Abstract
Antigen stimulation induces a rapid proliferation of B cells for expansion of specific B cell clones and their further differentiation into antibody-producing cells in germinal centers of T-dependent antigen-immunized mice. Previously, we identified a 210-kDa germinal center-associated nuclear protein (GANP) that is up-regulated selectively in germinal centers and carries an MCM-binding domain in the carboxyl-terminal side. In addition, here, we found a region (from 414 to 550 aa) in GANP molecule that is slightly similar to the known DNA-primase component p49. The recombinant GANP fragment covering this region synthesizes RNA primers for extension by DNA polymerase I with single-stranded DNA templates in vitro. GANP DNA-primase activity is controlled by phosphorylation at Ser502 that is induced by CD40-mediated signaling in vitro and in the germinal center B cells stimulated with antigen in vivo. Overexpression of ganp cDNA in Daudi B cells caused the increased DNA synthesis more than the levels of the mock-transfectants. These evidences suggested that the novel DNA-primase GANP is involved in regulation of cell proliferation of antigen-driven B cells in germinal centers.
Antigen stimulation induces a rapid proliferation of Ag-specific B cell clones to generate sufficient numbers in germinal centers (GCs) of the lymphoid follicles. Ag-driven B cells appear at the dark zone of GCs as rapidly proliferating centroblasts and then further differentiate during the transition to the light zone as centrocytes whose cell cycle is arrested abruptly (1, 2). Such Ag-driven B cells undergo various differentiation processes for the affinity maturation of the Abs, the isotype switching from IgM to IgG, and the selection of B cells with the expression of the effective and nonharmful B cell receptor during the maturation and differentiation in GCs (3, 4). In our search of the molecules involved in differentiation of GC-B cells, we currently identified a molecule, named germinal center-associated nuclear protein (GANP), that is up-regulated selectively in GC-B cells (5, 6). Ganp gene allele encodes two alternative RNA-spliced isoforms as an 80-kDa Map80 (7) and a larger 210-kDa protein GANP. Both proteins carry a domain that associates with a replication-licensing factor, MCM3 (8–10), that has suggested a functional association of the 210-kDa GANP with DNA replication of rapid proliferation of B cells.
The binding of the MCM complex to the origin-binding complex through Cdc6/Cdc18 and/or Ctd1 along with chromatin is essential for DNA replication in the late G1- and S-phases. This association is presumably because of the DNA-helicase activity of the MCM complex (11). After DNA replication, MCM3 is thought to be released from the chromatin and exported (8, 10). The binding and subsequent release of MCM3 from chromatin is considered a necessary event for DNA replication, but restricted once in cell division in accordance with cell cycle progression and the need to guarantee proper DNA content in daughter cells (12). Map80, a short form of the ganp gene product, binds to MCM3 through a Map-box as an import factor for MCM3 (7).
Here, we found a similarity of GANP with the p49 component of conventional DNA-primase complex (13). DNA-primase activity is known to exert a fundamental role for lagging strand synthesis as the Okazaki fragment with discontinuous DNA extension (14). DNA-primase activity is confined to a DNA-primase complex composed of the p49, p58, and p180 (14). The novel DNA-primase domain found in GANP suggested its potentially important function for DNA replication in rapid proliferating cells. We identified an RNA/DNA primase activity of the putative primase domain of the 210-kDa GANP, and therefore now have designated GANP as GC-Associated DNA Primase. GANP has two critical domains for DNA replication: with the binding activity to MCM3 containing DNA-helicase activity (11) and with the DNA-primase activity in the amino-terminal side, as reported here.
Materials and Methods
Animals.
Male C57BL/6 mice and Lewis rats were purchased from Charles River Breeding Laboratories. All animals were maintained in the Center for Animal Resources and Development (CARD), Kumamoto University.
Generation of Recombinant Primase Domain of GANP (GANP-PD).
