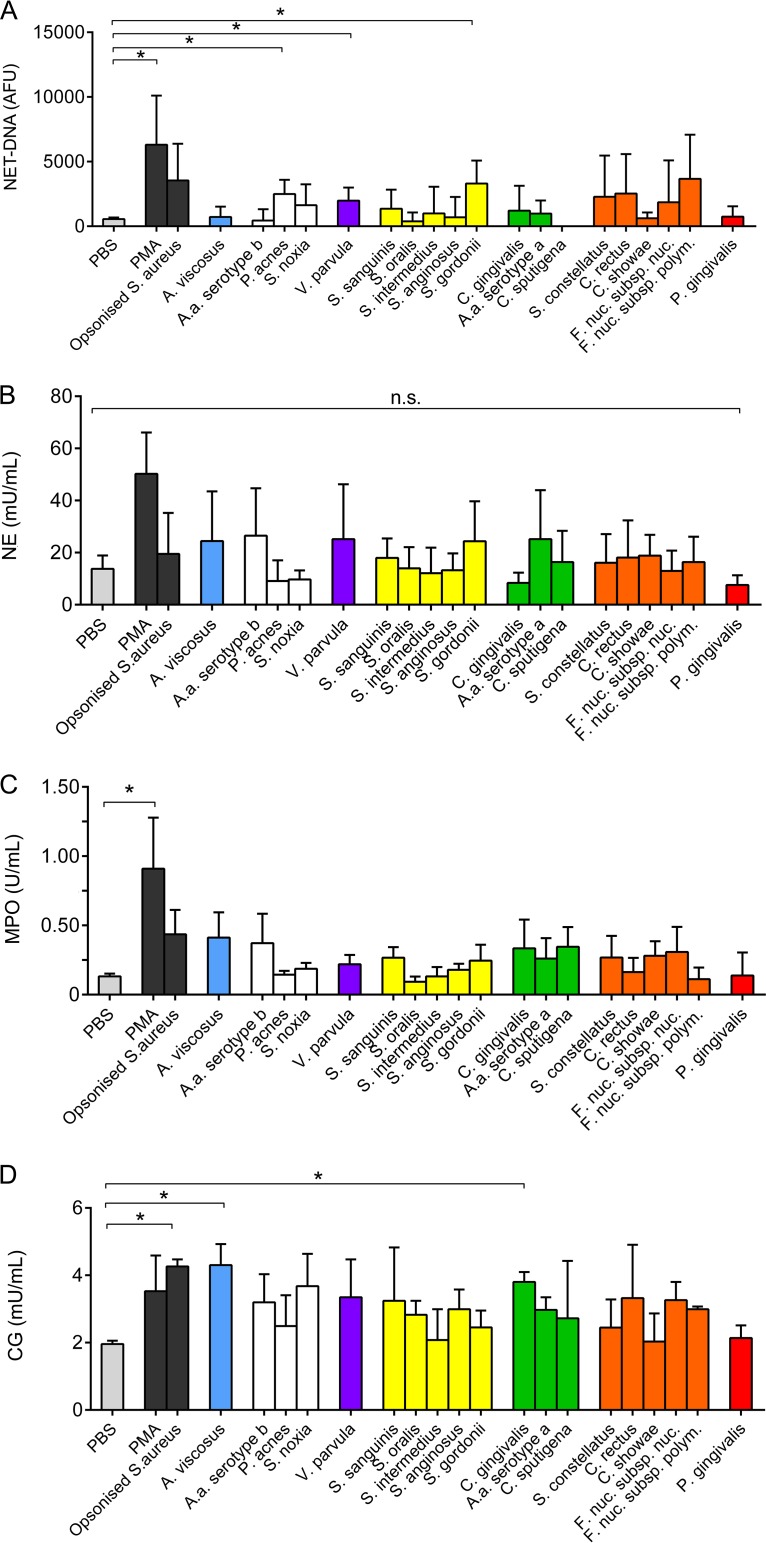
NET release in response to the bacterial challenge was quantified. Some bacteria led to enhanced NET DNA production, which was statistically significant for P. acnes, Veillonella parvula, and S. gordonii compared with the PBS control (Fig. 2A). NET-bound NE, MPO, and CG were quantified colorimetrically, and the data demonstrated that certain periodontal bacteria elicited increased levels of production of NET-bound proteins relative to the amounts elicited by PBS (Fig. 2B to D). Similarly, stimulation with PMA and opsonized S. aureus (positive controls) induced statistically significant elevations in MPO and CG expression (Fig. 2C and D).
FIG 2.
Quantification of neutrophil extracellular trap (NET) production in response to periodontal bacteria. NET production in response to periodontal bacteria and to PBS (unstimulated negative control), phorbol 12-myristate 13-acetate (PMA; 50 nM; positive control), and opsonized S. aureus (positive control) was quantified. NET DNA was quantified using a Sytox green assay (A), and NET-bound neutrophil elastase (B), myeloperoxidase (C), and cathepsin G (D) were quantified colorimetrically. Data are presented as arbitrary fluorescence units (AFU), units per milliliter, or milliunits per milliliter and represent the results for neutrophils from 10 different donors assessed in triplicate wells. *, P < 0.05; n.s., not significant.
NET entrapment of bacteria does not associate with Socransky complexes or with bacterial cell death.
