Abstract
Hoxa5 is a member of the Hox gene family that plays critical roles in successive steps of the central nervous system formation during embryonic and fetal development. In the mouse, Hoxa5 was recently shown to be expressed in the medulla oblongata and the pons from fetal stages to adulthood. In these territories, Hoxa5 transcripts are enriched in many precerebellar neurons and several nuclei involved in autonomic functions, while the HOXA5 protein is detected mainly in glutamatergic and GABAergic neurons. However, whether HOXA5 is functionally required in these neurons after birth remains unknown. As a first approach to tackle this question, we aimed at determining the molecular programs downstream of the HOXA5 transcription factor in the context of the postnatal brainstem. A comparative transcriptomic analysis was performed in combination with gene expression localization, using a conditional postnatal Hoxa5 loss-of-function mouse model. After inactivation of Hoxa5 at postnatal days (P)1–P4, we established the transcriptome of the brainstem from P21 Hoxa5 conditional mutants using RNA-Seq analysis. One major finding was the downregulation of several genes associated with synaptic function in Hoxa5 mutant specimens including different actors involved in glutamatergic synapse, calcium signaling pathway, and GABAergic synapse. Data were confirmed and extended by reverse transcription quantitative polymerase chain reaction analysis, and the expression of several HOXA5 candidate targets was shown to co-localize with Hoxa5 transcripts in precerebellar nuclei. Together, these new results revealed that HOXA5, through the regulation of key actors of the glutamatergic/GABAergic synapses and calcium signaling, might be involved in synaptogenesis, synaptic transmission, and synaptic plasticity of the cortico-ponto-cerebellar circuitry in the postnatal brainstem.
Keywords: RNA sequencing, conditional mutagenesis, calcium signaling, glutamatergic synapse, GABAergic synapse, brainstem
Introduction
The HOX family of transcription factors gathers key regulators of embryo patterning, organ development, and cell differentiation during animal development, but also throughout adult life (Mallo et al., 2010; Rezsohazy et al., 2015). In the central nervous system (CNS) Hox genes play critical roles in successive developmental steps, from neurulation to the establishment of neuronal networks (Nolte and Krumlauf, 2007; Dasen and Jessell, 2009; Narita and Rijli, 2009; Di Bonito et al., 2013a; Karmakar et al., 2017). During neurulation, combinatorial expression of Hox genes provides a segmental identity and antero-posterior patterning information to neural progenitors (Nolte and Krumlauf, 2007). Some Hox genes remain expressed in the CNS after neurulation and they are implicated in normal tangential migration of pontine neurons, in axonal connection and arborization, in topographic connectivity, in synaptic refinement and in neuron survival (Oury et al., 2006; Pasqualetti et al., 2007; Geisen et al., 2008; Di Bonito et al., 2013b; Philippidou and Dasen, 2013; Bechara et al., 2015; Catela et al., 2016; Karmakar et al., 2017). Finally, recent evidences show that Hox genes from paralog groups 3–8 (PG3–8) display differential anterior limits of expression in the perinatal and adult hindbrain derivatives suggesting a continuing role in differentiation of neuronal derivatives after birth (Hutlet et al., 2016; Tomas-Roca et al., 2016).
Hoxa5, a member of Hox PG5, is known for its numerous activities during development (Jeannotte et al., 2016) and, together with its paralog genes, for its implication in different processes of fetal CNS development (Philippidou et al., 2012; Di Meglio et al., 2013; Catela et al., 2016). In the hindbrain at fetal stages, Hoxa5 expression is detected in the caudal part of the medulla oblongata and in the posterior pontine region, where it participates in the tangential migration of precerebellar neurons from the rhombic lip toward the pontine gray (PG) nucleus (Di Meglio et al., 2013; Tomas-Roca et al., 2016; Lizen et al., 2017). Indeed, PG5 HOX proteins are shown to be functionally implicated in the topographic organization of migrating precerebellar pontine neurons, through the negative regulation of Unc5b expression, a netrin repulsive receptor (Di Meglio et al., 2013). In the spinal cord, lack of Hoxa5 and Hoxc5 gene functions in mutant mice results in an important reduction and disorganization of motor neurons from the phrenic motor column, severe defects in diaphragm innervation, and death at birth (Philippidou et al., 2012). PG5 genes also contribute with other Hox genes to neural fate, organization, and peripheral connectivity of spinal cord motor neurons targeting the forelimb, through regulation of Ret and Gfra genes (Catela et al., 2016). Recently, our detailed analysis of Hoxa5 expression pattern in the mouse brain revealed that Hoxa5 is still expressed after birth until adulthood. A key feature of this analysis was the enrichment of Hoxa5 transcripts in pontine and medullary precerebellar neurons. They are also detected in different nuclei implicated in the control of autonomic functions (Lizen et al., 2017). In these territories, the HOXA5 protein is specifically present in neurons, and principally in glutamatergic and GABAergic neurons. This mapping analysis led us to hypothesize Hoxa5 functions in CNS at fetal and postnatal stages, such as having a potential role in the establishment, refinement, or plasticity of precerebellar circuits during postnatal and adult life. To test these hypotheses and to provide insight into the HOXA5 functions in the brain after birth, we aimed in the present paper at identifying the transcriptional programs and main biological processes downstream of HOXA5 in the postnatal brainstem.
To achieve this objective, we performed a RNA-Seq transcriptomic analysis using a mouse model carrying a conditional postnatal loss-of-Hoxa5 function mutation. Using this approach, we bypassed the early requirement for HOXA5 in the fetal hindbrain and directly tackled HOXA5 functions at the postnatal phase of brainstem development. This paradigm allowed to identify genes that are differentially expressed in Hoxa5-inactivated brainstem when compared with Hoxa5-expressing brainstem. We expect that understanding of the transcriptional programs downstream of HOXA5 will serve as a stepstone for further functional analysis of this transcription factor in the postnatal hindbrain at a critical period for remodeling and maturation of neuronal circuits.
Materials and Methods
Animals
Mice were maintained in a conventional facility and fed in standard conditions (mice maintenance and mice breeding diets, Carfil Quality, Belgium) on a 14 h light/10 h dark cycle. Experimental procedures on animals were performed in accordance with the Belgian national guidelines and in agreement with the European directive 2010/63/UE. The protocol was approved by the animal ethic committee of the Université catholique de Louvain (approval 122803). For all gas euthanasia, CO2 was administered by progressive delivery in the cage volume in accordance with guidelines. The transgenic CMV-CreERT2 mice and the characterization of Cre activity with reporter mice have been described elsewhere (Santagati et al., 2005; Lizen et al., 2015). We used the Hoxa5tm1.1Ljea mouse line carrying a Hoxa5 floxed allele as described (Tabaries et al., 2007). The characterization of Cre efficiency on the Hoxa5 floxed allele after tamoxifen induction has already been described (Lizen et al., 2015). Genotyping of animals was accomplished by polymerase chain reaction (PCR) using the primers and the PCR program described in Supplementary Table S1.
CMV-CreERT2 transgenic mice were bred with mice carrying the conditional Hoxa5 floxed allele to generate double transgenic animals homozygous for the Hoxa5 floxed allele (Hoxa5flox/flox) and heterozygous for the CMV-CreERT2 transgene. Compound transgenic Hoxa5flox/flox;CMV-CreERT2+/- males were bred with Hoxa5flox/flox females. These crosses had the advantage of producing in the same litter pups that were either CreERT2 positive or CreERT2 negative, providing intra-litter controls. The Hoxa5flox/flox;CMV-CreERT2+/- mice were used in a mixed background composed principally of 129/Sv, with a minor contribution of C57BL/6 and DBA/2.
Tamoxifen Administration
For induction treatments, tamoxifen (Sigma–Aldrich, T5648) was dissolved in corn oil (Sigma–Aldrich, C8267). Intragastric injections of 50 μl of three different stock solutions were performed: we injected the first solution of 1 mg/ml at postnatal day (P)1, the second of 1.5 mg/ml was injected at P2 and P3, and the last one of 2 mg/ml was injected at P4 (Lizen et al., 2015). Pups stayed with their mother during the whole treatment until they were euthanized at P21. For euthanasia, CO2 was administered by progressive delivery in the cage volume in accordance with guidelines.
Sample Preparation
Brains were collected after euthanasia as described above. For RNA-Seq experiments, whole brainstems were collected while for reverse transcription quantitative PCR (RT-qPCR) assays, the pons and caudal part of the medulla oblongata were dissected as illustrated in Supplementary Figure S1. Tissues were then snap frozen in liquid nitrogen, and stored at -80°C until RNA was extracted. RNA was isolated using TRI Reagent (Sigma–Aldrich, T9424) and quantified using a NanoDrop ND-1000 fluorospectrometer (Thermo Fisher Scientific, Waltham, MA, United States).
RNA-Seq Analyses
RNA sequencing analysis was carried out on total RNA extracted from tamoxifen-treated individuals of two different genotypes: Hoxa5flox/flox;CMV-CreERT2+/- (Hoxa5 cKO) and Hoxa5flox/flox;CMV-CreERT2-/- (Hoxa5 control). To minimize sample variability caused by individual differences between animals linked to tamoxifen treatment, RNA from three brainstems was pooled, and three pools were used per genotype (for a total of nine samples per genotype) (Supplementary Figure S1). Library preparation and sequencing were performed at the Genomics Facility of the Interdisciplinary Cluster for Applied Genoproteomics (GIGA Genomics, ULg-Liège, Belgium). RNA integrity was verified on the Bioanalyzer 2100 with RNA 6000 Nano chips, RNA integrity number scores were more than 8 for all samples. Illumina truSeq stranded total RNA library prep kit with ribo-zero was used to prepare libraries from 1 μg of total RNA following Illumina’s protocol. Ribosomal RNA was depleted from total RNA. The remaining RNA was purified, fragmented, and then primed with random hexamers and reverse transcribed into first strand cDNA followed by second strand synthesis. A single A nucleotide was added to the 3′end of the fragments before ligation to multiple indexing adapters. DNA fragments with adapter molecules on both ends were enriched by PCR and purified with Ampure XP magnetic beads. Libraries were validated on Bioanalyzer DNA 1000 chip and quantified by qPCR with the KAPA library quantification kit. Sequencing was performed on Illumina NextSeq 500 with NextSeq 500 High Output V2 kit (2 × 75 cycles). For data analysis, Fastq files were trimmed for adaptor sequences. The reads were aligned with Tophat 2.0.91 to the mouse genome. Cufflinks 2.2.0 suite2 was used to generate fragments per kilobase of transcript per million fragments mapped values. CuffDiff was also used to identify significantly differentially expressed genes between the two genotypes. Statistical significance was defined by the Benjamini–Hochberg method. The q-value is a corrected p-value. The q-values below 0.05 were considered as a significant difference of gene expression between the two conditions. Gene ontology (GO) term analysis was performed using DAVID 6.7 Bioinformatics resource tools (Huang da et al., 2009). High-throughput sequencing data used in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE1033903.