The ganp cDNA corresponding to amino acids 138 to 560 was inserted into pET-29b (+) vector (Novagen; pET-ganp-PD). The recombinant GANP-PD was purified from bacteria harboring pET-ganp-PD after isopropyl-β-d(−)-thiogalactopyranoside (IPTG) induction using His-Bind Resin (Novagen). Subsequently, the buffer used to dissolve the recombinant protein was replaced with 50 mM potassium phosphate (pH 7.5), 3 mM 2-mercaptoethanol, 0.1 mM MnSO4, and 1 mM MgCl2 (15). For long storage, 10% glycerol was added. Mutagenesis was performed as described (16).
In Vitro DNA-Primase Assay.
The reaction mixture was prepared as 20 mM Tris⋅HCl (pH7.5), 10 mM MgCl2, 1 mM DTT, 0.5 μg M13 single-stranded DNA (ssDNA), 2 mM ATP, 400 μM GTP, 400 μM CTP, 400 μM UTP, 100 μM dNTP mixture, and 10 μM [3H]dTTP. After adding 0.1 unit DNA polymerase I (pol I; Klenow fragment) and recombinant GANP-PD, the reaction was performed at 37°C for 30 min. The samples were applied onto DE81 filters (Whatman), the filters were washed with 5% sodium phosphate buffer several times, and the radioactivity was measured by liquid scintillation counting.
In Vitro Kinase Assay.
The Cdk2 complex was immunoprecipitated using anti-Cdk2 Ab (Santa Cruz Biotechnology) from BAL17 cell lysates. After washing with the cell lysis buffer, the immunoprecipitates were resuspended in the kinase reaction buffer (17). The Cdk2 complex was radiolabeled with 5 μCi [γ-32P]ATP (Amersham Pharmacia) in the presence of 5 μg His-tagged recombinant GANP-PD. The radiolabeled proteins were analyzed on SDS/PAGE, and the gel was fixed, dried, and exposed to x-ray film.
Stable ganp-Transfectants.
Human Daudi B cells were harvested, and the cell number was adjusted to 1 × 107 cells per 1 ml in RPMI 1640 medium with 10% FCS as described elsewhere (17). A mouse ganp cDNA, introduced into a vector under the chicken β-actin promoter, was transfected into Daudi by electroporation and selected with G418 (1 mg/ml; Life Technologies, Grand Island, NY).
Cell-Cycle Analysis.
Cells were suspended in hypotonic propidium iodide (PI; Sigma) solution (50 μg/ml PI in 0.1% sodium citrate and 0.1% Triton X-100). After incubation for 4 h at 4°C, samples were analyzed by FACScan with modfit lt software (Becton Dickinson).
Establishment of mAb Against pSer502 of GANP.
Lewis rats were s.c. immunized with the synthetic peptides (CHKKKIpSPSKKL) coupled to keyhole limpet hemocyanin (KLH) in complete Freund's adjuvant (Difco), and the hybridomas secreting rat mAbs were performed as described (17). After hypoxanthine/aminopterin/thymidine (HAT) selection, the hybridomas were screened by ELISA, and the specificity was confirmed by immunoprecipitation and immunostaining method.
Immunocytostaining and Immunohistochemical Staining.
Spleen B cells from 7-wk-old C57BL/6 mice were prepared as described (18). After stimulation of purified B cells in vitro for 48 h with either one of reagents of lipopolysaccharide (LPS; 10 μg/ml; Sigma), anti-IgM Ab (10 μg/ml; ICN), and anti-CD40 mAb (10 μg/ml; LB429; ref. 19), cells were harvested and cytosmears were prepared on silanized slides. For the GCs, C57BL/6 mice were immunized with trinitrophenyl-keyhole limpet hemocyanin (TNP-KLH; Biosearch), and the spleen sections were prepared as described previously (18). For immunohistological staining, slides were incubated with the mAbs prepared in our laboratory as a control rat mAb of the same isotype recognizing glutathione S-transferase, anti-GANP mAb (29-15; ref. 5), and anti-pSer502 (PG/103) mAb, followed with alkaline phosphatase-conjugated goat anti-rat IgG Ab (ICN) to develop by Vector Red (Vector Laboratories). The serial 6-μm frozen sections were prepared from the Ag-immunized spleen and were stained with biotinylated reagent of peanut agglutinin (PNA; Vector Laboratories), anti-CR1 mAb (BD PharMingen), or anti-IgD mAb (5) in combination with streptavidin-peroxidase (Kirkegaard & Perry Laboratories), followed by 3,3′-diaminobenzidine tetrahydrochloride (Dojindo, Kumamoto, Japan). The double staining with rat anti-pSer502 (PG/103) mAb was also carried out, and only the double staining profiles were shown in the figures.
