Abstract
Background
Periodontitis is the major cause of tooth loss in adults and is linked to systemic illnesses, such as cardiovascular disease and stroke. The development of rapid point-of-care (POC) chair side diagnostics has the potential for the early detection of periodontal infection and progression to identify incipient disease and reduce health care costs. However, validation of effective diagnostics requires the identification and verification of biomarkers correlated with disease progression. This clinical study sought to determine the ability of putative host- and microbially derived biomarkers to identify periodontal disease status from whole saliva and plaque biofilm.
Methods
One hundred human subjects were equally recruited into a healthy/gingivitis group or a periodontitis population. Whole saliva was collected from all subjects and analyzed using antibody arrays to measure the levels of multiple proinflammatory cytokines and bone resorptive/turnover markers.
Results
Salivary biomarker data were correlated to comprehensive clinical, radiographic, and microbial plaque biofilm levels measured by quantitative polymerase chain reaction (qPCR) for the generation of models for periodontal disease identification. Significantly elevated levels of matrix metalloproteinase (MMP)-8 and -9 were found in subjects with advanced periodontitis with Random Forest importance scores of 7.1 and 5.1, respectively. The generation of receiver operating characteristic curves demonstrated that permutations of salivary biomarkers and pathogen biofilm values augmented the prediction of disease category. Multiple combinations of salivary biomarkers (especially MMP-8 and-9 and osteoprotegerin) combined with red-complex anaerobic periodontal pathogens (such as Porphyromonas gingivalis or Treponema denticola) provided highly accurate predictions of periodontal disease category. Elevated salivary MMP-8 and T. denticola biofilm levels displayed robust combinatorial characteristics in predicting periodontal disease severity (area under the curve = 0.88; odds ratio = 24.6; 95% confidence interval: 5.2 to 116.5).
Conclusions
Using qPCR and sensitive immunoassays, we identified host- and bacterially derived biomarkers correlated with periodontal disease. This approach offers significant potential for the discovery of biomarker signatures useful in the development of rapid POC chairside diagnostics for oral and systemic diseases. Studies are ongoing to apply this approach to the longitudinal predictions of disease activity.
Keywords: Diagnosis, periodontal disease, saliva
Periodontal disease is the leading cause of tooth loss in adults.1 Periodontitis is initiated by tooth-associated microbial biofilms triggering an altered host response leading to soft tissue inflammation and alveolar bone loss. Periodontal infections are implicated in a variety of other diseases, such as cardiovascular disease, stroke, and aspiration pneumonia, whereby the microbial biofilm serves as a “slow-delivery system” of oral pathogens adhering to teeth, leading to a chronic microbial challenge and downstream effects of an altered host response.2 Diagnostic methods used in clinical practice today lack the ability to detect the onset of inflammation and to identify those patients who are susceptible to future disease progression. Oral fluid–based point-of-care (POC) diagnostics are commonly used in medicine and, more recently, are being adapted for the potential “chairside” determination of oral diseases.3 The latest clinical applications use new “lab-on-a-chip” (LOC) technologies as rapid POC diagnostic tests for systemic infectious diseases4,5 and periodontal disease.6 The human salivary proteome project, supported by the United States National Institute of Dental and Craniofacial Research, Bethesda, Maryland, has generated further emphasis on the use of proteomic markers for disease diagnosis.7 The identification of the proteomic content of human saliva in diagnostic tests, assessing the fingerprint of different human illnesses, generally suggests the probability that multianalyte detection approaches will surpass conventional clinical diagnostic procedures using single biomarkers.