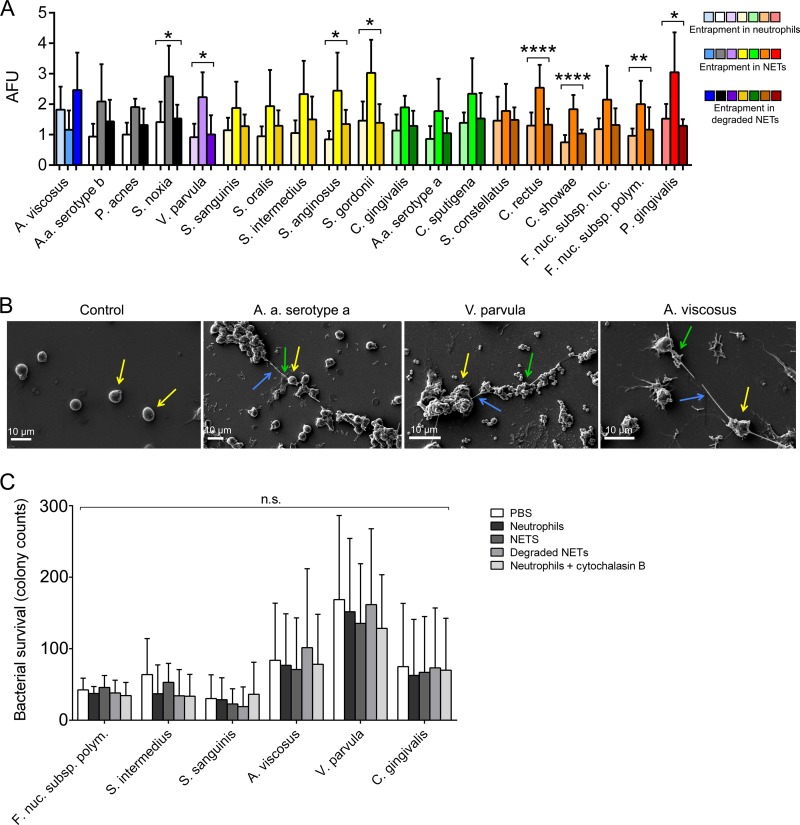
For clinical relevance, data are presented by grouping periodontal bacteria according to the Socransky complexes (4) (Fig. 3A). The members of the non-Socransky complex, consisting of Selenomonas noxia and V. parvula, were found to be entrapped within NET structures in higher numbers than the negative controls (unstimulated neutrophils or degraded NETs) However, Aggregatibacter actinomycetemcomitans (serotype b), P. acnes, and Actinomyces viscosus were significantly associated with NET entrapment. The yellow complex members S. anginosus and S. gordonii were significantly entrapped within NETs. However, the other yellow complex bacteria assayed, Streptococcus sanguinis, Streptococcus oralis, and Streptococcus intermedius, were not found within NET structures. None of the green complex bacteria assayed appeared within NETs at a significant level. Orange complex members C. rectus, Campylobacter showae, and F. nucleatum subsp. polymorphum were significantly entrapped within NETs relative to the negative controls, whereas Streptococcus constellatus and F. nucleatum subsp. nucleatum were not. The red complex member P. gingivalis was more significantly associated with NET structures than with unstimulated neutrophils or degraded NET structures. Scanning electron microscopy images of unstimulated neutrophils demonstrated spherical cells with no NET structures evident, whereas neutrophils incubated with A. actinomycetemcomitans serotype a, V. parvula, and A. viscosus revealed the release of NET structures (Fig. 3B). The strand-like filaments between the neutrophils appeared to associate with bacteria; for example, A. actinomycetemcomitans (serotype a) clustered along NET structures. The bacterial killing assays employed to detect the microbicidal properties of NETs revealed that the viability of the 6 periodontal bacteria tested was unaffected by NET trapping (Fig. 3C).
FIG 3.
Neutrophil extracellular trap (NET) entrapment of periodontal bacteria. (A) NET entrapment of bacteria that were not assigned to a Socransky complex (white, gray, black), as well as purple, yellow, green, orange, red, and blue complex bacteria. Results are normalized to those for fluorescein isothiocyanate-stained bacteria in PBS. The statistical significance of bacterial entrapment by NETs relative to that of bacterial entrapment by unstimulated neutrophils and degraded NETs is shown. Data are presented as arbitrary fluorescence units (AFU). (B) Representative images of bacterial entrapment by NETs. Neutrophils (yellow arrows) incubated with PBS (control), live A. actinomycetemcomitans serotype a, V. parvula, or A. viscosus were visualized by scanning electron microscopy. Blue arrows, NET strand structures; green arrows, NET-associated bacteria. Representative images from three experiments are shown. (C) Bacterial survival after exposure to neutrophils, NETs, degraded NETs, and neutrophils with cytochalasin B. All results shown represent those for neutrophils from five different donors assessed in triplicate wells. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; n.s., not significant.
Effect of NADPH oxidase pathway-modulating agents on ROS and NET production.
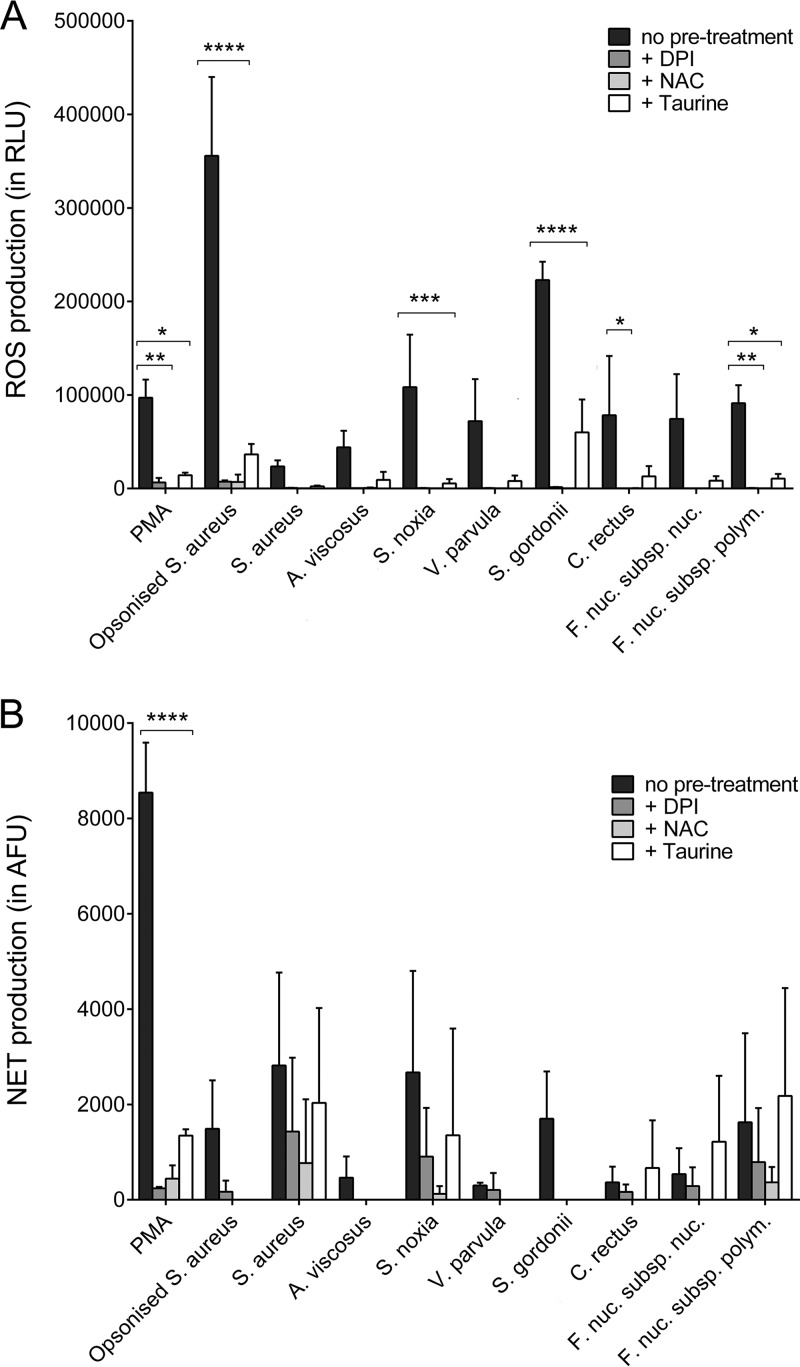
Components of the NADPH oxidase signaling pathway were targeted in order to assess whether NADPH oxidase is essential for neutrophil ROS and NET production in response to periodontal bacteria. The data showed that diphenyleneiodonium (DPI; an NADPH oxidase inhibitor), N-acetylcysteine (NAC; an H2O2 scavenger), and taurine (HOCl scavenger) treatment resulted in a reduction in total ROS release in response to all stimuli. This was statistically significant for PMA, opsonized S. aureus, S. gordonii, C. rectus, F. nucleatum subsp. polymorphum, and S. noxia (Fig. 4A). NET production was not significantly affected by these inhibitors, with the exception of PMA; however, a moderate reduction of NET release was visible in all samples (Fig. 4B).