RT-qPCR Analyses
Brain subregions were homogenized in 1 ml of TRI Reagent (Sigma–Aldrich, T9424) during 30 s using high-throughput tissue homogenizer (Precellys®24). RNA was isolated from the dissected brain subregions following manufacturer’s instructions. The precipitated RNA pellet was dissolved in 100 μl of nuclease free water and reverse-transcription was performed using a reverse transcription kit (Qiagen, 205311) according to the manufacturer’s instructions. Hoxa5 and target genes expression was assessed by qPCR on a StepOne+ apparatus (Applied Biosystems) using SYBR Green (Qiagen, 204143) as the detection method, and the geometric mean of two reference genes (H2A and Gapdh) as internal normalization reference. The relative transcripts abundance (fold change) was calculated using the 2-ΔΔCT calculation, where ΔCt = Ctgenetarget - Ctreferencegenes and ΔΔCt = ΔCtmutant - ΔCtcontrol. Supplementary Table S2 provides primers sequences and concentrations, as well as the efficiency of the amplification reaction.
Non-radioactive in Situ Hybridization
Eight sets of 14 μm thick serial coronal cryosections per brain were cut on a Leica CM 3050S cryostat. Gene expression was detected using digoxigenin-labeled RNA probes, as previously described, and as optimized for Hox genes analysis in the adult brain (Chotteau-Lelievre et al., 2006; Hutlet et al., 2016).
Plasmids allowing in vitro transcription of digoxigenin-labeled riboprobes were obtained from collaborators (En2: Dr A. Joyner; NeuroD2: M. Studer; Wnt7a: N. Bobbola). For other genes, probes were transcribed directly from PCR amplification products, following an adaptation of the Allen Brain Atlas procedures. For probes synthesis, cDNA of adult mouse brain was used to target genes of interest by a first PCR with designed gene-specific primers (NCBI primer blast). For practical purposes, the T7 universal primer sequence (5′-GCGTAATACGACTCACTATAGGG-3′) was directly attached to the forward and the Sp6 one (5′-ATTTAGGTGACACTATAG-3′) to the reverse primer. PCR products were then amplified by a second PCR reaction, purified using a PCR purification kit (Qiagen, 28104), and eluted with 30 μl of elution buffer (10 mM Tris, pH 8.5). In vitro transcription with SP6 and T7 polymerases was performed on the resulting eluate to generate both RNA sense and anti-sense probes. Probes size and quality were evaluated following in vitro transcription by migration on agarose gel. Prior use for in situ hybridization (ISH), all probes were further diluted to a concentration of 100 ng/μl of hybridization mix (Life Technologies, B8807G), and stored at -20°C. A detailed list of data regarding specific probe primers and sequences is available upon request.
Double ISH was performed on cryostat sections using a combination of digoxigenin- and fluorescein-labeled probes as described (Mayer et al., 2010). Anti-Fluorescein (Roche, 11426338910, 1:3000) immunodetection was used first to reveal candidate target genes, in combination with NBT-BCIP for the first staining reaction. The reaction was stopped with PBT/EDTA. Sections were washed 2 × 10 min in maleic acid buffer with 0.1% Tween 20 (MABT) (Sigma–Aldrich, 9005-64-5). Sections were then incubated in a blocking solution containing MABT with 2% of blocking reagent (Roche, 11 096 176 001) and 20% heat-inactivated goat serum (Sigma–Aldrich, G6767). To target the Hoxa5 probe, the sections were then incubated with anti-digoxigenin antibody (Roche, 11093274910, 1/1500) and INT-BCIP (Roche, 11 681 460 001) were used for the second staining reaction.
Hybridized sections were observed on a Leica DM2500 microscope, and pictures were captured with a Leica DFC420C camera. Annotation of expression thresholds versus background were established for each ISH taking into account the appropriate control and the duration of the staining reaction. A frequently used control for specific hybridization to mRNA is a sense strand probe which should not specifically anneal to the target RNA. In this study, we used as a negative control an antisense probe for a Hox gene not expressed in the brain (e.g., Hoxa10) and an antisense probe for Gad67 as a positive control.
Results
HOXA5 Regulates Multiple Genes Involved in Synaptic Function and Calcium Pathway in Postnatal Brainstem
To explore the HOXA5 transcriptional landscape in the postnatal brainstem, we carried out transcriptomic analyses by RNA-Seq using a model of postnatal Hoxa5 loss-of-function. We induced Hoxa5 inactivation after birth (P1–P4) using the tamoxifen-inducible CMV-CreERT2 mice and conditional Hoxa5 floxed allele mice (Hoxa5flox) (Santagati et al., 2005; Tabaries et al., 2007; Lizen et al., 2015). This treatment results in a severe reduction of Hoxa5 transcripts in brainstem subregions (8–12%, see below, RT-qPCR analysis) and of HOXA5-positive cells in the brain (7–26% depending on nuclei) (Lizen et al., 2015). RNA was extracted from the brainstem of P21 tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2+/- (herein called Hoxa5 cKO) pups and from tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2-/- (herein called Hoxa5 control) littermates. To identify differentially expressed genes, we performed three independent replicate experiments, in which each replicate contains RNA pooled from three brainstems (see section “Materials and Methods”; Supplementary Figure S1). Transcriptome analysis revealed 116 transcripts whose expression levels were significantly modified in brainstem from P21 mutants (q-value <0.05) and 318 transcripts with a p-value lower than 0.01 (Figure 1A and Supplementary Table S3). Among the 116 transcripts, 98 (84.5%) were downregulated and 18 (15.5%) were upregulated in mutants (Figure 1 and Supplementary Table S3). The relative abundance levels of transcripts that are significantly modified are illustrated for each of the three pooled samples in the heat map (Figure 1B).
FIGURE 1.
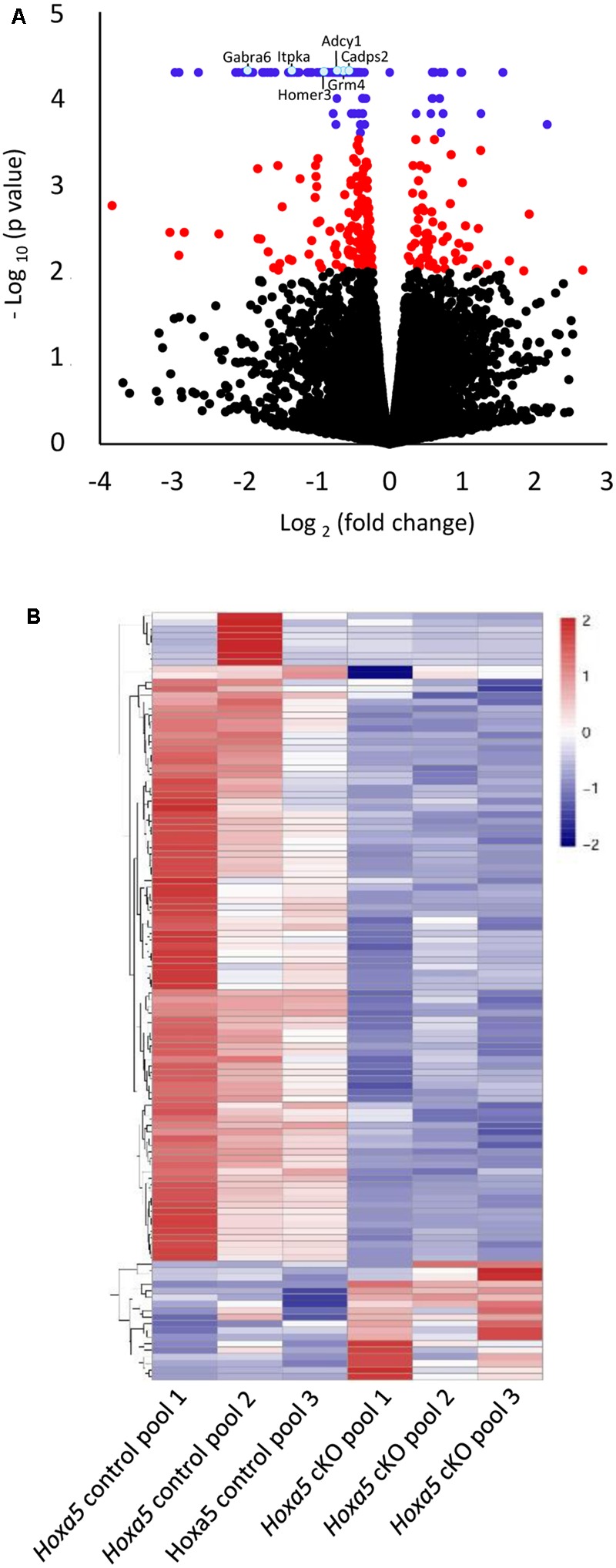
Transcriptome analysis of P21 brainstem in tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2-/- (Hoxa5 control) and tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2+/- (Hoxa5 cKO) (n = 3 pools of 3 individuals per genotype). (A) Volcano plot (p-value versus fold change) of differentially expressed transcripts. Transcripts whose abundance level is significantly modified in Hoxa5 cKO (q-value <0.05) are colored in blue, while those with a q-value >0.05 and a p-value <0.01 are colored in red. (B) Heatmap showing hierarchical clustering of significantly downregulated (blue) and upregulated (red) transcripts (q-value <0.05) in the Hoxa5 control pools and in the Hoxa5 cKO pools. A large proportion of significantly downregulated transcripts are detected in the Hoxa5 cKO pools. See Supplementary Table S3 for complete list of genes.