The use of oral fluids in oral-based diagnostics have proven to be easy to use for POC application8 in the detection of oral cancer9,10 or human immunodeficiency virus infection.11 Furthermore, the use of microfluidic devices as examples of LOC technology offers significant potential for rapid saliva diagnosis for widespread public health purposes.6,12 However, for periodontal disease determination, most research has focused primarily on gingival crevicular fluid (GCF) biomarkers that provide local disease status, but it represents a cumbersome, difficult-to-use approach for clinical application. 13 Easy-to-access saliva contains locally and systemically derived mediators of periodontal disease and, thus, offers significant potential for the assessment of periodontal disease status and risk.14
Although a single specific target biomarker for periodontal disease has not been identified, combinations of putative biomarkers of disease have been evaluated in GCF and demonstrated significant potential as panels of targets for the development of an oral fluid fingerprint of periodontal disease status. Given the multifaceted pattern of periodontal disease as a continuum of infection to inflammatory dysregulation and subsequent bone loss, specific biomarkers, such as matrix metalloproteinase (MMP)-8, interleukin (IL)-1β and -6, and type I collagen pyridinoline cross-linked telopeptide (ICTP), have been assessed in GCF singularly for disease identification.15 This approach of developing “biologic phenotypes” that consider the microbial and inflammatory response may be useful in the development of patient disease classifications with implications in targeted therapeutics.16,17
Here we demonstrate the validation of multiple proinflammatory and bone-specific biomarkers from whole saliva coupled with microbial biofilm pathogens for the identification of periodontal disease. This unique combinatorial approach resulted in robust predictions of periodontitis in human subjects.
MATERIALS AND METHODS
Subjects
This clinical study was approved by the University of Michigan Health Sciences Institutional Review Board and registered with the clinical trials database of the National Institutes of Health, Bethesda, Maryland. Research subjects were recruited from September 2005 through June 2006. Upon receiving written consent, 100 human subjects aged 18 years and older were evaluated at the Michigan Center for Oral Health Research. All subjects possessed ≥20 teeth and had received no periodontal treatment or antibiotic therapy for medical or dental reasons 3 months prior to the investigation. In addition, the subjects did not previously undergo any long-term use of medications affecting periodontal status, such as anti-inflammatory drugs.
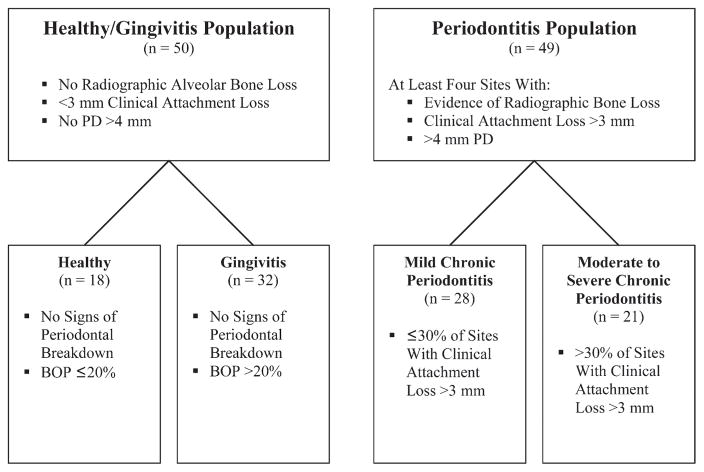
Subjects were enrolled into a healthy/gingivitis population (n = 50) or a periodontitis population (n = 49; one patient dropped out at experimental baseline). Subjects from the healthy and gingivitis population exhibited <3 mm of attachment loss, no periodontal probing depth (PD) >4 mm, and no radiographic alveolar bone loss. Periodontitis subjects exhibited at least four sites with evidence of radiographic bone loss, at least four sites with attachment loss >3 mm, and at least four sites with PD >4 mm (Fig. 1).
Figure 1.
Stratification of the low-risk population and the disease-susceptible population into four groups based on clinical attachment loss, PD, RBL, and BOP.
Subjects were excluded if they possessed a history of metabolic bone diseases, autoimmune diseases, unstable diabetes, or postmenopausal osteoporosis. Women who were pregnant were also excluded from the study.
Clinical Measures
All teeth except third molars were assessed for periodontal clinical measures by two calibrated examiners (CR and JK). Clinical parameters, including PD, clinical attachment level (CAL), and bleeding on probing (BOP), were measured at six sites per tooth. Other clinical assessments included dichotomous measures of plaque accumulation (PI) and gingival redness index (GRI), as previously described by Haffajee et al.18
Standardized periapical digital radiographs# were taken in the posterior dentition of all subjects using a parallel technique for the determination of alveolar bone height. Using a computer software measurement tool,** the interproximal alveolar bone levels of both premolars and first and second molars were measured on a digital computer screen by one calibrated examiner (LR). The distance from the alveolar bone crest to the cemento-enamel junction or the restorative margin reference was recorded as the radiographic alveolar bone level (RBL).