Results and Discussion
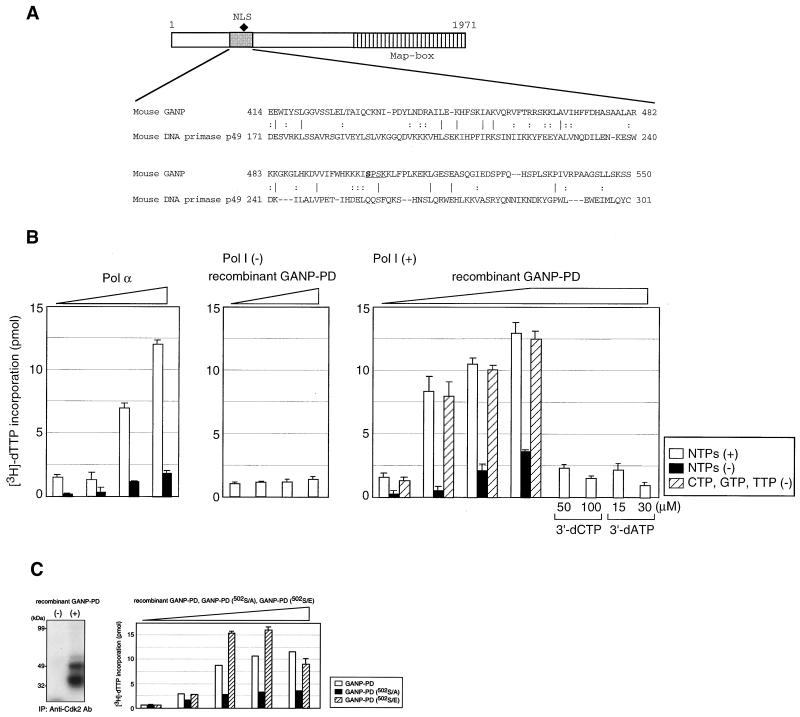
In addition to the MCM3 recognition region, GANP carries a motif slightly homologous to the mouse DNA-primase p49 subunit (Fig. 1A; homology region shown by shaded box with the comparison by nucleotide 11.5% and by amino acid with 9.5% identity or 24.8% similarity; ref. 20). DNA-primase comprises two subunits of p49 and p58, and both associated tightly with the 180-kDa DNA polymerase α (pol α). The p49 subunit contains the catalytic core of primase and binds to ssDNA and catalyses phosphodiesterase bond formation. It is thought that the p58 subunit tethers p49 to the p180 pol α and mediates the transfer of the newly generated primer-template to the pol α active site. Asp109, Asp111, and Asp306 of p49 are thought to form the metal-binding core of the active site. The sequence of primase, which can be compared with that of pol β, consists of two discrete domains, a 31-kDa domain that contains phosphodiester bond-forming activity and an 8-kDa domain that can bind DNA and contains deoxyribose lyase activity (14). We have not detected the similarity in the entire sequence of GANP molecule to these known residues as a DNA-primase member; therefore, it is necessary to determine whether GANP has a DNA-primase activity. Using a recombinant protein corresponding to the putative primase domain of GANP, DNA-primase activity was measured with an unprimed ssDNA in the presence of pol I Klenow fragment as primer for RNA synthesis-dependent DNA synthesis. Pol α associated with DNA-primase activity produced RNA primers that were then extended by pol α as a positive control (Fig. 1B Left). Pol I alone was used as a negative control and did not display comparable activity (Fig. 1B Right). GANP-PD did not drive DNA synthesis in the absence of pol I (Fig. 1B Center) but allowed the initiation of DNA extension by pol I (Fig. 1B Right). Dose-dependent DNA-primase activity was also detected with only one of the ribonucleotides and was inhibited by triphosphate derivatives, 5′-triphosphates of 3′-deoxynucleotides 3′-dCTP and 3′-dATP (Fig. 1B Right; ref. 21). These data indicate that GANP-PD provides RNA primers for initiation of DNA synthesis of the lagging strand as Okazaki fragments as well as by the conventional DNA-primase complex. A novel complex of DNA-primase activity might govern DNA replication potentially associated with DNA-helicase complex of MCMs in mammalian cells.