To gain insights into the molecular programs regulated by HOXA5, we applied a GO analysis on the 116 differentially expressed genes. The top 3 most significant enriched terms were respectively glutamatergic synapse (p-value = 2.6 × 10-7), calcium (p-value = 5.6 × 10-7), and synapse (p-value = 5.7 × 10-6), and included only downregulated genes (Figure 2A and Supplementary Table S4). A striking feature of this analysis is the importance of enriched terms associated with synapse such as glutamatergic synapse, calcium, synapse, calcium signaling pathway, long-term memory, cell junction, GABAergic synapse, long-term potentiation, postsynaptic membrane (Figure 2A). Further analyses through NCBI4 allowed to identify a total of 39 genes with evidence of synaptic function among the 98 downregulated genes (∼40%). In contrast, we did not identify any genes with evidence of function at the synapse among the upregulated transcripts in mutant brains.
FIGURE 2.
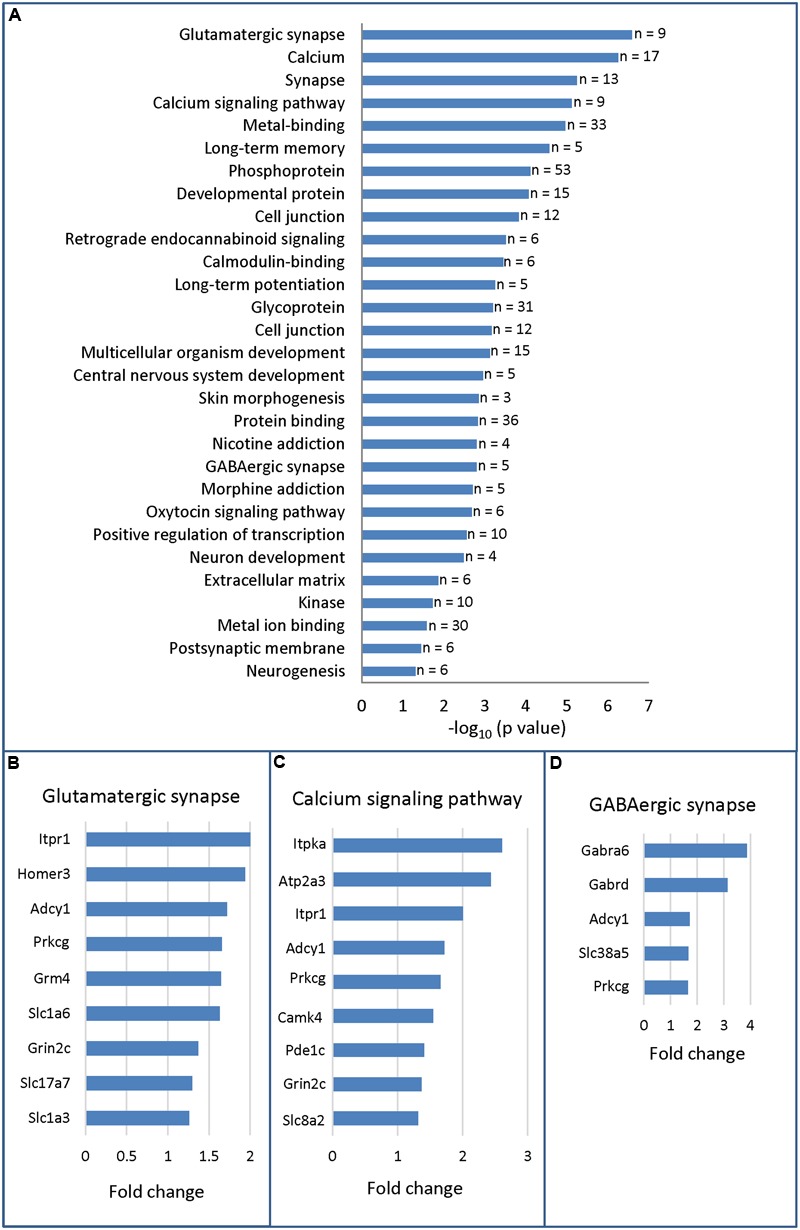
Gene ontology analysis (DAVID 6.7) of RNA-sequencing data from P21 brainstem in Hoxa5 control and Hoxa5 cKO. (A) Functional annotation chart. Significantly enriched terms (p < 0.05) associated to differentially expressed transcripts are presented in descending order (–log10 of p-value in x-axis). As detailed in Supplementary Table S4, p-values of the significantly enriched terms vary between 2.6 × 10-7 and 4.6 × 10-3. The number of genes for each term is indicated on the right. (B) Genes identified in the glutamatergic synapse term. (C) Genes identified in the calcium signaling pathway term. (D) Genes identified in the GABAergic synapse term. Fold change (x-axis) in (B–D) is the ratio between transcript abundance value measured in Hoxa5 control and transcript abundance value measured in Hoxa5 cKO in RNA-Seq analysis (see Supplementary Table S3 for detailed values).
Detailed analysis of candidate target genes involved in glutamatergic synapse revealed downregulation of several receptors and transporters, such as the metabotropic glutamate receptor Grm4, the subunit of the N-methyl-D-aspartate (NMDA) receptor Grin2c, the L-glutamate transmembrane transporters Slc1a6 (Eaat4) and Slc1a3 (Eaat1, Glast) and the vesicular glutamate transporter Slc17a7 (Vglut1) (Figure 2B). Downstream effectors, such as Prkcg, the member of the PKC family that regulates NMDA receptors trafficking (Lan et al., 2001), and the synaptic plasticity modulator Adcy1 were also downregulated (Wang and Zhang, 2012). Other candidate targets include members of the inositol 1,4,5-trisphosphate (IP3) pathway: Itpr1 (inositol 1,4,5-trisphosphate receptor), which mediates calcium release from the endoplasmic reticulum, and its interactor Homer3 (Figure 2B) (Kammermeier, 2006; Mizutani et al., 2008; Goto and Mikoshiba, 2011). Several of these targets are also influenced by calcium or involved in calcium signaling: Itpr1, Adcy1, Prkcg, and Grin2c (Figure 2C) (Oancea and Meyer, 1998; Wong et al., 1999; Goto and Mikoshiba, 2011). Additional downregulated targets involved in calcium signaling pathway are: Camk4, the calcium/calmodulin-dependent protein kinase IV involved in neural calcium signaling; Atp2a3, the ATPase endoplasmic reticulum Ca2+ transporting 3; Itpka, the inositol-trisphosphate 3-kinase; and Car8, the carbonic anhydrase 8 which regulates calcium homeostasis through ITPR1 inhibition (Zhuang et al., 2015). Finally, downregulated transcripts involved in GABAergic synapse included the two GABA receptor subunits Gabra6 and Gabrd, which were highly downregulated in mutants (fold change >3) and the System N Glutamine Transporter Slc38a5 (SNAT5) (Figure 2D). As indicated by the two enriched GO terms postsynaptic membrane and postsynaptic cell membrane (Figure 2A and Supplementary Table S4), most of the downregulated transcripts involved in glutamatergic synapse and GABAergic signal are mainly postsynaptic.
In summary, deletion of Hoxa5 gene function early after birth causes downregulation of numerous genes involved in synaptic function in the brain few weeks after. Notably, transcripts of multiple genes coding major players of glutamate and GABA neurotransmission, as well as key actors of calcium signaling, were downregulated. Due to the absence of highly enriched terms associated to upregulated genes (Supplementary Table S5), further analysis was carried out only for downregulated genes.
RT-qPCR Analysis of HOXA5 Candidate Targets in the Pons and Medulla Oblongata
A subset of candidate HOXA5 target genes identified by RNA-Seq analysis was analyzed by real-time RT-qPCR. Selection of genes was based on GO functional annotations, expression fold changes, and literature. Genes that were downregulated at least 1.5-fold in Hoxa5 cKO brainstem (Supplementary Table S4) and known to be functionally related to synapse and calcium signaling (Figures 2B–D) were favored. Zic genes encoding transcription factors were also selected based on their known expression and critical function in mossy fibers precerebellar neurons (Dipietrantonio and Dymecki, 2009). For quantitative analysis through RT-qPCR, we improved the resolution of the analysis by measuring RNA levels of the selected genes in dissected pons and caudal part of the medulla oblongata since HOXA5 expression was shown to be enriched in these restricted areas (Supplementary Figure S1) (Lizen et al., 2017).
RT-qPCR confirmed downregulation of Cadps2, Wnt7a, Zic2, Neurod2, Zic1, Prkcg, Grm4, Adcy1, and Homer3 in the dissected medulla oblongata of Hoxa5 cKO specimens (p-value <0.05; Table 1). For Gabra6, Cbln3, En2, and Slc1a6, transcript levels were reduced in Hoxa5 cKO samples compared to Hoxa5 control but did not reach the significance (p-value between 0.1 and 0.3) (Figure 3A and Table 1). In the pons of Hoxa5 cKO, Calb1 was the only candidate for which the downregulation was close to the level of significance (p-value 0.0519), but the level of Zic1 was also reduced in Hoxa5 cKO samples compared to Hoxa5 control, although it did not reach the significance (p-value 0.169) (Figure 3B and Table 1). Indeed, although we observed reduced transcription levels for several other candidate genes, the variability between the samples, possibly explained by the absence of precise landmarks for macro-dissection, prevented us from concluding.
Table 1.