FIG 4.
Effect of NADPH oxidase pathway-modulating agents on reactive oxygen species (ROS) and neutrophil extracellular trap (NET) production. Total ROS (A) and NET (B) production by neutrophils in response to selected periodontal bacteria, as well as to phorbol 12-myristate 13-acetate (PMA; 50 nM) and opsonized S. aureus (positive controls), was quantified following preincubation (30 min) with diphenyleneiodonium (DPI; 25 μM), N-acetylcysteine (NAC; 10 mM), and taurine (100 mM). Data are presented as relative light units (RLU) and arbitrary fluorescence units (AFU). Experiments were conducted in duplicate using neutrophils from three different donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Effect of TLR inhibition on ROS and NET production.
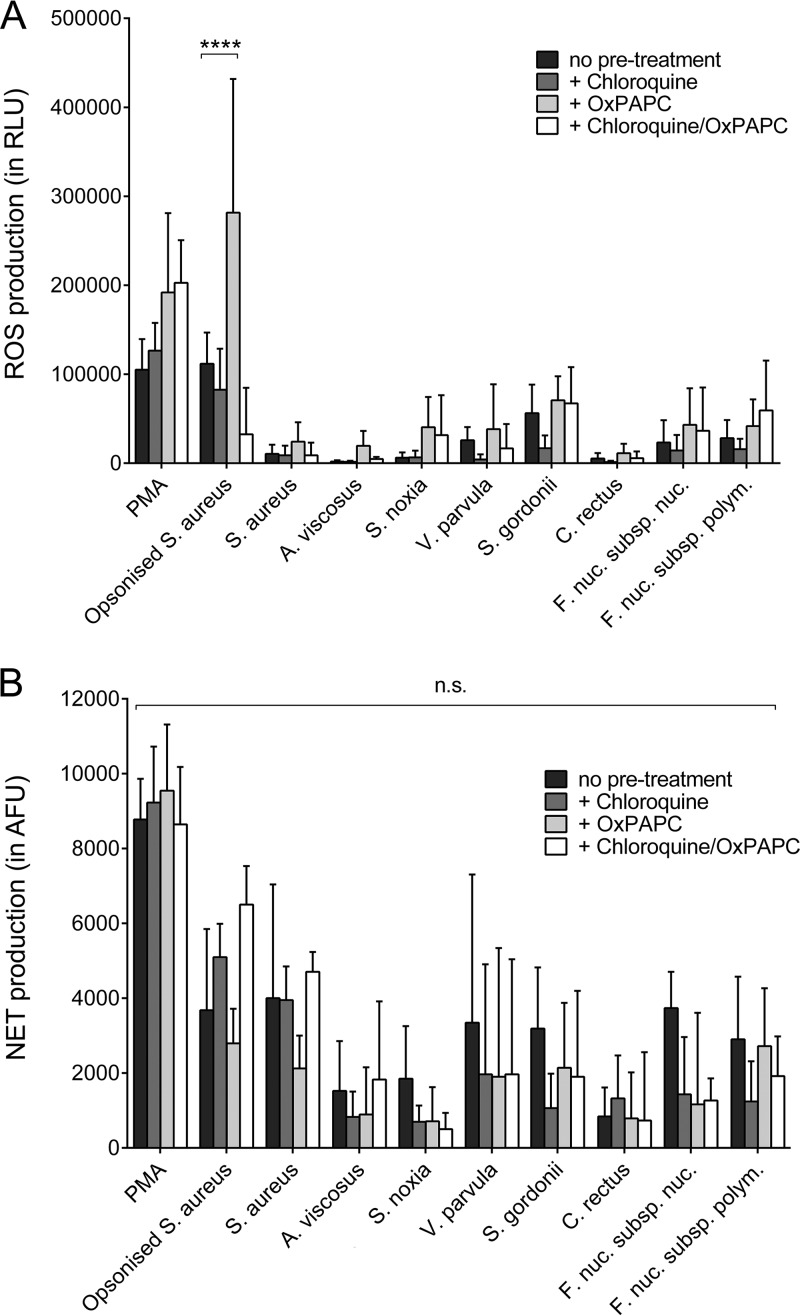
The role of Toll-like receptor (TLR) signaling in neutrophil ROS and NET responses to periodontal bacteria was investigated by using specific inhibitors. Chloroquine (a TLR3, TLR7, and TLR9 inhibitor) or oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (OxPAPC; a TLR2 and TLR4 inhibitor) treatment as well as treatment with both components did not reduce the level of ROS production by neutrophils. However, a significant increase in ROS release from neutrophils treated with OxPAPC and opsonized S. aureus was seen (Fig. 5A). Similarly, NET release in response to bacterial stimulation was not affected by the TLR inhibitors (Fig. 5B).
FIG 5.
Effect of Toll-like receptor inhibition on reactive oxygen species (ROS) and neutrophil extracellular trap (NET) production. Total ROS (A) and NET (B) production by neutrophils in response to selected periodontal bacteria, as well as to phorbol 12-myristate 13-acetate (PMA; 50 nM) and opsonized S. aureus (positive controls), was quantified following preincubation (30 min) with chloroquine (100 μM), oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (OxPAPC; 30 μg/ml), or chloroquine and OxPAPC. Experiments were conducted in duplicate wells using neutrophils from three different donors. ****, P < 0.0001; n.s., not significant.
DISCUSSION
Neutrophil ROS production is a vital component of the innate immune response, which enables killing and clearance of pathogens. Neutrophils are the predominant immune cell in periodontitis (18), and the results presented here support the suggestion that their stimulation with periodontal bacteria promotes extracellular, intracellular, and superoxide ROS release; however, data indicate that this may be species specific. Indeed, some species consistently elicited higher levels of neutrophil ROS production, while other bacteria, such as P. gingivalis or S. sanguinis, were not found to significantly promote ROS release. Bacteria like P. gingivalis, F. nucleatum, and oral streptococci can scavenge neutrophil-derived ROS, which is attributed to a range of oxidative stress response genes encoding proteins like rubrerythrin, glutathione peroxidase, glutaredoxin, NADH oxidase, and superoxide dismutase (19–23). It is possible that these bacterial defense mechanisms may function to afford protection to other biofilm organisms that are less resistant to ROS.