Figure 1.
Intrinsic DNA-primase activity in GANP. (A) A putative GANP-PD. The GANP sequence (from amino acid 414 to 550) is compared with a DNA-primase p49 subunit (from amino acid 171 to 301). (B) In vitro DNA-primase assay. The GANP-PD was tagged with 6 × His and expressed in Escherichia coli to purify the recombinant GANP-PD. DNA-primase activity is measured by the incorporation of [3H]dTTP into M13 ssDNA with DNA polymerase. As a positive control, pol α that contains endogenous primase activity shows efficient DNA synthesis in the presence of four ribonucleotides to initiate RNA primers at first. As a negative control, the primase activity is measured with the recombinant GANP-PD in the absence of pol I. Activity of GANP-PD is measured in the presence of pol I with or without four ribonucleotides, or with only ATP. Effects of the RNA polymerase, including primase inhibitors, are examined to confirm the primase activity. (C Left) In vitro kinase assay with Cdk2. The recombinant GANP-PD was phosphorylated with cellular Cdk2 complex immunoprecipitated with anti-Cdk2 Ab. Incorporation of [γ-32P]ATP was visualized by autoradiography after SDS/PAGE. The 49-kDa GANP-PD and the degradation products are phosphorylated. (Right) Mutant GANP-PD was prepared by the substitution of Ser502 to Ala (502S/A) or Glu (502S/E). Primase activity was measured in the presence of pol I with the recombinant protein as in Fig. 1B. All data were confirmed by the repeated experiments.
Because GANP expression is selective in GC-B cells, the inducible nature of DNA-primase activity is particular in comparison with the conventional DNA-primase complex that appears in every kind of cell. To study the inducible regulation of GANP DNA-primase during cell activation, we examined phosphorylation induced by various kinds of kinases. We observed that the cell cycle-associated kinase Cdks induced phosphorylation of GANP in vitro. Examination of immunoprecipitates of Cdk2 from B cells revealed phosphorylation of GANP-PD at a consensus sequence of Cdk phosphorylation at Ser502 (S/T-P-X-K/R) (Fig. 1C Left; ref. 22). The amino acid substitution of Ser502 to Ala, a nonphosphorylatable mutant, caused loss of the DNA-primase activity, which also confirmed the presence of DNA-primase activity of this region. A substitution of Ser502 with Glu, providing molecular mimicry to phosphorylated Ser, showed higher DNA-primase activity (Fig. 1C Right). Ser502 is in the nuclear localization signal sequence of GANP and is phosphorylated, suggesting that GANP DNA-primase may change cellular localization and alter the activity. The mutant analysis clearly suggested that GANP DNA-primase activity is strictly regulated by the phosphorylation at Ser502. The conventional DNA-primase complex exerts rather a stable and universal function for DNA replication of various cells with cell cycle progression. The similar phosphorylation site is not found in the primase domain of the conventional DNA-primase of various evolutionarily developed cells.