RT-qPCR analysis of selected candidate target genes of HOXA5 in the medulla oblongata and pons.
| Target genes | Fold change | p-value | |
|---|---|---|---|
| Medulla | Cadps2 | 1.52 | 0.0052∗∗ |
| Oblongata | Wnt7a | 3.21 | 0.0076∗∗ |
| Zic2 | 1.61 | 0.0061∗∗ | |
| NeuroD2 | 1.45 | 0.0063∗∗ | |
| Zic1 | 1.37 | 0.0088∗∗ | |
| Prkcg | 1.30 | 0.0091∗∗ | |
| Grm4 | 1.56 | 0.0243∗ | |
| Adcy1 | 1.60 | 0.0381∗ | |
| Homer3 | 1.26 | 0.0463∗ | |
| Gabra6 | 2.49 | 0.0672 | |
| En2 | 1.42 | 0.2089 | |
| Slc1a6 | 1.49 | 0.2856 | |
| Cbln3 | 1.27 | 0.2662 | |
| Calb1 | 0.95 | 0.6538 | |
| Pons | Calb1 | 1.30 | 0.0519 |
| Zic1 | 1.22 | 0.1649 |
p-values are from ANOVA test. n = 3–6 for Hoxa5 control brains, n = 3–8 for Hoxa5 cKO brains. Asterisks depict statistically significant differences in Hoxa5 cKO brains compared with Hoxa5 control brains: ∗∗p < 0.01 and ∗p < 0.05.
FIGURE 3.
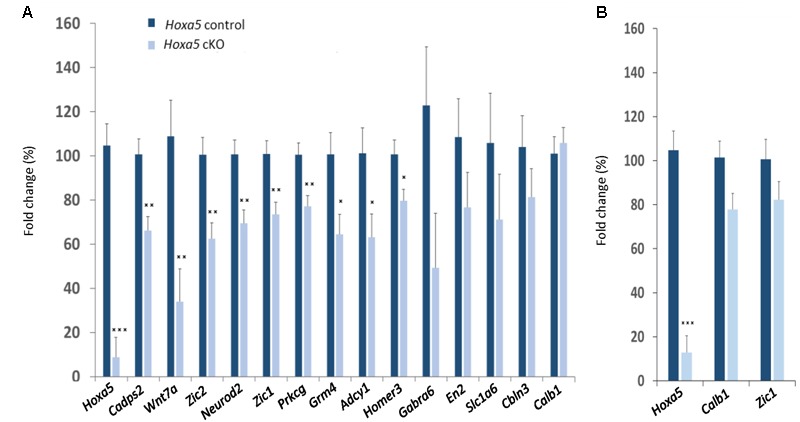
RT-qPCR analysis of selected candidate target genes of HOXA5 in the medulla oblongata (A) and pons (B) of tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2-/- (Hoxa5 control; dark blue) and tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2+/- (Hoxa5 cKO; light blue) at P21. Data are reported as means ± SEM. Asterisks depict statistically significant differences compared with Hoxa5 control brains: ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. p-Values are from ANOVA test and are provided in Table 1. n = 3–6 for Hoxa5 control brains, n = 3–8 for Hoxa5 cKO brains. The relative transcripts abundance is calculated using the 2-ΔΔCt method as detailed in Section “Materials and Methods,” where normalized values measured in Hoxa5 cKO are reported to normalized values measured in Hoxa5 control. The fold change is presented as a percentage of this value: (2-ΔΔCt) × 100.
Thus, RT-qPCR data confirmed the RNA-Seq results, thereby validating the robustness of the analysis. Moreover, this analysis suggested region-specific regulation of HOXA5 downstream genes. This prompted us to evaluate the neuroanatomical expression profiles of HOXA5 candidate target genes.
Anatomical and Cellular Localization of HOXA5 Candidate Targets Expression in the Postnatal Brainstem
To gain insights into the expression pattern of candidate target genes in brainstem, ISH was performed on the brain of wild-type mice at P21. Probes were designed for the subset of target genes analyzed by RT-qPCR (see above for criteria). Reliable labeling was detected for Calb1, Cadps2, Camk4, Cbln1, Grin2c, Itpr1, Neurod2, Vglut1, Wnt7a, Zic1, and Zic2 in Hoxa5-expressing nuclei (Table 2).
Table 2.
Neuroanatomical localization of HOXA5 candidate target genes expression in Hoxa5-expressing nuclei, as evaluated by in situ hybridization analysis.
| HOXA5 positive nuclei | Detected transcripts | |
|---|---|---|
| Medulla | XII (hypoglossal nucleus) | Calb1, Itpr1, Vglut1, Wnt7a |
| Oblongata | X (nucleus x) | Cbln1, Grin2c, Vglut1, Zic1 |
| SPIV (spinal vestibular nucleus) | Cadps2 | |
| MV (medial vestibular nucleus) | Cadps2, Grin2c, Zic1 | |
| PRP (nucleus prepositus) | Cadps2, Grin2c, Zic1 | |
| PARN (parvicellular reticular nucleus) | Calb1, Cbln1 | |
| MDRN (medullary reticular nucleus) | Calb1 | |
| IRN (intermediate reticular nucleus) | Itpr1 | |
| GRN (gigantocellular reticular nucleus) | Cadps2, Cbln1 | |
| DMX (dorsal motor nucleus of the vagus nerve) | Cadps2, Calb1 | |
| LRN (lateral reticular nucleus) | Camk4, Cbln1, NeuroD2, Vglut1, Zic1, Zic2 | |
| SPVI (spinal nucleus of the trigeminal, interpolar part) | Cbln1, Itpr1, Vglut1 | |
| SPVC (spinal nucleus of the trigeminal, caudal part) | Adcy1, Calb1, Cbln1 | |
| NTS (nucleus of the solitary tract) | Calb1, Cbln1, Zic1 | |
| ECU (external cuneate nucleus) | Camk4, Cbln1, Grin2c, Vglut1, Zic1, Zic2 | |
| GR (gracile nucleus) | Cbln1 | |
| CU (cuneate nucleus) | Cbln1 | |
| AP (area postrema) | Zic1 | |
| Pons | IF5 (interfascicular trigeminal nucleus) | Cbln1, NeuroD2, Vglut1, Zic1 |
| TRN (tegmental reticular nucleus) | Camk4, NeuroD2, Vglut1, Zic1, Zic2 | |
| PG (pontine gray) | Camk4, NeuroD2, Vglut1, Zic1, Zic2 | |
| KF (koelliker-fuse subnucleus) | Calb1 |
Nuclei that showed the highest enrichment in candidate target genes are the PG, the tegmental reticular nucleus (TRN), the lateral reticular nucleus (LRN) and the external cuneate nucleus (Parys et al., 2012), which are all major precerebellar nuclei providing inputs almost exclusively to the cerebellum. By forming mossy fibers that synapse onto the cerebellar granule cells, those nuclei are essential for coordinated motor activity, motor learning, and procedural memory (Rodriguez and Dymecki, 2000; Fu et al., 2013). Expression of Camk4, Neurod2, Vglut1, Zic1, and Zic2 was observed in the PG and TRN, in a pattern overlapping with Hoxa5 expression (Figure 4 and Table 2). Cbln1, Neurod2, Vglut1, and Zic1 transcripts were also detected in the interfascicular trigeminal nucleus (IF5), another Hoxa5-expressing major precerebellar pontine nucleus (Figure 4 and Table 2). Co-labeling by double ISH revealed that all Hoxa5-positive cells were also positive for Camk4, Zic1, and Zic2 in these nuclei (Figures 4C,F,I,L). In the medulla oblongata, Camk4, Cbln1, NeuroD2, Vglut1, Zic1, and Zic2 expression was detected in the LRN as Hoxa5 (Figure 5 and Table 2). As observed in the pons, all the Hoxa5-positive cells in the caudal part of LRN were also Camk4, NeuroD2, Zic1, and Zic2-positive, while only a portion of Hoxa5-positive cells were Cbln1-positive (Figures 5C,F,I and data not shown). In the external cuneate nucleus, signal for Cbln1, Grin2c, Vglut1, Zic1, and Zic2 was detected (Table 2). For Zic1 and Zic2, the signal seemed to completely co-localize with Hoxa5 expression, while only a portion of Hoxa5-positive cells were Cbln1-positive (data not shown). In addition to major precerebellar nuclei, transcripts of target genes were also detected in minor precerebellar nuclei, which have a less prominent projection to cerebellum, as illustrated in the caudal part of the spinal nucleus of the trigeminal (SPVC) for Adcy1 and Cbln1 (Figures 5K,N and Table 2). In this nucleus, co-localization of Adcy1 and Cbln1 with Hoxa5 was partial (Figures 5L,O).
FIGURE 4.
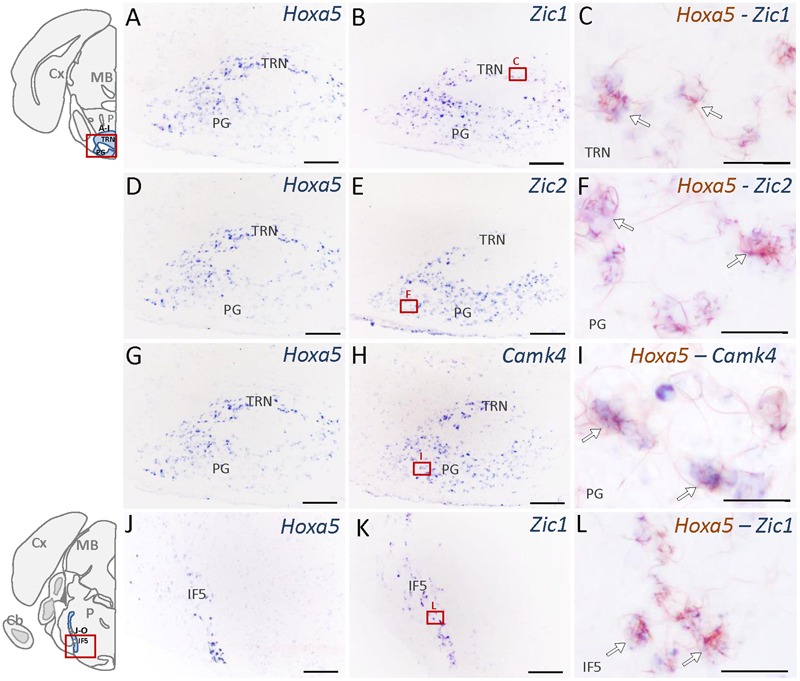
Expression of HOXA5 candidate target genes in the pons analyzed by single and double in situ hybridization (ISH) on coronal cryosections of P21 mouse brains. Schematic views of coronal brain sections illustrate brain structures while square red boxes on the schemes refer to the localization of single ISH pictures. In double ISH (right column) Hoxa5 signal is in red/brown precipitate while target gene signal is in blue. Localization of double ISH pictures is illustrated by square red boxes on the single ISH pictures (middle column). (A–C) Signal of Hoxa5 and Zic1 in the pontine gray (PG) and tegmental reticular nucleus (TRN). (D–F) Signal of Hoxa5 and Zic2 in PG and TRN. (G–I) Signal of Hoxa5 and Camk4 in PG and TRN. (J–L) Signal of Hoxa5 and Zic1 in the interfascicular trigeminal nucleus (IF5). As illustrated in the right column, many Hoxa5-positive cells also show labeling for target gene transcripts (white arrows). n = 2–4 brains hybridized for Hoxa5 and each target genes in single and double ISH. Cb, cerebellum; Cx, cortex; MB, mid brain; P, pons. Medial is on the right, lateral is on the left. Scale bar 200 μm in (A,B,D,E,G,H,J,K); 100 μm in (C,F,I,L).