Periodontitis is known to arise from an exaggerated inflammatory response to microbial plaque (6). While it is recognized that ROS facilitate microbial killing, ROS do not discriminate between pathogens and host tissues, and therefore, tissue injury can arise from excess plaque-induced extracellular ROS release. ROS are reported to contribute to periodontitis progression by direct and indirect mechanisms, including tissue damage (24, 25), lipid peroxidation (26), DNA strand breakage (27), increased osteoclast differentiation (28), and initiation of a self-perpetuating cycle that activates chronic immune cell-derived ROS production (29). Notably, Matthews et al. showed increased ROS production by peripheral blood neutrophils in chronic periodontitis (9, 10). In patients susceptible to the deleterious effects of ROS, a discordance between oxidant and antioxidant levels may also play a role. This is supported by the work of Chapple et al., who demonstrated that total antioxidant activity is lower in the saliva of periodontitis patients (30). It has also been reported that neutrophil chemotaxis is compromised in patients with chronic periodontitis and that these patients' neutrophils produce the chemoattractant interleukin-8 in excess when stimulated, potentially creating distracted chemotaxis (31). Such processes may increase neutrophil tissue transit times and thereby potentially exacerbate ROS-mediated collateral tissue damage (18).
Quantification of NET DNA and NET-bound antimicrobial proteins demonstrated differential NET production in response to the periodontal bacteria tested. DNA is released during other forms of neutrophil cell death, such as necrosis, and the quantification of NET-bound components (NE, MPO, and CG) therefore provides a DNA-independent measure of NETs. It is noteworthy that differences between individuals have been reported, such as neutrophil responsiveness to stimuli, which can also affect NET quantification results, regardless of the analytical method employed (32, 33). Significant NET production in response to individual periodontal bacteria, however, indicates that these events likely occur in vivo. Notably, NETs have previously been shown to exist in purulent exudate from periodontal pockets, where they are postulated to entrap invading microbes and prevent their dissemination (34, 35). Recently, many periodontal bacterial species have been shown to release DNases, which, in addition to regulating biofilm formation (36), can potentially disassemble NET structures to enable NET evasion (37). Thus, bacterial DNase expression may explain why some periodontal species, such as S. constellatus, which reportedly releases large quantities of DNase, showed less entrapment (37).
Following bacterial entrapment, the high local concentration of antimicrobial proteins associated with NETs is thought to disable and kill pathogens (38). In the present study, the incubation of NETs with periodontal bacteria did not impede bacterial growth or survival, a finding in accordance with data reported by Menegazzi et al. (39). Cytochalasin B was applied in our study to exclude the possibility of bacterial killing through phagocytosis, and inhibition by cytochalasin B occurs via blocking of actin polymerization (40). As functional actin filaments may play a role in NET formation (41), it is possible that cytochalasin B interfered with NET and antimicrobial protein release in our study and thus prevented bacterial killing. However, other known inhibitors of phagocytosis and endocytosis, such as latrunculin A or CK666, also exert their effects by disturbing actin polymerization (42, 43). Future experiments may be directed at investigating differences among such inhibitors regarding their interference with NET release.
Treatment of neutrophils with the NADPH oxidase inhibitor DPI, the glutathione peroxidase precursor substrate NAC, and the HOCl scavenger taurine abrogated total ROS release, a finding that is in accordance with data previously reported (44–46). NET release was inhibited only marginally in response to the bacterial challenge. At the same time, NET production was significantly affected by the inhibitors in neutrophils stimulated with PMA. This may be explained by the fact that PMA induces NETs via protein kinase C (PKC), which then activates the NADPH oxidase and thus can elicit NETs only via the generation of ROS (44). These findings indicate that NADPH oxidase-independent NET formation may play a role in host defense against periodontal bacteria (47, 48).
Further experiments aimed to establish the role of TLR activation in NET production. Pretreatment of neutrophils with the intracellular TLR3, TLR7, TLR8, and TLR9 inhibitor chloroquine and with the TLR2 and TLR4 inhibitor OxPAPC, separately or combined, did not lead to significant reductions in ROS or NET release. Previous findings have suggested that ROS release is both TLR2 and TLR4 dependent (49); however, Gould et al. recently demonstrated that the blocking of TLR2 and TLR4 did not abolish NET release (50), a finding that is in line with the findings of this study. Notably, neutrophils are not responsive to TLR3 ligands (51); therefore, the involvement of TLR7, TLR8, and TLR9 was investigated by using chloroquine. Similar to our results, Salmon et al. found that chloroquine had no effect on oxidative metabolism in neutrophils (52). Thus, a lack of inhibition of ROS and NET generation by chloroquine and OxPAPC indicates that other signaling pathways may have played a role in this study. For example, C-type lectin receptors and NOD-like receptors can be activated by bacterial triggers, and both have been reported to induce immune activation in neutrophils, including the release of ROS (53, 54). Moreover, signaling via TLR coreceptors may have bypassed the inhibited pathways (55). Importantly, although it is widely used as a TLR inhibitor, chloroquine is thought to directly interfere with multiple physiological cell functions, including chemotaxis, phagocytosis, and ROS release, by alkalizing lysosomes and phagolysosomes (56, 57). Our results do not support an inhibitory effect of chloroquine on these functions at the concentrations applied in the present study, as no significant differences between neutrophils treated with chloroquine and negative controls were seen. However, results from functional cell assays using chloroquine as an inhibitor should be interpreted with care.
Future experiments should target these receptors to further elucidate their specific role in ROS and NET release. Interestingly, in OxPAPC-treated neutrophils stimulated with opsonized S. aureus, a significant increase in the amount of ROS was seen. Previous studies reported that OxPAPC has the potential to increase ROS release in endothelial cells via activating the NADPH oxidase (58, 59). Moreover, Fc gamma receptor (FcγR) signaling is known to trigger ROS release (60). It is therefore possible that OxPAPC may act as a cotrigger of FcγR-mediated ROS release in neutrophils challenged with opsonized bacteria; however, further experiments are required to confirm this hypothesis.
As a limitation of this study, planktonic single-species preparations were used to stimulate neutrophils. In vivo, however, neutrophils are challenged by multispecies biofilms. These biofilms produce metabolites and extracellular matrix components that may lead to a response pattern different from that observed under our experimental conditions. The variability in these extracellular products generated by naturally or artificially grown biofilms is high, and the reproduction of experiments involving such biofilms is difficult (13, 61). Moreover, natural dental biofilms are highly variable in their composition, and it is difficult to attribute their activation of neutrophils to certain species or biofilm components. Therefore, little is known about the interactions between host cells and mixed-species biofilms. Further efforts aimed at creating a reproducible neutrophil-biofilm interaction model in vitro are currently being carried out by our group. Nevertheless, in order to understand the interaction of neutrophils with oral bacterial species, such microorganisms playing key roles in neutrophil activation need to be identified and investigated separately. Insights from these experiments may subsequently allow a better understanding of neutrophil responses to oral biofilms.