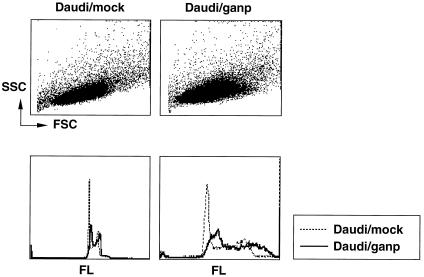
If GANP DNA-primase works in cells, it is expected to have an excess amount of DNA content in each cell, as was displayed in Saccharomyces mutant cells by flow cytometric analysis (23). The effect of GANP overexpression was studied by introduction of ganp cDNA into the Daudi B cell line. Most of the cells did not survive long-term, but some transfectants expressing the introduced ganp gene were obtained. DNA content in these ganp-transfectants showed increases in DNA content as compared with the diploid and tetraploid profile of mock-transfectants (Fig. 2). The proportions of G1-, S-, G2-, and M-phases were almost the same with mock-transfectants, indicating that ganp-transfectants had increased DNA in each cell cycle but a normal cycling period. Such increase of DNA contents was demonstrated by the destruction of p34cdc2 in fission yeast cells that disrupt the dependency and undergo an extra round of DNA replication without an intervening mitosis (24). Considering the observation from others (23) that also showed the similar oversynthesis of DNA contents, these results suggest that GANP DNA-primase activity would support DNA synthesis in rapidly proliferating cells. It would be important to study the interaction of GANP with the molecules involved in cell cycle regulation.
Figure 2.
Effect of over-expressed ganp in cell cycling. Ganp- and mock-transfectants were treated with the PI solution and characterized for cell size and DNA ploidity. Flow cytometric analysis was carried out with side scatter (SSC) and forward scatter (FSC) or with PI staining.
To study whether phosphorylation of GANP DNA-primase is stimulation dependent, we prepared a mAb specific to the phospho-Ser at 502 of GANP (pSer502) and determined that it recognizes GANP but not the short Map80 form in wild-type mice (data not shown). We compared the GANP expression in B cells after stimulation in vitro. Resting spleen B cells were not positive with both anti-GANP and anti-pSer502 mAb, but anti-CD40 stimulation or LPS induced the expression of GANP in B cells in vitro (Fig. 3A; stained with 29-15). CD40-stimulation induced a marked phosphorylation of Ser502 in vitro; however, LPS could not induce such phosphorylation response (Fig. 3A; stained with anti-pSer502 mAb). LPS stimulation is not sufficient enough to result in the induction of phosphorylation, suggesting that the phosphorylation at Ser502 requires CD40-dependent signal(s) in B cells. Next, we examined whether the phosphorylation of GANP-Ser502 occurs in GC-B cells after Ag-immunization. Spleens immunized with T-dependent Ag, TNP-KLH, demonstrated that GC-B cells up-regulated the pSer502 of GANP as well as the increase of GANP expression in the same area of PNA+ GC-B cells as demonstrated previously (Fig. 3B Left; ref.5). The expression of pSer502 GANP is up-regulated in the PNA+ area surrounded by IgD+ cells (Fig. 3B Center) and appears in both CR1+ and CR1− areas. The pSer502 GANP is also expressed in cells surrounded by the CR1+ follicular dendritic cell network, suggesting that the phosphorylation of GANP DNA-primase is induced in most of GC-B cells, including centroblasts and centrocytes. These results clearly suggested that Ag stimulation induces GANP expression and that the active form of DNA-primase is selectively up-regulated in GC-B cells with the costimulatory signal mediated through CD40/CD40L. GANP DNA-primase would be recruited to the origin-binding complex with DNA-helicase of MCMs, which is presumably efficient for rapid cell growth. The conventional DNA-primase, composed of p49, p58, and p180 pol α subunit, first synthesizes RNA primers up to 10 mers (14). However, the activity might not be sufficient to coordinate the enormous rate of proliferation that takes place in GC-B cells.
Figure 3.