FIGURE 5.
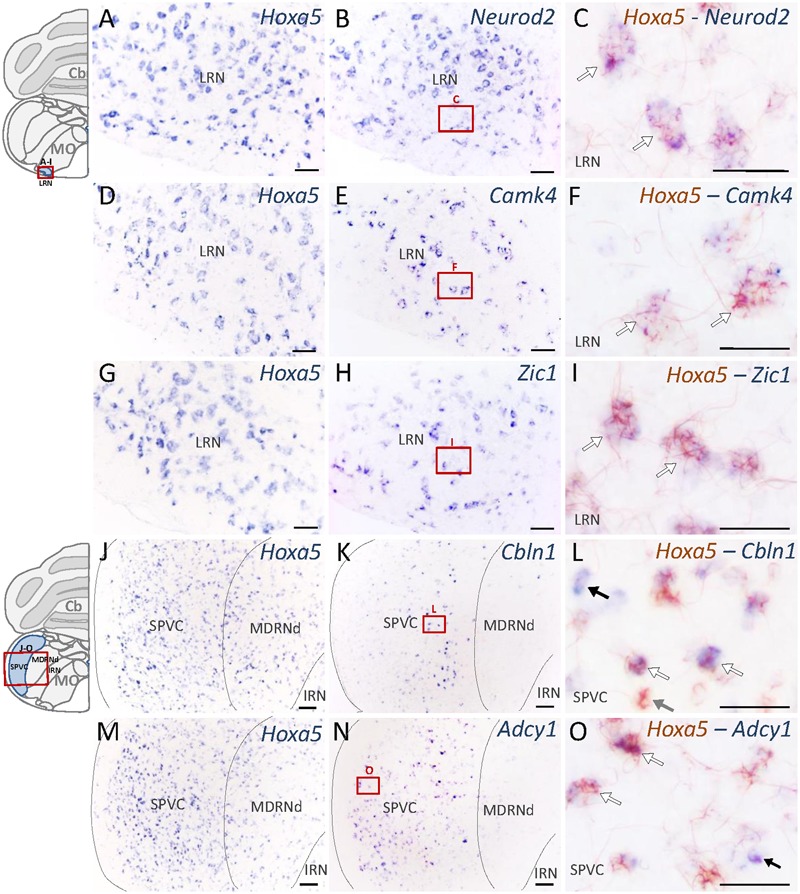
Expression of HOXA5 candidate target genes in the medulla oblongata analyzed by single and double in situ hybridization (ISH) on coronal cryosections of P21 mouse brains. Schematic views of coronal brain sections illustrate brain structures while square red boxes on the schemes refer to the localization of single ISH pictures. In double ISH (right column) Hoxa5 signal is in red/brown precipitate while target gene signal is in blue. Localization of double ISH pictures is illustrated by square red boxes on the single ISH pictures (middle column). (A–C) Signal of Hoxa5 and Neurod2 in the lateral reticular nucleus (LRN). (D–F) Signal of Hoxa5 and Camk4 in LRN. (G–I) Signal of Hoxa5 and Zic1 in LRN. (J–L) Signal of Hoxa5 and Cbln1 in the caudal part of the spinal nucleus of the trigeminal (SPVC). (M–O) Signal of Hoxa5 and Adcy1 in SPVC. As illustrated in the right column, many Hoxa5-positive cells also show labeling for target gene transcripts. White arrows point to examples of co-localization, while gray arrows show example of cells only positive for Hoxa5, and black arrows show example of cells only positive for target genes. n = 2–4 brains hybridized for Hoxa5 and each target genes in single and double ISH. Cb, cerebellum; MO, medulla oblongata. Medial is on the right, lateral is on the left. Scale bar 200 μm in (A,B,D,E,G,H); 500 μm in (J,K,M,N); 100 μm in (C,F,I,L,O).
Altogether, the expression data revealed regionalized expression of several HOXA5 candidate targets in the brainstem of P21 wild-type mice, in patterns that highly overlapped with the Hoxa5-expression pattern. Double ISH showed cellular co-localization of Hoxa5 and candidate targets transcripts in many precerebellar nuclei, although in PG, TRN, IF5, and LRN candidate targets displayed broader expression profiles than Hoxa5 (data not shown). This supports a regulation of these candidate target genes by HOXA5 in the precerebellar system, regulation that could be either direct or indirect.
Discussion
To tackle HOXA5 functions in the pons and medulla oblongata after birth, we analyzed the transcriptional programs downstream of HOXA5 in the brainstem. We found that HOXA5 regulates many genes associated to synaptic function. Indeed, GO analysis revealed that numerous transcripts downregulated in Hoxa5 mutants are involved in glutamate synapse, GABAergic synapse and calcium signaling. Using RT-qPCR, we confirmed and extended these data, validating the robustness of the analysis and suggesting region-specific regulation of HOXA5 on target genes. By ISH, we also revealed the co-expression of Hoxa5 with its targets, especially in nuclei of the precerebellar system.
Functions of Genes with Downregulated Expression in Hoxa5 Mutant Brainstem
Strikingly, very few candidate target genes are components or essential regulators of the Wnt/β-catenin, FGF, and TGF-β/Activin/BMP signaling pathways that have been identified as downstream pathways of several HOX proteins in embryonic development, neural differentiation and tumorigenesis (Chen et al., 2005; Schiedlmeier et al., 2007; Bami et al., 2011; Makki and Capecchi, 2011; Donaldson et al., 2012; Carroll and Capecchi, 2015; Duan et al., 2015; Hrycaj et al., 2015; Roux et al., 2015; Karmakar et al., 2017; Zhang et al., 2017). Moreover, except from Pcp2 (see Supplementary Table S3, line 17), none of the already identified direct or indirect targets of HOXA5 was significantly downregulated in our analysis (e.g., Ptn, Fslt1) (Sanlioglu et al., 1998; Chen et al., 2005; Boucherat et al., 2013). Our data support a model in which HOX transcription factors regulate the expression of different target genes depending on the ontogenetic context. In the postnatal brainstem, HOXA5 seems to specifically participate in the regulation of synaptic function.
Indeed, GO analysis and literature mining revealed that about 40% of the candidate genes are associated to the synapse, notably the glutamatergic synapse. A tentative synthesis of the functional links between proteins encoded by the HOXA5 candidate target genes associated to the glutamatergic synapse and calcium/calcium signaling is illustrated in Figure 6A, highlighting the importance of partnership between these molecules. Although the GABAergic synapse was not among the most significant enriched terms, the associated genes were among those whose fold change was the highest, such as Gabra6 and Gabrd (Figure 6B).
FIGURE 6.
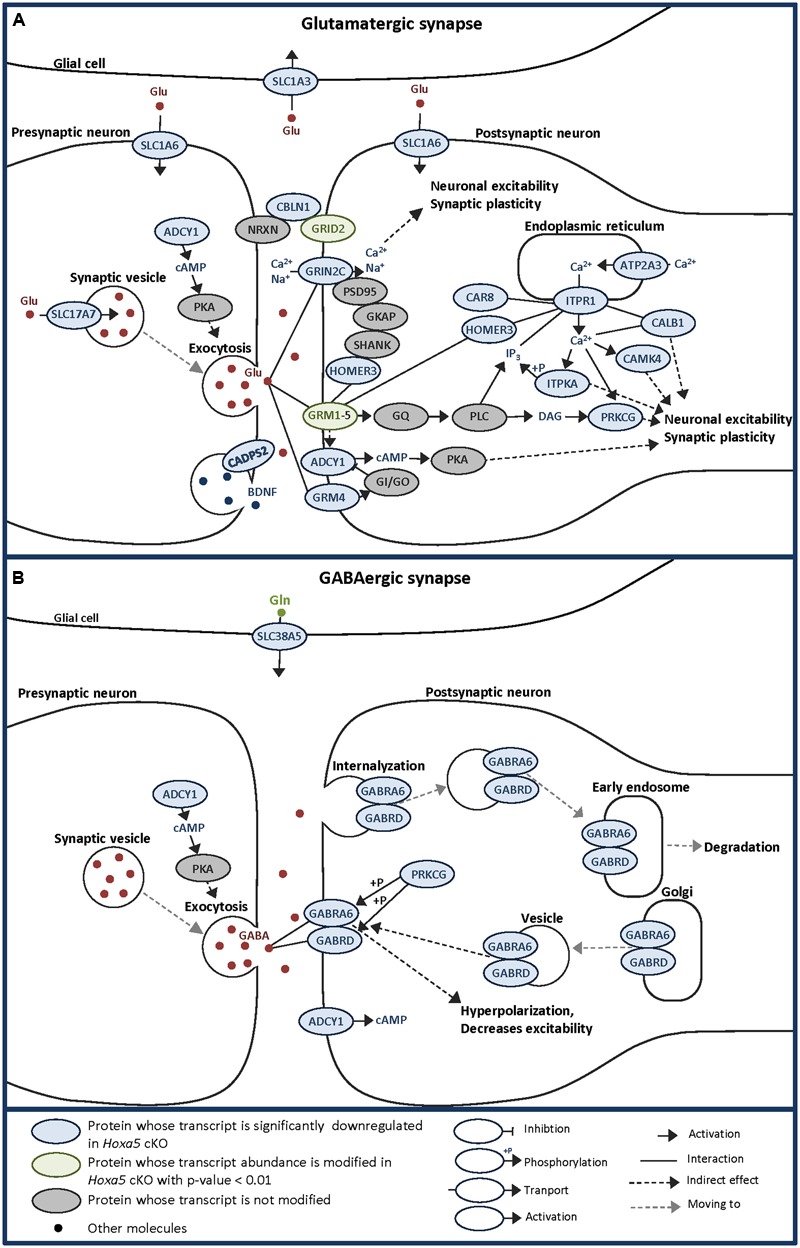
Schematic representations of glutamatergic (A) and GABAergic (B) synapses allowing to visualize the localization and potential links between HOXA5 potential targets identified by transcriptome analysis of P21 brainstem in this study. Adapted from KEGG pathway database. Hoxa5 cKO: tamoxifen-treated Hoxa5flox/flox;CMV-CreERT2+/-; Hoxa5 control: tamoxifen-treated Hoxa5flox/flox; CMV-CreERT2-/-.