Quantitative Polymerase Chain Reaction (qPCR) Microbial Plaque Biofilm Analysis
Plaque biofilm collection
Subgingival plaque biofilm was collected from the mesio-buccal surfaces of all teeth and immediately placed into labeled vials containing 500 μl stabilizing buffer to prevent mRNA degradation,†† as previously described.19 After vortexing for 30 seconds, the samples were stored at 4°C until they were sent to the laboratory for analysis.
Detection of oral bacteria colonization in plaque biofilm samples
The detection of Aggregatibacter actinomycetemcomitans (previously Actinobacillus actinomycetemcomitans), Campylobacter rectus, Fusobacterium nucleatum, Prevotella intermedia, Porphyromonas gingivalis, Tannerella forsythia (previously T. forsythensis), and Treponema denticola in pooled plaque samples was evaluated by real-time qPCR, as described,20,21 using primers specific for the hypervariable segments of the 16S rRNA genes of each bacterium (Table 1). The percentage of the total flora for each species was calculated by dividing the number of target organisms by the total number of bacteria as determined by qPCR using 16S rRNA primers that reacted with all bacterial species. Data were represented using a patient-based assessment.
Table 1.
Primers for qPCR Analysis of Plaque Biofilm Bacteria
| Bacterial Species | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
|
| ||
| A. actinomycetemcomitans | GGCACGTAGGCGGACCTT | ACCAGGGCTAAAGCCCAATC |
| C. rectus | TTTCGGAGCGTAAACTCCTTTTC | TTTCTGCAAGCAGACACTCTT |
| F. nucleatum | ACCAGCGTTTGACATCTTAGGAATG | AGCCATGCACCTGTCTTTAG |
| P. intermedia | AGATTGACGGCCCTATGGGT | CCGGTCCTTATTCGAAGGGTA |
| P. gingivalis | CATAGATATCACGAGGAACTCCGATT | AAACTGTTAGCAACTACCGATGTGG |
| T. forsythia | GGGTGAGTAACGCGTATGTAACCT | ACCCATCCGCAACCAATAAA |
| T. denticola | CGTTCCTGGGCCTTGTACA | TAGCGACTTCAGGTACCCTCG |
| Universal | CCATGAAGTCGGAATCGCTAG | GCTTGACGGGCGGTGT |
Whole Saliva Collection
Unstimulated whole saliva was collected with passive drooling into sterile plastic tubes from all subjects at the beginning of the screening appointment.22 The collection was completed as soon as 2 ml whole saliva was collected or 15 minutes of sampling time had elapsed. Subsequently, the samples were placed on ice, aliquotted, and supplemented with a proteinase inhibitor combination of 1% aprotinin and 0.5% phenylmethylsulphonyl fluoride prior to storage at −80°C.
Protein Biomarker Assays
Protein biomarker levels were determined using colorimetric-based enzyme-linked immunosorbent assays (ELISAs), fluorescence-based protein microarrays, and radioimmunoassay (RIA), run according to manufacturer protocols. ELISAs‡‡ were used for measurement of MMP-8 and -9, calprotectin, and osteoprotegerin (OPG). Detection of the cytokines IL-1β, -2, -4, -5, -6, -10, and -13, tumor necrosis factor-alpha (TNF-α), and interferon (IFN)-γ was accomplished using a protein microarray.§§ The concentration of ICTP was determined using an RIA.||||
Prior to each assay, whole saliva samples were thawed at room temperature and microcentrifuged for 5 minutes to obtain cell-free supernatant for analysis. For ELISAs, absorbance measurements were collected using a primary signal at 450 nm with background subtraction of the 540-nm signal. A fluorescence scanner¶¶ was operated to collect Cy5 fluorescence signal from the cytokine protein micro-arrays. Data collection of the protein microarray signals was performed using software.##
Statistical Analysis
Basic demographics were summarized with means and proportions for each subject group; between-group comparisons were made with a one-way analysis of variance. Biomarker levels were summarized with medians for each group; between-group comparisons were made with a Kruskal-Wallis test. Areas under the curve (AUCs) for receiver operating characteristic (ROC) curves were estimated non-parametrically. 23 Thresholds for biomarkers were preselected as those values for which sensitivity and specificity were as equal as possible. ROC curves and corresponding AUCs for multiple biomarker combinations were based upon predicted probabilities of diseased subjects from a logistic regression model in which a subject’s biomarker levels were dichotomized as being above or below their corresponding thresholds. Furthermore, the biomarkers and microbial gene biofilm levels were ranked in importance via Random Forest methods.24 Statistical significance was defined as P ≤0.05.