Induction of phosphorylated form of GANP DNA-primase in activated B cells in vitro and in vivo. (A) Normal spleen B cells were stimulated in vitro for 48 h with the stimulatory reagents as indicated. GANP expression was detected by anti-GANP mAb, and the induction of phosphorylation at Ser502 was detected by anti-pSer502 (PG/103) mAb in comparison with a control anti-glutathione S-transferase mAb. (B) The spleen sections from the TNP-KLH-immunized mice were stained histochemically as described in Materials and Methods. (Left) Most of pSer502 signal (blue) overlaps with PNA+ cells (brown). (Center) pSer502 signal (blue) appears up-regulated selectively in GC-B cells surrounded with IgD+ cells (brown). (Right) The up-regulation of pSer502 signal (blue) is detected in both CR1+ (brown) and CR1− area of GCs. The sections were prepared by the serial manner so that the profiles of the multiple markers are to be superimposed.
The conventional DNA-primase complex is evolutionarily conserved and considered as a universal component involved in the DNA replication. The counterpart of GANP DNA-primase was found in a computer search of the Saccharomyces Genome Database. Because the GANP homologue SAC3 in Saccharomyces cerevisiae does not carry a DNA-primase domain (25), we predict that GANP DNA-primase activity would be necessary in organs with extraordinary rapid cell proliferation. A recent study demonstrated that Map80, alternatively called MCM3AP, has an acetyltransferase activity on MCM3 in vivo (26). GANP has a stimulation-dependent DNA-primase domain and an acetyltransferase domain against MCM-complex of DNA-helicase activity (11). Both might interact for DNA replication of mammalian cells.
GANP bears two molecular functions of DNA-primase and the association with DNA-helicase complex of MCMs, both of which will presumably participate in DNA replication of Ag-driven B cells. B cells at this stage proliferate so rapidly that conventional DNA-primase α may not supply sufficient amount of RNA primers in a short period. GC-B cells generate a high frequency somatic hypermutation of V region genes through the processes of clonal expansion and the subsequent differentiation. Recent observations demonstrated that the generation of somatic hypermutation accompanies double-strand breaks (DSBs) of V region genes (27, 28). A model suggested that three molecular events are necessary for generation of V-region somatic hypermutation (29). The first molecular event is an insertion of nicks at the sequence-selective sites into the V region gene. This DNA injury is spontaneously or B cell specifically converted to DSBs, which will be then repaired by error-prone DNA polymerase(s). To confirm this model, it is necessary to determine the molecular members for generation of site-specific nicks, DSBs, and the error-prone repair mechanism that undergo in GC-B cells. Although there is no direct interaction with GANP, it would be important to study the involvement of B cell DNA-primase activity in such molecular events of GC-B cells.
The DNA-primase complex is evolutionarily conserved, and the mechanism of DNA replication and repair has been studied in various cells. However, GANP DNA-primase activity is a novel component of DNA replication that appears related to a cell type-specific function. GANP might be involved in the acceleration of DNA replication of precursor cells to achieve the clonal expression. This could be the case for Ag-driven B cells at a particular differentiation stage in peripheral lymphoid tissues. It may be related to fate specification of lymphoid cells after cell cycle exit. A thorough understanding of GANP DNA-primase complex-mediated mechanisms should be informative about many aspects of cell differentiation, DNA replication, and DNA repair in highly developed mammalian cells.
Acknowledgments
This work was supported by the Ministry of Education, Science, Culture, Sports and Technology in Japan.