Most of the downregulated transcripts in Hoxa5 mutants involved in glutamatergic synapse are mainly postsynaptic while only a few are presynaptic (Figure 6A). Among the postsynaptic actors, we identified the two glutamate receptors: GRM4, a G protein-coupled receptor involved in the inhibition of cyclic AMP cascade, and GRIN2C, a NMDA receptor subunit that forms a channel permeable to calcium, potassium, or sodium depending on the other subunits. In the downstream pathway, we detected the calcium release mediator ITPR1, together with three of its interacting proteins, namely HOMER3, CAR8 and CALB1 (Tu et al., 1998; Hirota et al., 2003). Many proteins whose activity is influenced by calcium were identified, such as CAMK4, ADCY1, PRKCG, and ITPKA. We also detected the glutamatergic synapse organizer CBLN1, which interacts with NRXN and GRID2 to form a trimolecular trans-synaptic organizer (Hirai et al., 2005; Matsuda et al., 2010; Uemura et al., 2010; Ryu et al., 2012). Interestingly, GRID2 was also found downregulated in Hoxa5 mutant specimens with a p-value <0.01. Through the regulation of these postsynaptic targets, HOXA5 could actively influence neuronal excitability and synaptic plasticity. Among the presynaptic actors of the glutamatergic synapse, we detected Slc17a7 (VGLUT1), which has an essential role during postnatal development by loading presynaptic vesicle with glutamate, thereby regulating the efficacy of neurotransmission (Wojcik et al., 2004). Another presynaptic target of HOXA5 is the Ca2+-dependent activator protein for secretion 2 (CADPS2), which enhances release of the neurotrophic factor BDNF involved in the functional maturation of the glutamatergic synapses and essential for normal postnatal cerebellar development (Gottmann et al., 2009; Sadakata and Furuichi, 2009). By regulating Vglut1 and Cadps2, HOXA5 could thus control postnatal development of the cerebellum through the maturation of mossy fiber connections with the granular cells. This hypothesis is also supported by the downregulation of Zic1 transcripts in Hoxa5 cKO brainstem, which encode a Zinc-finger transcription factor involved in mossy fiber development (Dipietrantonio and Dymecki, 2009).
Although few members of GABAergic synapses are downregulated in Hoxa5 mutants, they are specific actors of cerebellar function. Indeed Gabra6 has been reported to have a very restricted expression in granule cells within the cerebellum and in a subset of precerebellar nuclei (Funfschilling and Reichardt, 2002; Maejima et al., 2013). This GABAa receptor subunit was shown to associate to the δ subunit encoded by Gabrd in the cerebellar granule cells, where they could impact on processes underlying motor coordination (Hanchar et al., 2005).
Together, these results support a new role for HOXA5 as a critical regulator of synaptic function, such as glutamatergic synapse integrity, excitability and/or plasticity, during the first three postnatal weeks.
A Role for Hoxa5 in the Development of Postnatal Cerebellar Pathways
A key feature of Hoxa5 expression in the postnatal and adult hindbrain is its enrichment in precerebellar nuclei in the pons and medulla oblongata (Lizen et al., 2017). Neurons of the precerebellar system, which relay both peripheral sensation and cortical input to the cerebellum, derive from the caudal part of the rhombic lip (r6–r8), using tangential migration to reach their final position. It has been suggested that a rhombomere-specific HOX code controls appropriate precerebellar neuron position during tangential migration as well as appropriate cortical input on pontine nuclei. Notably, PG5 HOX proteins were shown to be functionally implicated in the topographic organization of migrating precerebellar pontine neurons (Di Meglio et al., 2013).
Strikingly, a high proportion of significantly downregulated genes identified in the brainstem of Hoxa5 cKO mice at P21 are associated to the cerebellum. The ISH analysis revealed that these candidate target genes are also expressed in the precerebellar nuclei, where several co-localize at the cellular level with Hoxa5. These results suggest a postnatal requirement of HOXA5 in these nuclei for processes beyond the neuronal migration, such as activity-dependent refinement of neuronal pre-established circuits. As cerebellum grows and matures essentially during the postnatal period, cerebellar and precerebellar circuits are particular vulnerable to environmental signals (Wang and Zoghbi, 2001), as shown for pontine neurons that carry on their maturation during the first weeks after birth. Indeed, around P1, pyramidal tract fibers from the cortex grow toward the PG where they enter and establish synapses. Refinement of cortico-pontine projections is then observed by the decreasing of collateral branches around P16 (Inoue et al., 1991; Leergaard et al., 1995). During this postnatal refinement, both presynaptic (cortico-pontine axons) and postsynaptic (pontine neurons) elements play an active role. Our data support a role for HOXA5 in cortico-pontine projections refinement through regulation of transcripts involved in postsynaptic ending on pontine neurons. Of note, calcium and CAMK4 are part of signaling mechanisms involved in the regulation of dendrite growth, branching, and elongation (Nagendran and Hardy, 2011). A role for ADCY1 in the regulation of synapse stabilization/elimination during the late stage of fine topography of sensory circuits has also been demonstrated (Nicol and Gaspar, 2014). Finally, through its regulation of Wnt7a, HOXA5 could influence synapse development, remodeling, and maintenance at both presynaptic and postsynaptic levels (Dickins and Salinas, 2013). This hypothesis is further supported by the involvement of HOX factors in refinement of connectivity in brainstem cochlear neurons (Karmakar et al., 2017), where PG2 HOX proteins present in the postsynaptic neurons, the glutamatergic bushy cells, orchestrate the tonotopic precision of pre-synaptic input.
HOXA5 could also influence the postnatal maturation of pontine output in the granular layer of the cerebellum through regulating presynaptic actors such as VGLUT1 and CADPS2 mentioned above. Involvement of HOXA5 in the postnatal maturation of cortico-ponto-cerebellar connections could affect the different functions in which these circuits are involved such as procedural memory, motor learning, and motor coordination. In this context, it is worth mentioning that our GO analysis revealed enrichment in associated terms (e.g., long-term memory and long-term potentiation).
As PG2 to PG5 Hox genes are expressed in pontine nuclei from fetal to postnatal stages (Hutlet et al., 2016), they may share different functions in postnatal maturation of cortico-ponto-cerebellar circuits. Expression of candidate target genes in anterior part of pontine nuclei, where Hoxa5 is not expressed, suggests co-regulation by HOX proteins expressed more anteriorly in pontine nuclei. These proteins could regionally specify cortico-ponto-cerebellar projections during postnatal maturation through regulation of specific transcriptional codes in each pontine nuclei sub-population. As such, Hox genes could be involved in postnatal activity-dependent refinement of topographic precision of connectivity. In support of this hypothesis, a few of our candidate targets genes, such as Cadps2, Camk4, NeuroD1, Zic1, Cbln1, were identified downstream of other HOX proteins (Bami et al., 2011; Makki and Capecchi, 2011; Anderson et al., 2013; Karmakar et al., 2017).
HOXA5 in Synaptic Function and Dysfunction
Glutamatergic synapse dysfunction has profound effects both in neurological diseases and psychiatric disorders such as Parkinson and Alzheimer diseases, depressive disorders, schizophrenia, and autism spectrum disorder (Ravasi et al., 2010). Dysfunction of these synapses in the first week postnatally could impact on ASD. In this context, several genes downregulated in the brainstem of Hoxa5 cKO mouse at P21, such as Grm4, Cbln1, En2, Camk4, and Cadps2, have been associated to autism traits either in humans or in mouse models (Sadakata and Furuichi, 2009; Becker et al., 2014; Waltes et al., 2014; Genestine et al., 2015; Krishnan et al., 2017). Moreover, several HOXA5 candidate target genes identified such as Cbln1, Cbln3, Homer3, Grm4, En2, Atp2a3, Fat2, and Car8, are also downregulated in a recent mouse model of ASD, generated by increasing dosage of E3 ubiquitin ligase UBE3A, which recapitulates a highly penetrant type of autism in humans (Smith et al., 2011; Krishnan et al., 2017). Although HOXA5 protein has not been detected in the forebrain-derived territories, which have been mostly investigated in deciphering ASD underlying pathways, recent analysis highlights a role for the cerebellum in the perinatal risks for autism (Wang et al., 2014). Therefore, these data point to HOXA5 as a potential regulator in pathways that are affected in ASD.