As a further limitation, heat-killed microorganisms were employed in our study. Although heat killing may lead to the denaturation of surface antigens and pathogen-associated molecular patterns (PAMPs), this is thought to be reversible at temperatures below 80°C (62). Moreover, previous studies using live bacteria (A. actinomycetemcomitans serotype b or S. gordonii, F. nucleatum subsp. polymorphum, and V. parvula) showed similar NET formation outcomes regarding arbitrary fluorescence unit (AFU) measurements or relative differences in NET production, respectively (14, 63). Another restriction in our study was the limited number of different bacterial species used to investigate neutrophil activation. Future studies may need to include further species, particularly of the red complex, such as Treponema denticola and Tannerella forsythia.
In the present study, neutrophils from periodontally and systemically healthy donors were used. Overall, these neutrophils were responsive to some health-associated species and opportunistic pathogens rather than disease-associated species. In contrast, our previous investigations of periodontally diseased patients have shown that their neutrophils are hyperactive and hyperreactive toward F. nucleatum and P. gingivalis in terms of ROS and cytokine release. On the other hand, NET production in response to various stimuli was not altered and was similar in periodontitis patients and nonperiodontitis control subjects. However, these studies did not compare the effects of health- and disease-associated bacteria on neutrophils (9, 31, 64, 65). It is possible that neutrophils from periodontitis patients may show a higher reactivity toward periodontal bacteria than those from healthy subjects, as these neutrophils may be primed in the circulation by bacterial components, such as lipopolysaccharide, accessing the bloodstream through periodontal microlesions (7, 66). Further studies examining the responses of neutrophils from healthy and periodontally diseased individuals to different oral bacteria may shed light on possible mechanisms of immune tolerance in health and disease.
In summary, the data presented here demonstrate variability between periodontal bacteria in their ability to stimulate neutrophil ROS production and NET responses. This may contribute to the pathogenesis of periodontitis by mechanisms such as bacterial avoidance of host defense mechanisms and, thus, the persistence of infection or excess ROS release with associated tissue damage. Moreover, our results indicate that innate immune receptors other than the TLRs investigated here may be involved in bacterium-triggered ROS and NET release and that NADPH oxidase-independent NET formation may occur in response to periodontal pathogens. Comprehensive studies are required to fully elucidate the role of NETs and ROS in periodontitis, in particular with regard to the receptors, activation pathways, and intracellular responses triggered by different bacteria. Also, investigation of the possible activation of protective mechanisms, such as glutathione upregulation, or of the anti-inflammatory signaling routes by these bacteria may improve our understanding of their differential effects seen in this study.
MATERIALS AND METHODS
Neutrophil isolation.
Neutrophils were isolated from the peripheral venous blood of periodontally and systemically healthy volunteers (University of Birmingham ethics reference ERN_13-0325) using discontinuous Percoll gradients (GE Healthcare, Amersham, UK) as previously described (67). The medical history was taken from each donor, and periodontal examinations were conducted to ensure periodontal and systemic health. Cell viability and purity were confirmed by trypan blue exclusion and flow cytometry, respectively, and these were typically >98%.
Bacterial culture.
A panel of 19 periodontal bacteria and opsonized S. aureus were employed to stimulate neutrophils. Bacterial stocks were originally obtained from the Forsyth Institute (Boston, MA, USA) or purchased from the American Type Culture Collection (ATCC). Blood agar plates (base no. 2 with 7% horse blood) were purchased from Oxoid (Basingstoke, UK) and used for growing most bacterial strains. P. gingivalis (strain W83) was cultured on anaerobic 20% blood agar plates (Wilkins-Chalgren; Oxoid), and S. aureus was cultured on tryptone soy agar (TSA) plates. Trypticase soy broth (TSB; Oxoid), brain heart infusion (BHI) broth (Oxoid), or fastidious anaerobe broth (Lab M, Heywood, UK) was used for planktonic growth of the microorganisms. The optical density at 600 nm of the bacterial cell suspensions was measured spectrophotometrically to estimate bacterial numbers, and the bacteria were heat killed at 80°C for 30 min. Bacterial cells were washed with PBS and centrifuged, and the pellet was resuspended to produce a stock solution, which was stored at −20°C prior to use. The bacteria used, their growth conditions, and their assignments to Socransky complexes are listed in Table 1.
Opsonization of S. aureus.
S. aureus was grown planktonically in TSB. Following 48 h of aerobic growth, the bacteria were washed and pelleted by centrifugation for 15 min at a relative centrifugal force of 1,800 and 4°C. Bacteria were opsonized with Vigam liquid (5 mg/ml IgG; Bio Products Laboratory, Borehamwood, UK). This mixture was agitated overnight at room temperature and, after it was washed, stored at −20°C until needed.
Stimuli employed to activate neutrophils.
Neutrophils were stimulated using a range of stimuli. PMA targets NET production via the activation of protein kinase C (PKC). Our previous findings demonstrated that the concentration of PMA required for NET release is 50 nM, whereas 25 nM is sufficient to stimulate ROS production (44). Both Gram-positive and Gram-negative bacteria were used to activate neutrophils via TLR2 and TLR4 in ROS and NET assays. In addition, ROS and NETs were produced in response to stimulation with opsonized S. aureus, which activates neutrophils via the FcγR. For some supplementary assays, a smaller panel of bacteria was employed, where the selection was made on the basis of the variable characteristics of the microorganisms: Gram-positive, Gram-negative, aerobic, anaerobic, and microaerophilic bacteria from different complexes with FcγR and TLR activation properties were chosen, and both health- and disease-associated bacteria were represented. Moreover, variable DNase production was a selection criterion, as DNases have the ability to disassemble NETs (e.g., F. nucleatum and S. aureus are known to produce DNase, whereas A. viscosus and V. parvula produce little or no DNase) (37). In addition, the following bacteria reported to interfere with ROS scavenging were included: F. nucleatum and S. noxia, which can metabolize the antioxidants glutathione and l-cysteine (68), as well as A. viscosus, which produces the ROS scavenger catalase (69).
Quantification of NET production in response to periodontal bacteria.
NET release was determined using a NET quantification assay previously described (17). Neutrophils were stimulated with the positive controls PMA (50 nM) and opsonized S. aureus as well as with 19 heat-killed periodontal bacteria (multiplicity of infection [MOI], 1,000 [70]) after being equilibrated for a 30-min baseline period. Unstimulated (PBS-treated) neutrophils were employed as negative controls. Neutrophils from 10 periodontally and systemically healthy donors were used to perform NET quantification in triplicate wells per donor.
Chemiluminescence protocol for ROS assay.
ROS production in response to the periodontal bacteria (MOI, 1,000) was determined using enhanced chemiluminescence. Neutrophils (1 × 105) from five different donors were added to a 96-well plate (using triplicate wells per donor) precoated with 1% bovine serum albumin (BSA). ROS release following exposure to PBS (unstimulated negative control), PMA (25 nM; positive control), and opsonized S. aureus (MOI, 500; positive control) was quantified. Neutrophils were stimulated after being equilibrated for a 30-min baseline period, and then the amount of ROS was measured over the subsequent 100 min. To measure the amounts of total ROS, extracellular ROS, and superoxide, luminol (3 mM), isoluminol (3 mM) with 1.5 units of horseradish peroxidase (HRP), and lucigenin (0.25 mg/ml), respectively, were added to the samples and the light output was read for 130 min in a luminometer (Berthold Tristar2; Berthold Technologies, Harpenden, UK). All readings were expressed as relative light units (RLUs) and obtained at 37°C (MikroWin2000; Informer Technologies, Madrid, Spain). All reagents for chemiluminescence were purchased from Sigma-Aldrich (Dorset, UK).