Abbreviations
- LPS
lipopolysaccharide
- PD
primase domain
- PI
propidium iodide
- PNA
peanut agglutinin
- pol α
DNA polymerase α
- pol I
DNA polymerase I
- ssDNA
single-stranded DNA
- TNP-KLH
trinitrophenyl-keyhole limpet hemocyanin
- GC
germinal center
- GANP
GC-associated nuclear protein or GC-associated DNA primase
References
- 1.MacLennan I C M. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Kelsoe G. Immunity. 1996;4:107–111. doi: 10.1016/s1074-7613(00)80675-5. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 4.Kosco-Vilbois M H, Zentgraf H, Gerdes J, Bonnefoy J-H. Immunol Today. 1997;18:225–230. doi: 10.1016/s0167-5699(97)01048-7. [DOI] [PubMed] [Google Scholar]
- 5.Kuwahara K, Yoshida M, Kondo E, Sakata A, Watanabe Y, Abe E, Kouno Y, Tomiyasu S, Fujimura S, Tokuhisa T, Kimura H, Ezaki T, Sakaguchi N. Blood. 2000;95:2321–2328. [PubMed] [Google Scholar]
- 6.Abe E, Kuwahara K, Yoshida M, Suzuki M, Terasaki H, Matsuo Y, Takahashi E, Sakaguchi N. Gene. 2000;255:219–222. doi: 10.1016/s0378-1119(00)00336-x. [DOI] [PubMed] [Google Scholar]
- 7.Takei Y, Tsujimoto G. J Biol Chem. 1998;273:22177–22180. doi: 10.1074/jbc.273.35.22177. [DOI] [PubMed] [Google Scholar]
- 8.Kimura H, Nozaki N, Sugimoto K. EMBO J. 1994;13:4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong J P, Thoemmes P, Blow J J. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 10.Ritzi M, Knippers R. Gene. 2000;245:13–20. doi: 10.1016/s0378-1119(00)00020-2. [DOI] [PubMed] [Google Scholar]
- 11.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 12.Tye B K. Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 13.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 14.Arezi B, Kuchta R D. Trends Biochem Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 15.Schneider A, Smith R W P, Kautz A R, Weisshart K, Grosse F, Nasheuer H-P. J Biol Chem. 1998;273:21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- 16.Imai Y, Matsushima Y, Sugimura T, Terada M. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara K, Matsuo T, Nomura J, Igarashi H, Inui S, Kimoto M, Sakaguchi N. J Immunol. 1994;152:2742–2752. [PubMed] [Google Scholar]
- 18.Igarashi H, Kuwata N, Kiyota K, Sumita K, Suda T, Ono S, Bauer S R, Sakaguchi N. Blood. 2001;97:2680–2687. doi: 10.1182/blood.v97.9.2680. [DOI] [PubMed] [Google Scholar]
- 19.Nomura J, Inui S, Yamasaki T, Kataoka S, Maeda K, Nakanishi K, Sakaguchi N. Immunol Lett. 1995;45:195–203. doi: 10.1016/0165-2478(95)00006-q. [DOI] [PubMed] [Google Scholar]
- 20.Miyazawa H, Izumi M, Tada S, Takada R, Masutani M, Ui M, Hanaoka F. J Biol Chem. 1993;268:8111–8122. [PubMed] [Google Scholar]
- 21.Izuta S, Kohsaka-Ichikawa M, Yamaguchi T, Saneyoshi M. J Biochem. 1996;119:1038–1044. doi: 10.1093/oxfordjournals.jbchem.a021345. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa M, Higashi H, Jung H K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths D J, Liu V F, Nurse P, Wang T S. Mol Biol Cell. 2001;12:115–128. doi: 10.1091/mbc.12.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broek D, Bartlett R, Crawford K, Nurse P. Nature (London) 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- 25.Bauer A, Koelling R. J Cell Sci. 1996;109:1575–1583. doi: 10.1242/jcs.109.6.1575. [DOI] [PubMed] [Google Scholar]
- 26.Takei Y, Swietlik M, Tanoue A, Tsujimoto G, Kouzarides T, Laskey R. EMBO Rep. 2001;2:119–123. doi: 10.1093/embo-reports/kve026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 28.Papavasiliou F N, Schatz D G. Nature (London) 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs H, Bross L. Curr Opin Immunol. 2001;13:208–218. doi: 10.1016/s0952-7915(00)00206-5. [DOI] [PubMed] [Google Scholar]