Conclusion
Our molecular phenotyping of Hoxa5 cKO brainstem at P21 highlights a downregulation of many genes associated with synaptic function in Hoxa5 mutant. GO analysis and literature mining allowed us to point key actors involved in glutamatergic synapse, calcium signaling pathway, and GABAergic synapse. In agreement with our previous hypothesis, this study supports a new role for HOXA5 in the establishment and refinement/plasticity of precerebellar circuits after birth. Indeed, HOXA5 could orchestrate the cortico-ponto-cerebellar circuit formation and maturation during the postnatal period. As such it could modulate important cerebellar functions such as motor coordination, motor learning, and procedural memory, and its alteration could impact on the development of synaptopathies such as ASD.
Author Contributions
BL and FG designed the research. LJ provided the mouse transgenic line. BL, CM, JM, TS, M-TA, and AS performed the research. BL, CM, JM, AS, and FG analyzed the data. BL and FG wrote the article with all other authors and LJ revised it critically.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the GIGA-Genomics core facility (GIGA, University of Liège), notably Benoît Hennuy and Benoît Charloteaux, for their technical assistance on RNA sequencing experiments and their support for data analysis and presentation. We wish to thank Coralie Piget for animal husbandry. The authors would like to thank Bernard Knoops, René Rezsohazy, and Maryse Hermant for helpful comments on the manuscript.
Funding. This work was supported by the “Communauté Française de Belgique-Actions de Recherche Concertées” (12/17-041). BL is a FRIA fellow of the Fonds de la Recherche Scientifique-FNRS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2017.00369/full#supplementary-material
Primers and parameters of quantitative real-time RT-qPCR analysis.
References
- Anderson M., Amin S., Luise F., Zeef L., Bobola N. (2013). Differential distribution of the Ca (2+) regulator Pcp4 in the branchial arches is regulated by Hoxa2. PLOS ONE 8:e63160. 10.1371/journal.pone.0063160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bami M., Episkopou V., Gavalas A., Gouti M. (2011). Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors. PLOS ONE 6:e20197. 10.1371/journal.pone.0020197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara A., Laumonnerie C., Vilain N., Kratochwil C. F., Cankovic V., Maiorano N. A., et al. (2015). Hoxa2 selects barrelette neuron identity and connectivity in the mouse somatosensory brainstem. Cell Rep. 13 783–797. 10.1016/j.celrep.2015.09.031 [DOI] [PubMed] [Google Scholar]
- Becker J. A., Clesse D., Spiegelhalter C., Schwab Y., Le Merrer J., Kieffer B. L. (2014). Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacology 39 2049–2060. 10.1038/npp.2014.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherat O., Montaron S., Berube-Simard F. A., Aubin J., Philippidou P., Wellik D. M., et al. (2013). Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. Am. J. Physiol. Lung Cell. Mol. Physiol. 304 L817–L830. 10.1152/ajplung.00006.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. S., Capecchi M. R. (2015). Hoxc8 initiates an ectopic mammary program by regulating Fgf10 and Tbx3 expression and Wnt/beta-catenin signaling. Development 142 4056–4067. 10.1242/dev.128298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C., Shin M. M., Lee D. H., Liu J. P., Dasen J. S. (2016). Hox proteins coordinate motor neuron differentiation and connectivity programs through Ret/Gfralpha genes. Cell Rep. 14 1901–1915. 10.1016/j.celrep.2016.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Rubin E., Zhang H., Chung S., Jie C. C., Garrett E., et al. (2005). Identification of transcriptional targets of HOXA5. J. Biol. Chem. 280 19373–19380. 10.1074/jbc.M413528200 [DOI] [PubMed] [Google Scholar]
- Chotteau-Lelievre A., Dolle P., Gofflot F. (2006). Expression analysis of murine genes using in situ hybridization with radioactive and nonradioactively labeled RNA probes. Methods Mol. Biol. 326 61–87. [DOI] [PubMed] [Google Scholar]
- Dasen J. S., Jessell T. M. (2009). Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 88 169–200. 10.1016/S0070-2153(09)88006-X [DOI] [PubMed] [Google Scholar]
- Di Bonito M., Glover J. C., Studer M. (2013a). Hox genes and region-specific sensorimotor circuit formation in the hindbrain and spinal cord. Dev. Dyn. 242 1348–1368. 10.1002/dvdy.24055 [DOI] [PubMed] [Google Scholar]
- Di Bonito M., Narita Y., Avallone B., Sequino L., Mancuso M., Andolfi G., et al. (2013b). Assembly of the auditory circuitry by a Hox genetic network in the mouse brainstem. PLOS Genet. 9:e1003249. 10.1371/journal.pgen.1003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio T., Kratochwil C. F., Vilain N., Loche A., Vitobello A., Yonehara K., et al. (2013). Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science 339 204–207. 10.1126/science.1229326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins E. M., Salinas P. C. (2013). Wnts in action: from synapse formation to synaptic maintenance. Front. Cell. Neurosci. 7:162 10.3389/fncel.2013.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipietrantonio H. J., Dymecki S. M. (2009). Zic1 levels regulate mossy fiber neuron position and axon laterality choice in the ventral brain stem. Neuroscience 162 560–573. 10.1016/j.neuroscience.2009.02.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson I. J., Amin S., Hensman J. J., Kutejova E., Rattray M., Lawrence N., et al. (2012). Genome-wide occupancy links Hoxa2 to Wnt-beta-catenin signaling in mouse embryonic development. Nucleic Acids Res. 40 3990–4001. 10.1093/nar/gkr1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R., Han L., Wang Q., Wei J., Chen L., Zhang J., et al. (2015). HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget 6 27778–27793. 10.18632/oncotarget.4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Tvrdik P., Makki N., Machold R., Paxinos G., Watson C. (2013). The interfascicular trigeminal nucleus: a precerebellar nucleus in the mouse defined by retrograde neuronal tracing and genetic fate mapping. J. Comp. Neurol. 521 697–708. 10.1002/cne.23200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funfschilling U., Reichardt L. F. (2002). Cre-mediated recombination in rhombic lip derivatives. Genesis 33 160–169. 10.1002/gene.10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisen M. J., Di Meglio T., Pasqualetti M., Ducret S., Brunet J. F., Chedotal A., et al. (2008). Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLOS Biol. 6:e142. 10.1371/journal.pbio.0060142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genestine M., Lin L., Durens M., Yan Y., Jiang Y., Prem S., et al. (2015). Engrailed-2 (En2) deletion produces multiple neurodevelopmental defects in monoamine systems, forebrain structures and neurogenesis and behavior. Hum. Mol. Genet. 24 5805–5827. 10.1093/hmg/ddv301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto J., Mikoshiba K. (2011). Inositol 1,4,5-trisphosphate receptor-mediated calcium release in Purkinje cells: from molecular mechanism to behavior. Cerebellum 10 820–833. 10.1007/s12311-011-0270-5 [DOI] [PubMed] [Google Scholar]
- Gottmann K., Mittmann T., Lessmann V. (2009). BDNF signaling in the formation, maturation and plasticity of glutamatergic and GABAergic synapses. Exp. Brain Res. 199 203–234. 10.1007/s00221-009-1994-z [DOI] [PubMed] [Google Scholar]
- Hanchar H. J., Dodson P. D., Olsen R. W., Otis T. S., Wallner M. (2005). Alcohol-induced motor impairment caused by increased extrasynaptic GABA(A) receptor activity. Nat. Neurosci. 8 339–345. 10.1038/nn1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Pang Z., Bao D., Miyazaki T., Li L., Miura E., et al. (2005). Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci. 8 1534–1541. 10.1038/nn1576 [DOI] [PubMed] [Google Scholar]
- Hirota J., Ando H., Hamada K., Mikoshiba K. (2003). Carbonic anhydrase-related protein is a novel binding protein for inositol 1,4,5-trisphosphate receptor type 1. Biochem. J. 372(Pt 2) 435–441. 10.1042/BJ20030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycaj S. M., Dye B. R., Baker N. C., Larsen B. M., Burke A. C., Spence J. R., et al. (2015). Hox5 genes regulate the Wnt2/2b-Bmp4-signaling axis during lung development. Cell Rep. 12 903–912. 10.1016/j.celrep.2015.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hutlet B., Theys N., Coste C., Ahn M. T., Doshishti-Agolli K., Lizen B., et al. (2016). Systematic expression analysis of Hox genes at adulthood reveals novel patterns in the central nervous system. Brain Struct. Funct. 221 1223–1243. 10.1007/s00429-014-0965-8 [DOI] [PubMed] [Google Scholar]
- Inoue K., Terashima T., Inoue Y. (1991). Postnatal development of the pontine projections from the visual cortex of the mouse. Okajimas Folia Anat. Jpn. 67 479–492. 10.2535/ofaj1936.67.6_479 [DOI] [PubMed] [Google Scholar]
- Jeannotte L., Gotti F., Landry-Truchon K. (2016). Hoxa5: a key player in development and disease. J. Dev. Biol. 4:13 10.3390/jdb4020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier P. J. (2006). Surface clustering of metabotropic glutamate receptor 1 induced by long Homer proteins. BMC Neurosci. 7:1. 10.1186/1471-2202-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar K., Narita Y., Fadok J., Ducret S., Loche A., Kitazawa T., et al. (2017). Hox2 genes are required for tonotopic map precision and sound discrimination in the mouse auditory brainstem. Cell Rep. 18 185–197. 10.1016/j.celrep.2016.12.021 [DOI] [PubMed] [Google Scholar]
- Krishnan V., Stoppel D. C., Nong Y., Johnson M. A., Nadler M. J., Ozkaynak E., et al. (2017). Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 543 507–512. 10.1038/nature21678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J. Y., Skeberdis V. A., Jover T., Grooms S. Y., Lin Y., Araneda R. C., et al. (2001). Protein kinase C modulates NMDA receptor trafficking and gating. Nat. Neurosci. 4 382–390. 10.1038/86028 [DOI] [PubMed] [Google Scholar]