NET entrapment and quantification of NET-mediated killing of periodontal bacteria.
To assess the ability of NETs to immobilize periodontal bacteria, fluorescein isothiocyanate (FITC)-stained live bacteria (MOI, 100) were incubated for 1 h with unstimulated neutrophils, intact NETs (produced by prior 0.75 mM HOCl stimulation with a subsequent washing step [44]), or NET structures degraded with micrococcal nuclease (MNase; New England BioLabs, Hitchin, UK) in a 96-well plate precoated with 1% BSA, using neutrophils from five different donors and triplicate wells for the neutrophils from each donor. In vitro NETs formed within 2 to 3 h; therefore, the possibility of a relevant induction of NET release from otherwise unstimulated neutrophils by bacteria could be excluded. Following multiple wash steps to remove any unbound bacteria, the amount of bacteria entrapped was fluorometrically quantified and normalized to the amount of FITC-stained bacteria incubated with PBS (cell-free control). To determine whether NETs are capable of killing entrapped bacteria, six strains (F. nucleatum subsp. polymorphum, S. intermedius, S. sanguinis, A. viscosus, V. parvula, and C. gingivalis) were incubated at an MOI of 100 with PBS (negative control) or unstimulated neutrophils, intact NETs, or degraded NETs from five different donors in triplicate wells per donor. Additionally, samples containing neutrophils were treated with the phagocytosis inhibitor cytochalasin B (Sigma-Aldrich, Harpenden, UK) at a concentration of 10 μg/ml (71). Following 1 h of incubation, bacteria were released from NETs by MNase digestion, diluted, and inoculated onto agar plates and cultured for 24 h prior to performing colony counts.
Effect of NADPH oxidase pathway-modulating agents on ROS and NET production.
To further understand the importance of NADPH oxidase and downstream products in bacterium-induced ROS and NET production, specific components of the NADPH oxidase signaling pathway were targeted. Neutrophils isolated from three different donors were incubated with DPI (25 μM), an inhibitor of NADPH oxidase; NAC (10 mM), a synthetic glutathione precursor that scavenges H2O2; or taurine (100 mM), which scavenges HOCl to produce taurine chloramine (duplicate wells per donor). Neutrophil total ROS and NET production was measured following preincubation with the modulating agent for 30 min prior to stimulation with PMA (50 nM), opsonized S. aureus (MOI, 500), and 8 selected bacteria (MOI, 1,000; S. aureus, V. parvula, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, S. gordonii, C. rectus, A. viscosus, and S. noxia). NET DNA was quantified with Sytox green following enzymatic degradation of NET structures with MNase. All reagents were purchased from Sigma-Aldrich (Dorset, UK).
Effect of TLR inhibition on ROS and NET production.
To better understand the signaling involved in ROS and NET activation, the effect of TLR inhibitors was investigated. Neutrophils isolated from three different donors were incubated in duplicate wells per donor with chloroquine (100 μM; InvivoGen, Toulouse, France), an intracellular inhibitor of endosomal TLR3, TLR7, TLR8, and TLR9, or OxPAPC (30 μg/ml; InvivoGen, Toulouse, France), which inhibits intracellular signaling of activated TLR2 and TLR4, or both TLR inhibitors were used simultaneously. Neutrophil total ROS and NET production was measured following preincubation with the inhibitor for 30 min prior to stimulation with PMA (50 nM), opsonized S. aureus (MOI, 500), and 8 selected bacteria (MOI, 1,000; S. aureus, V. parvula, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, S. gordonii, C. rectus, A. viscosus, and S. noxia).
Statistical analysis.
All statistical analyses were performed in the GraphPad Prism (version 5) software package for Windows (San Diego, CA, USA). The distribution of data and, thus, whether the data were considered parametric or nonparametric were determined by Kolmogorov-Smirnov tests. Statistical tests employed for the purpose of this study were at a significance of 0.05. The specific level of significance is indicated in the figure legends. Kruskal-Wallis and Dunn's multiple-comparison tests were performed for quantification of ROS and NET release. One-way analysis of variance (ANOVA) and Dunnett's post hoc tests were employed for NET entrapment assays. Two-way ANOVA and Bonferroni post hoc tests were applied to calculate the significance of pathway modulation and inhibition assays. All quantitative data are shown as mean values ± standard deviations, and all statistical tests were performed by comparison of the results for different donors.
ACKNOWLEDGMENTS
We thank Helen Wright and Naomi Hubber for their technical support. We are also grateful to Iru Dias, who helped with the flow cytometry.
This research was funded by the University of Birmingham Dental School, the Medical Research Council (MR/J500434/1), the British Society of Periodontology, and the Oral & Dental Research Trust (14945).
We declare that there is no conflict of interest regarding the publication of this paper.