RESULTS
Fifty-seven female (74% white) and 42 male (81% white) subjects, ranging in age from 20 to 77 years, were enrolled in the study. Following the recording of periodontal clinical and radiographic parameters, the 99 subjects were stratified and subdivided into four groups, according to previously described disease categories, prior to the analysis of the data (Fig. 1).25,26
From the healthy and gingivitis population, 18 subjects were stratified as healthy (group A), with no signs of periodontal breakdown and with BOP ≤20%. Thirty-two subjects were categorized as having gingivitis (BOP >20%) and no alveolar bone loss (group B). From the periodontitis population, 28 subjects exhibiting ≤30% of sites with CAL >3 mm were classified as having mild chronic periodontitis (group C), and 21 subjects were labeled as having moderate to severe chronic periodontitis (group D); CAL >3 mm was found in >30% of sites.
Dental and periodontal data (Table 2) were significantly different among the four groups for the mean number of teeth (25 to 28; P <0.001), BOP (12% to 64%; P <0.001), GRI (13% to 56%; P <0.001), accumulation of plaque (13% to 56%; P <0.001), mean PD (1.49 to 3.03 mm; P <0.001), sites with PD >4 mm (0% to 20%; P <0.001), mean CAL (0.59 to 2.93 mm; P <0.001), and mean RBL (1.89 to 4.33 mm; P < 0.001). Additionally, the prevalence of smoking was significantly higher in groups C and D (36% and 81%, respectively; P <0.001). The demographics for gender and ethnicity were balanced among the four groups. However, mean age was statistically significantly different among the four groups (range, 42 to 53 years; P = 0.02).
Table 2.
Patient Demographics and Clinical Parameters Stratified by Level of Disease
| Group A (healthy) | Group B (gingivitis) | Group C (mild chronic periodontitis) | Group D (moderate to severe chronic periodontitis) |
P Values Comparing A Through D
|
P Values Comparing A and B Versus C and D | ||
|---|---|---|---|---|---|---|---|
| Overall | Trend | ||||||
|
| |||||||
| Subjects (n) | 18 | 32 | 28 | 21 | NA | NA | NA |
| Males (%) | 56 | 41 | 39 | 38 | 0.67 | 0.32 | 0.47 |
| Whites (%) | 78 | 78 | 68 | 86 | 0.54 | 0.80 | 0.78 |
| Smokers (%) | 0 | 19 | 36 | 81 | <0.001 | <0.001 | <0.001 |
| Mean number of teeth | 28 | 27 | 26 | 25 | <0.001 | <0.001 | <0.001 |
| Mean age (years) | 45 | 42 | 53 | 50 | 0.02 | 0.03 | 0.002 |
| Sites with BOP (%) | 12 | 31 | 52 | 64 | <0.001 | <0.001 | <0.001 |
| Sites with gingival redness (%) | 13 | 22 | 49 | 56 | <0.001 | <0.001 | <0.001 |
| Sites with plaque (%) | 24 | 26 | 57 | 61 | <0.001 | <0.001 | <0.001 |
| Mean PD (mm) | 1.49 | 1.65 | 2.29 | 3.03 | <0.001 | <0.001 | <0.001 |
| Sites with PD >4 mm (%) | 0 | 0 | 7 | 20 | <0.001 | <0.001 | <0.001 |
| Mean CAL (mm) | 0.59 | 0.72 | 1.69 | 2.93 | <0.001 | <0.001 | <0.001 |
| Mean RBL (mm) | 1.89 | 2.00 | 3.13 | 4.33 | <0.001 | <0.001 | <0.001 |
NA = not applicable.
Data from our analysis of putative biomarkers of periodontal disease are shown in Table 3. Because the majority (>70%) of the subjects did not have detecable protein levels of IL-5 and IFN-γ in their whole saliva, these proteins were not included (data not shown).
Table 3.