- Leergaard T. B., Lakke E. A., Bjaalie J. G. (1995). Topographical organization in the early postnatal corticopontine projection: a carbocyanine dye and 3-D computer reconstruction study in the rat. J. Comp. Neurol. 361 77–94. 10.1002/cne.903610107 [DOI] [PubMed] [Google Scholar]
- Lizen B., Claus M., Jeannotte L., Rijli F. M., Gofflot F. (2015). Perinatal induction of Cre recombination with tamoxifen. Transgenic Res. 24 1065–1077. 10.1007/s11248-015-9905-5 [DOI] [PubMed] [Google Scholar]
- Lizen B., Hutlet B., Bissen D., Sauvegarde D., Hermant M., Ahn M. T., et al. (2017). HOXA5 localization in postnatal and adult mouse brain is suggestive of regulatory roles in postmitotic neurons. J. Comp. Neurol. 525 1155–1175. 10.1002/cne.24123 [DOI] [PubMed] [Google Scholar]
- Maejima T., Wollenweber P., Teusner L. U., Noebels J. L., Herlitze S., Mark M. D. (2013). Postnatal loss of P/Q-type channels confined to rhombic-lip-derived neurons alters synaptic transmission at the parallel fiber to Purkinje cell synapse and replicates genomic Cacna1a mutation phenotype of ataxia and seizures in mice. J. Neurosci. 33 5162–5174. 10.1523/JNEUROSCI.5442-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki N., Capecchi M. R. (2011). Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol. 357 295–304. 10.1016/j.ydbio.2011.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M., Wellik D. M., Deschamps J. (2010). Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344 7–15. 10.1016/j.ydbio.2010.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K., Miura E., Miyazaki T., Kakegawa W., Emi K., Narumi S., et al. (2010). Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328 363–368. 10.1126/science.1185152 [DOI] [PubMed] [Google Scholar]
- Mayer M., Bercsenyi K., Geczi K., Szabo G., Lele Z. (2010). Expression of two type II cadherins, Cdh12 and Cdh22 in the developing and adult mouse brain. Gene Expr. Patterns 10 351–360. 10.1016/j.gep.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Mizutani A., Kuroda Y., Futatsugi A., Furuichi T., Mikoshiba K. (2008). Phosphorylation of Homer3 by calcium/calmodulin-dependent kinase II regulates a coupling state of its target molecules in Purkinje cells. J. Neurosci. 28 5369–5382. 10.1523/JNEUROSCI.4738-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendran T., Hardy L. R. (2011). Calcium/calmodulin-dependent protein kinase IV mediates distinct features of basal and activity-dependent dendrite complexity. Neuroscience 199 548–562. 10.1016/j.neuroscience.2011.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y., Rijli F. M. (2009). Hox genes in neural patterning and circuit formation in the mouse hindbrain. Curr. Top. Dev. Biol. 88 139–167. 10.1016/S0070-2153(09)88005-8 [DOI] [PubMed] [Google Scholar]
- Nicol X., Gaspar P. (2014). Routes to cAMP: shaping neuronal connectivity with distinct adenylate cyclases. Eur. J. Neurosci. 39 1742–1751. 10.1111/ejn.12543 [DOI] [PubMed] [Google Scholar]
- Nolte C., Krumlauf R. (2007). “Expression of Hox genes in the nervous system of vertebrates,” in HOX Gene Expression ed. Papageorgio S. (Berlin: Springer Science+Business Media; ) 14–41. 10.1007/978-0-387-68990-6_2 [DOI] [Google Scholar]
- Oancea E., Meyer T. (1998). Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95 307–318. 10.1016/S0092-8674(00)81763-8 [DOI] [PubMed] [Google Scholar]
- Oury F., Murakami Y., Renaud J. S., Pasqualetti M., Charnay P., Ren S. Y., et al. (2006). Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science 313 1408–1413. 10.1126/science.1130042 [DOI] [PubMed] [Google Scholar]
- Parys J. B., Decuypere J. P., Bultynck G. (2012). Role of the inositol 1,4,5-trisphosphate receptor/Ca2+-release channel in autophagy. Cell Commun. Signal. 10:17. 10.1186/1478-811X-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualetti M., Diaz C., Renaud J. S., Rijli F. M., Glover J. C. (2007). Fate-mapping the mammalian hindbrain: segmental origins of vestibular projection neurons assessed using rhombomere-specific Hoxa2 enhancer elements in the mouse embryo. J. Neurosci. 27 9670–9681. 10.1523/JNEUROSCI.2189-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Dasen J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80 12–34. 10.1016/j.neuron.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Walsh C. M., Aubin J., Jeannotte L., Dasen J. S. (2012). Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 15 1636–1644. 10.1038/nn.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Cannistraci C. V., Katayama S., Bajic V. B., Tan K., et al. (2010). An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140 744–752. 10.1016/j.cell.2010.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezsohazy R., Saurin A. J., Maurel-Zaffran C., Graba Y. (2015). Cellular and molecular insights into Hox protein action. Development 142 1212–1227. 10.1242/dev.109785 [DOI] [PubMed] [Google Scholar]
- Rodriguez C. I., Dymecki S. M. (2000). Origin of the precerebellar system. Neuron 27 475–486. 10.1016/S0896-6273(00)00059-3 [DOI] [PubMed] [Google Scholar]
- Roux M., Laforest B., Capecchi M., Bertrand N., Zaffran S. (2015). Hoxb1 regulates proliferation and differentiation of second heart field progenitors in pharyngeal mesoderm and genetically interacts with Hoxa1 during cardiac outflow tract development. Dev. Biol. 406 247–258. 10.1016/j.ydbio.2015.08.015 [DOI] [PubMed] [Google Scholar]
- Ryu K., Yokoyama M., Yamashita M., Hirano T. (2012). Induction of excitatory and inhibitory presynaptic differentiation by GluD1. Biochem. Biophys. Res. Commun. 417 157–161. 10.1016/j.bbrc.2011.11.075 [DOI] [PubMed] [Google Scholar]
- Sadakata T., Furuichi T. (2009). Developmentally regulated Ca2+-dependent activator protein for secretion 2 (CAPS2) is involved in BDNF secretion and is associated with autism susceptibility. Cerebellum 8 312–322. 10.1007/s12311-009-0097-5 [DOI] [PubMed] [Google Scholar]
- Sanlioglu S., Zhang X., Baader S. L., Oberdick J. (1998). Regulation of a Purkinje cell-specific promoter by homeodomain proteins: repression by engrailed-2 vs. synergistic activation by Hoxa5 and Hoxb7. J. Neurobiol. 36 559–571. [DOI] [PubMed] [Google Scholar]
- Santagati F., Minoux M., Ren S. Y., Rijli F. M. (2005). Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development 132 4927–4936. 10.1242/dev.02078 [DOI] [PubMed] [Google Scholar]
- Schiedlmeier B., Santos A. C., Ribeiro A., Moncaut N., Lesinski D., Auer H., et al. (2007). HOXB4’s road map to stem cell expansion. Proc. Natl. Acad. Sci. U.S.A. 104 16952–16957. 10.1073/pnas.0703082104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. E., Zhou Y. D., Zhang G., Jin Z., Stoppel D. C., Anderson M. P. (2011). Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 3:103ra197. 10.1126/scitranslmed.3002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaries S., Lemieux M., Aubin J., Jeannotte L. (2007). Comparative analysis of Hoxa5 allelic series. Genesis 45 218–228. 10.1002/dvg.20292 [DOI] [PubMed] [Google Scholar]
- Tomas-Roca L., Corral-San-Miguel R., Aroca P., Puelles L., Marin F. (2016). Crypto-rhombomeres of the mouse medulla oblongata, defined by molecular and morphological features. Brain Struct. Funct. 221 815–838. 10.1007/s00429-014-0938-y [DOI] [PubMed] [Google Scholar]
- Tu J. C., Xiao B., Yuan J. P., Lanahan A. A., Leoffert K., Li M., et al. (1998). Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron 21 717–726. 10.1016/S0896-6273(00)80589-9 [DOI] [PubMed] [Google Scholar]
- Uemura T., Lee S. J., Yasumura M., Takeuchi T., Yoshida T., Ra M., et al. (2010). Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141 1068–1079. 10.1016/j.cell.2010.04.035 [DOI] [PubMed] [Google Scholar]
- Waltes R., Duketis E., Knapp M., Anney R. J., Huguet G., Schlitt S., et al. (2014). Common variants in genes of the postsynaptic FMRP signalling pathway are risk factors for autism spectrum disorders. Hum. Genet. 133 781–792. 10.1007/s00439-013-1416-y [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang M. (2012). The role of Ca2+-stimulated adenylyl cyclases in bidirectional synaptic plasticity and brain function. Rev. Neurosci. 23 67–78. 10.1515/revneuro-2011-0063 [DOI] [PubMed] [Google Scholar]
- Wang S. S., Kloth A. D., Badura A. (2014). The cerebellum, sensitive periods, and autism. Neuron 83 518–532. 10.1016/j.neuron.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V. Y., Zoghbi H. Y. (2001). Genetic regulation of cerebellar development. Nat. Rev. Neurosci. 2 484–491. 10.1038/35081558 [DOI] [PubMed] [Google Scholar]
- Wojcik S. M., Rhee J. S., Herzog E., Sigler A., Jahn R., Takamori S., et al. (2004). An essential role for vesicular glutamate transporter 1 (VGLUT1) in postnatal development and control of quantal size. Proc. Natl. Acad. Sci. U.S.A. 101 7158–7163. 10.1073/pnas.0401764101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. T., Athos J., Figueroa X. A., Pineda V. V., Schaefer M. L., Chavkin C. C., et al. (1999). Calcium-stimulated adenylyl cyclase activity is critical for hippocampus-dependent long-term memory and late phase LTP. Neuron 23 787–798. 10.1016/S0896-6273(01)80036-2 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao J. H., Suo Z. M. (2017). Knockdown of HOXA5 inhibits the tumorigenesis in esophageal squamous cell cancer. Biomed. Pharmacother. 86 149–154. 10.1016/j.biopha.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Zhuang G. Z., Keeler B., Grant J., Bianchi L., Fu E. S., Zhang Y. P., et al. (2015). Carbonic anhydrase-8 regulates inflammatory pain by inhibiting the ITPR1-cytosolic free calcium pathway. PLOS ONE 10:e0118273. 10.1371/journal.pone.0118273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and parameters of quantitative real-time RT-qPCR analysis.