REFERENCES
- 1.Paster BJ, Dewhirst FE. 2009. Molecular microbial diagnosis. Periodontol 2000 51:38–44. doi: 10.1111/j.1600-0757.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, Tonetti MS, Wade WG, Zaura E. 2016. The oral microbiome—an update for oral healthcare professionals. Br Dent J 221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 3.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 5.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol 35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott DA, Krauss J. 2012. Neutrophils in periodontal inflammation. Front Oral Biol 15:56–83. doi: 10.1159/000329672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajishengallis G, Lamont RJ. 2014. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol 44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews JB, Wright HJ, Roberts A, Cooper PR, Chapple IL. 2007. Hyperactivity and reactivity of peripheral blood neutrophils in chronic periodontitis. Clin Exp Immunol 147:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews JB, Wright HJ, Roberts A, Ling-Mountford N, Cooper PR, Chapple IL. 2007. Neutrophil hyper-responsiveness in periodontitis. J Dent Res 86:718–722. doi: 10.1177/154405910708600806. [DOI] [PubMed] [Google Scholar]
- 11.Shapira L, Borinski R, Sela MN, Soskolne A. 1991. Superoxide formation and chemiluminescence of peripheral polymorphonuclear leukocytes in rapidly progressive periodontitis patients. J Clin Periodontol 18:44–48. doi: 10.1111/j.1600-051X.1991.tb01118.x. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. 2007. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirschfeld J. 2014. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol 6:26102. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschfeld J, Dommisch H, Skora P, Horvath G, Latz E, Hoerauf A, Waller T, Kawai T, Jepsen S, Deschner J, Bekeredjian-Ding I. 2015. Neutrophil extracellular trap formation in supragingival biofilms. Int J Med Microbiol 305:453–463. doi: 10.1016/j.ijmm.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Palmer LJ, Damgaard C, Holmstrup P, Nielsen CH. 2016. Influence of complement on neutrophil extracellular trap release induced by bacteria. J Periodont Res 51:70–76. doi: 10.1111/jre.12284. [DOI] [PubMed] [Google Scholar]
- 16.Guentsch A, Puklo M, Preshaw PM, Glockmann E, Pfister W, Potempa J, Eick S. 2009. Neutrophils in chronic and aggressive periodontitis in interaction with Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. J Periodont Res 44:368–377. doi: 10.1111/j.1600-0765.2008.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White PC. 2016. The role of neutrophil extracellular traps in the pathogenesis of periodontal diseases. PhD dissertation University of Birmingham, Birmingham, United Kingdom. [Google Scholar]
- 18.Roberts HM, Ling MR, Insall R, Kalna G, Spengler J, Grant MM, Chapple IL. 2015. Impaired neutrophil directional chemotactic accuracy in chronic periodontitis patients. J Clin Periodontol 42:1–11. doi: 10.1111/jcpe.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC III, Kurtz DM Jr, Travis J, Collins LV, Nguyen KA, Genco CA, Potempa J. 2006. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog 2:e76. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S-N, Cho E, Kim H-S, Kim D-S, Jung J, Baek J-H, Kyong Lim Y, Jo E, Chang Y-H, Hwan Shin J, Choi S-H, Kang J, Choi Y, Park H-S, Kim H, Kook J-K. 2013. Draft genome sequence of Fusobacterium nucleatum subsp. nucleatum ChDC F316, isolated from a human peri-implantitis lesion in the Republic of Korea. Genome Announc 1:e01041-13. doi: 10.1128/genomeA.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch MC, Kuramitsu HK. 1999. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun 67:3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen PT, Abranches J, Phan TN, Marquis RE. 2002. Repressed respiration of oral streptococci grown in biofilms. Curr Microbiol 44:262–266. doi: 10.1007/s00284-001-0001-0. [DOI] [PubMed] [Google Scholar]
- 23.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol 36:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Sharma S. 2011. Reactive oxygen species and antioxidants in periodontics: a review. Int J Dent Clin 3:44–47. [Google Scholar]
- 25.Chapple IL. 1996. Role of free radicals and antioxidants in the pathogenesis of the inflammatory periodontal diseases. Clin Mol Pathol 49:M247–M255. doi: 10.1136/mp.49.5.M247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dix TA, Aikens J. 1993. Mechanisms and biological relevance of lipid peroxidation initiation. Chem Res Toxicol 6:2–18. doi: 10.1021/tx00031a001. [DOI] [PubMed] [Google Scholar]
- 27.Dix TA, Hess KM, Medina MA, Sullivan RW, Tilly SL, Webb TL. 1996. Mechanism of site-selective DNA nicking by the hydrodioxyl (perhydroxyl) radical. Biochemistry 35:4578–4583. doi: 10.1021/bi952010w. [DOI] [PubMed] [Google Scholar]
- 28.Waddington RJ, Moseley R, Embery G. 2000. Reactive oxygen species: a potential role in the pathogenesis of periodontal diseases. Oral Dis 6:138–151. [DOI] [PubMed] [Google Scholar]
- 29.Dahiya P, Kamal R, Gupta R, Bhardwaj R, Chaudhary K, Kaur S. 2013. Reactive oxygen species in periodontitis. J Indian Soc Periodontol 17:411–416. doi: 10.4103/0972-124X.118306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapple IL, Brock GR, Milward MR, Ling N, Matthews JB. 2007. Compromised GCF total antioxidant capacity in periodontitis: cause or effect? J Clin Periodontol 34:103–110. [DOI] [PubMed] [Google Scholar]
- 31.Ling MR, Chapple IL, Matthews JB. 2015. Peripheral blood neutrophil cytokine hyper-reactivity in chronic periodontitis. Innate Immun 21:714–725. doi: 10.1177/1753425915589387. [DOI] [PubMed] [Google Scholar]
- 32.Barrientos L, Marin-Esteban V, de Chaisemartin L, Le-Moal VL, Sandre C, Bianchini E, Nicolas V, Pallardy M, Chollet-Martin S. 2013. An improved strategy to recover large fragments of functional human neutrophil extracellular traps. Front Immunol 4:166. doi: 10.3389/fimmu.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halverson TW, Wilton M, Poon KK, Petri B, Lewenza S. 2015. DNA is an antimicrobial component of neutrophil extracellular traps. PLoS Pathog 11:e1004593. doi: 10.1371/journal.ppat.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitkov L, Klappacher M, Hannig M, Krautgartner WD. 2009. Extracellular neutrophil traps in periodontitis. J Periodont Res 44:664–672. doi: 10.1111/j.1600-0765.2008.01175.x. [DOI] [PubMed] [Google Scholar]
- 35.Vitkov L, Klappacher M, Hannig M, Krautgartner WD. 2010. Neutrophil fate in gingival crevicular fluid. Ultrastruct Pathol 34:25–30. doi: 10.3109/01913120903419989. [DOI] [PubMed] [Google Scholar]
- 36.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 37.Palmer LJ, Chapple IL, Wright HJ, Roberts A, Cooper PR. 2012. Extracellular deoxyribonuclease production by periodontal bacteria. J Periodont Res 47:439–445. doi: 10.1111/j.1600-0765.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 38.Brinkmann V, Zychlinsky A. 2012. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol 198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menegazzi R, Decleva E, Dri P. 2012. Killing by neutrophil extracellular traps: fact or folklore? Blood 119:1214–1216. doi: 10.1182/blood-2011-07-364604. [DOI] [PubMed] [Google Scholar]
- 40.MacLean-Fletcher S, Pollard TD. 1980. Mechanism of action of cytochalasin B on actin. Cell 20:329–341. doi: 10.1016/0092-8674(80)90619-4. [DOI] [PubMed] [Google Scholar]