Median Levels (ranges) and Diagnostic Ability of Salivary Biomarkers and Plaque Biofilm Pathogens
| Biomarker | Group A Healthy (median [range]) | Group B Gingivitis (median [range]) | Group C Mild Chronic Periodontitis (median [range]) | Group D Moderate to Severe Chronic Periodontitis (median [range]) | P Values Comparing A Through D | P Values Comparing A and B Versus C and D | AUC | Importance Score via Random Forest |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| MMP-8 (ng/ml) | 23.6 (2.5 to 322.5) | 54.1 (1 to 473.9) | 129.9 (8.5 to 978.9) | 203.8 (10.1 to 2,681.1) | <0.001 | <0.001 | 0.75 | 7.1 |
| OPG (pg/ml) | 2.3 (1.4 to 6.6) | 2.7 (1.2 to 6.2) | 1.9 (0.2 to 10.1) | 1.6 (0.5 to 11.8) | 0.056 | 0.007 | 0.62 | 6.3 |
| MMP-9 (ng/ml) | 106.4 (10 to 1,185.7) | 225.8 (4.9 to 1,732.2) | 301.6 (4.6 to 3,348.1) | 780.8 (10.4 to 9,778.2) | 0.002 | 0.001 | 0.72 | 5.1 |
| Calprotectin (ng/ml) | 3.0 (1.3 to 10) | 3.5 (0 to 24.6) | 4.3 (0 to 17.8) | 5.4 (1.7 to 97.6) | 0.082 | 0.023 | 0.68 | 4.7 |
| IL-1β (pg/ml) | 158.6 (0 to 6,000) | 206.7 (0 to 3,856.8) | 247.5 (24.1 to 3,120) | 462.2 (15.7 to 6,000) | 0.157 | 0.059 | 0.72 | 3.7 |
| ICTP (ng/ml) | 0.9 (0 to 4) | 0.8 (0 to 4) | 0.6 (0 to 5.4) | 0.9 (0 to 13.9) | 0.195 | 0.185 | 0.58 | 3.2 |
| IL-6 (pg/ml) | 0.0 (0 to 1,915) | 22.1 (0 to 8,784.9) | 14.6 (0 to 5,259.7) | 88.7 (0 to 10,816.9) | 0.127 | 0.092 | 0.71 | 2.2 |
| IL-10 (pg/ml) | 881.4 (0 to 11,088.8) | 120.6 (0 to 45,488.9) | 1,153.1 (0 to 24,581.4) | 1,445.1 (0 to 30,633.1) | 0.618 | 0.329 | 0.68 | 1.9 |
| TNF-α (pg/ml) | 9.8 (0 to 1,788.3) | 0.0 (0 to 3,720.5) | 8.1 (0 to 4,370.2) | 0.0 (0 to 8,212.7) | 0.483 | 0.954 | 0.64 | 1.8 |
| IL-13 (pg/ml) | 14.3 (0 to 83,151.1) | 0.0 (0 to 92,423.8) | 0.0 (0 to 76,046) | 169.9 (0 to 75,445.2) | 0.780 | 0.783 | 0.64 | 1.5 |
| IL-4 (pg/ml) | 0.0 (0 to 5,315.1) | 0.0 (0 to 6,579.3) | 54.4 (0 to 14,588) | 69.5 (0 to 11,714.3) | 0.377 | 0.086 | 0.71 | 1.3 |
| IL-2 (pg/ml) | 0.0 (0 to 3,718.1) | 0.0 (0 to 6,000) | 8.0 (0 to 6,205.5) | 0.0 (0 to 14,400.1) | 0.421 | 0.178 | 0.69 | 1.2 |
| T. denticola (%) | 0.11 (0 to 0.54) | 0.10 (0 to 2.95) | 1.53 (0 to 5.25) | 2.34 (0.79 to 6.63) | <0.001 | <0.001 | 0.86 | 13.7 |
| P. gingivalis (%) | 0.05 (0 to 0.9) | 0.04 (0 to 0.66) | 0.53(0 to 2.36) | 1.00 (0.43 to 3.24) | <0.001 | <0.001 | 0.84 | 9.6 |
| T. forsythia (%) | 0.09 (0 to 0.88) | 0.07 (0 to 0.8) | 0.71 (0 to 3.16) | 1.26 (0.11 to 3.55) | <0.001 | <0.001 | 0.85 | 8.4 |
| P. intermedia (%) | 0.11 (0 to 1.17) | 0.20 (0 to 1.99) | 0.82 (0 to 3.77) | 1.85 (0 to 3.5) | <0.001 | <0.001 | 0.79 | 6.7 |