- 41.Neeli I, Dwivedi N, Khan S, Radic M. 2009. Regulation of extracellular chromatin release from neutrophils. J Innate Immun 1:194–201. doi: 10.1159/000206974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumida GM, Yamada S. 2015. Rho GTPases and the downstream effectors actin-related protein 2/3 (Arp2/3) complex and myosin II induce membrane fusion at self-contacts. J Biol Chem 290:3238–3247. doi: 10.1074/jbc.M114.612168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Oliveira CA, Mantovani B. 1988. Latrunculin A is a potent inhibitor of phagocytosis by macrophages. Life Sci 43:1825–1830. doi: 10.1016/0024-3205(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 44.Palmer LJ, Cooper PR, Ling MR, Wright HJ, Huissoon A, Chapple IL. 2012. Hypochlorous acid regulates neutrophil extracellular trap release in humans. Clin Exp Immunol 167:261–268. doi: 10.1111/j.1365-2249.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchner T, Hermann E, Möller S, Klinger M, Solbach W, Laskay T, Behnen M. 2013. Flavonoids and 5-aminosalicylic acid inhibit the formation of neutrophil extracellular traps. Mediators Inflamm 2013:710239. doi: 10.1155/2013/710239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirchner T, Möller S, Klinger M, Solbach W, Laskay T, Behnen M. 2012. The impact of various reactive oxygen species on the formation of neutrophil extracellular traps. Mediators Inflamm 2012:849136. doi: 10.1155/2012/849136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uriarte SM, Edmisson JS, Jimenez-Flores E. 2016. Human neutrophils and oral microbiota: a constant tug-of-war between a harmonious and a discordant coexistence. Immunol Rev 273:282–298. doi: 10.1111/imr.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, Sibley CD, Robbins SM, Green FH, Surette MG, Sugai M, Bowden MG, Hussain M, Zhang K, Kubes P. 2010. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 185:7413–7425. doi: 10.4049/jimmunol.1000675. [DOI] [PubMed] [Google Scholar]
- 49.Sabroe I, Dower SK, Whyte MK. 2005. The role of Toll-like receptors in the regulation of neutrophil migration, activation, and apoptosis. Clin Infect Dis 41(Suppl 7):S421–S426. doi: 10.1086/431992. [DOI] [PubMed] [Google Scholar]
- 50.Gould TJ, Vu TT, Swystun LL, Dwivedi DJ, Mai SH, Weitz JI, Liaw PC. 2014. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler Thromb Vasc Biol 34:1977–1984. doi: 10.1161/ATVBAHA.114.304114. [DOI] [PubMed] [Google Scholar]
- 51.Hayashi F, Means TK, Luster AD. 2003. Toll-like receptors stimulate human neutrophil function. Blood 102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 52.Salmon D, Verdier F, Malhotra K, Pussard E, Clavier F, Le Bras J, Vilde JL, Pocidalo JJ. 1990. Absence of effect of chloroquine in vivo on neutrophil oxidative metabolism in human subjects. J Antimicrob Chemother 25:367–370. doi: 10.1093/jac/25.3.367. [DOI] [PubMed] [Google Scholar]
- 53.Graham LM, Gupta V, Schafer G, Reid DM, Kimberg M, Dennehy KM, Hornsell WG, Guler R, Campanero-Rhodes MA, Palma AS, Feizi T, Kim SK, Sobieszczuk P, Willment JA, Brown GD. 2012. The C-type lectin receptor CLECSF8 (CLEC4D) is expressed by myeloid cells and triggers cellular activation through Syk kinase. J Biol Chem 287:25964–25974. doi: 10.1074/jbc.M112.384164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekman A-K, Cardell LO. 2010. The expression and function of Nod-like receptors in neutrophils. Immunology 130:55–63. doi: 10.1111/j.1365-2567.2009.03212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, Buwitt-Beckmann U, Röschmann K, Jung G, Wiesmüller K-H, Ulmer AJ. 2008. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukoc Biol 83:692–701. [DOI] [PubMed] [Google Scholar]
- 56.Labro MT, Babin-Chevaye C. 1988. Effects of amodiaquine, chloroquine, and mefloquine on human polymorphonuclear neutrophil function in vitro. Antimicrob Agents Chemother 32:1124–1130. doi: 10.1128/AAC.32.8.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Bari MA. 2015. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother 70:1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rouhanizadeh M, Hwang J, Clempus RE, Marcu L, Lassegue B, Sevanian A, Hsiai TK. 2005. Oxidized-1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphorylcholine induces vascular endothelial superoxide production: implication of NADPH oxidase. Free Radic Biol Med 39:1512–1522. doi: 10.1016/j.freeradbiomed.2005.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starosta V, Wu T, Zimman A, Pham D, Tian X, Oskolkova O, Bochkov V, Berliner JA, Birukova AA, Birukov KG. 2012. Differential regulation of endothelial cell permeability by high and low doses of oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine. Am J Respir Cell Mol Biol 46:331–341. doi: 10.1165/rcmb.2011-0153OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi T, Butchar JP, Tridandapani S. 2006. Fcgamma receptor signaling in phagocytes. Int J Hematol 84:210–216. doi: 10.1532/IJH97.06140. [DOI] [PubMed] [Google Scholar]
- 61.Lebeaux D, Chauhan A, Rendueles O, Beloin C. 2013. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuura Y, Takehira M, Joti Y, Ogasahara K, Tanaka T, Ono N, Kunishima N, Yutani K. 2015. Thermodynamics of protein denaturation at temperatures over 100 °C: CutA1 mutant proteins substituted with hydrophobic and charged residues. Sci Rep 5:15545. doi: 10.1038/srep15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirschfeld J, Roberts HM, Chapple IL, Parcina M, Jepsen S, Johansson A, Claesson R. 2016. Effects of Aggregatibacter actinomycetemcomitans leukotoxin on neutrophil migration and extracellular trap formation. J Oral Microbiol 8:33070. doi: 10.3402/jom.v8.33070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White P, Sakellari D, Roberts H, Risafi I, Ling M, Cooper P, Milward M, Chapple I. 2016. Peripheral blood neutrophil extracellular trap production and degradation in chronic periodontitis. J Clin Periodontol 43:1041–1049. doi: 10.1111/jcpe.12628. [DOI] [PubMed] [Google Scholar]
- 65.Ling MR, Chapple IL, Matthews JB. 2016. Neutrophil superoxide release and plasma C-reactive protein levels pre- and post-periodontal therapy. J Clin Periodontol 43:652–658. doi: 10.1111/jcpe.12575. [DOI] [PubMed] [Google Scholar]
- 66.Hirschfeld J, Kawai T. 2015. Oral inflammation and bacteremia: implications for chronic and acute systemic diseases involving major organs. Cardiovasc Hematol Disord Drug Targets 15:70–84. doi: 10.2174/1871529X15666150108115241. [DOI] [PubMed] [Google Scholar]
- 67.Roberts H, White P, Dias I, McKaig S, Veeramachaneni R, Thakker N, Grant M, Chapple I. 2016. Characterization of neutrophil function in Papillon-Lefevre syndrome. J Leukoc Biol 100:433–444. doi: 10.1189/jlb.5A1015-489R. [DOI] [PubMed] [Google Scholar]
- 68.Persson S, Edlund MB, Claesson R, Carlsson J. 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol 5:195–201. doi: 10.1111/j.1399-302X.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 69.Gerencser MA, Slack JM. 1969. Identification of human strains of Actinomyces viscosus. Appl Microbiol 18:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White PC, Chicca IJ, Ling MR, Wright HJ, Cooper PR, Milward MR, Chapple IL. 2017. Characterization, quantification, and visualization of neutrophil extracellular traps. Methods Mol Biol 1537:481–497. doi: 10.1007/978-1-4939-6685-1_29. [DOI] [PubMed] [Google Scholar]
- 71.Malawista SE, Gee JB, Bensch KG. 1971. Cytochalasin B reversibly inhibits phagocytosis: functional, metabolic, and ultrastructural effects in human blood leukocytes and rabbit alveolar macrophages. Yale J Biol Med 44:286–300. [PMC free article] [PubMed] [Google Scholar]