| C. rectus (%) | 0.00 (0 to 1.22) | 0.00 (0 to 1.18) | 0.66 (0 to 2.82) | 1.32 (0 to 3.34) | 0.001 | <0.001 | 0.74 | 4.7 |
| F. nucleatum (%) | 2.96 (0 to 8.27) | 2.33 (0 to 7.32) | 3.29 (0 to 10.74) | 3.30 (0 to 9.56) | 0.251 | 0.196 | 0.59 | 3.9 |
| E. corrodens (%) | 0.00 (0 to 0.96) | 0.00 (0 to 1.04) | 0.00 (0 to 1.32) | 0.00 (0 to 0.1) | 0.697 | 0.259 | 0.56 | 0.3 |
Compared to the healthier individuals, the median levels of protein concentrations of MMP-8 (P <0.001), MMP-9 (P = 0.001), and calprotectin (P = 0.023) were increased in subjects with advancing stages of periodontal disease. Increased levels of OPG demonstrated a significant ability to predict health (P = 0.007; Table 3). Various trends were noted for other biomarkers, including ICTP and IL-1 and -6, but these were used to rank significant. Random Forest methods were ranking the importance of MMP-8 with a score of 7.1 and OPG with a score of 6.3, reflecting the highest importance level among the biomarkers in this dataset.
Further analysis was done using a subset of biomarkers demonstrating high Random Forest importance scores, relatively low P values, and high AUCs. The diagnostic properties of specific thresholds that gave nearly equal levels of sensitivity and specificity for our selection of biomarkers were selected as cutoff values. MMP-8 and -9 and calprotectin demonstrated significant abilities to predict disease category (odds ratios [ORs] were 5.3 for MMP-8 and -9 and 2.7 for calprotectin) (Table 4).
Table 4.
Diagnostic Properties of Specific Thresholds of Selected Salivary Biomarkers and Plaque Biofilm Pathogens
| Biomarker | Threshold | Above Threshold | Periodontitis
|
Sensitivity | Specificity | OR | 95% CI | |
|---|---|---|---|---|---|---|---|---|
| No (n) | Yes (n) | |||||||
|
| ||||||||
| MMP-8 (ng/ml) | 87.0 | − | 28 | 12 | 0.69 | 0.70 | 5.3 | 2.0 to 13.7 |
| + | 12 | 27 | ||||||
|
| ||||||||
| MMP-9 (ng/ml) | 240.0 | − | 28 | 12 | 0.69 | 0.70 | 5.3 | 2.0 to 13.7 |
| + | 12 | 27 | ||||||
|
| ||||||||
| Calprotectin (ng/ml) | 3.6 | − | 25 | 15 | 0.62 | 0.63 | 2.7 | 1.1 to 6.6 |
| + | 15 | 24 | ||||||
|
| ||||||||
| IL-6 (pg/ml) | 22.4 | − | 24 | 16 | 0.59 | 0.60 | 2.2 | 0.9 to 5.3 |
| + | 16 | 23 | ||||||
|
| ||||||||
| IL-1β (pg/ml) | 235.8 | − | 22 | 18 | 0.54 | 0.55 | 1.4 | 0.6 to 3.5 |
| + | 18 | 21 | ||||||
|
| ||||||||
| IL-10 (pg/ml) | 520.9 | − | 22 | 18 | 0.54 | 0.55 | 1.4 | 0.6 to 3.5 |
| + | 18 | 21 | ||||||
|
| ||||||||
| OPG (pg/ml) | 2.0 | − | 17 | 22 | 0.44 | 0.43 | 0.6 | 0.2 to 1.4 |
| + | 23 | 17 | ||||||
|
| ||||||||
| ICTP (ng/ml) | 0.7 | − | 16 | 23 | 0.41 | 0.40 | 0.5 | 0.2 to 1.1 |
| + | 24 | 16 | ||||||
|
| ||||||||
| T. denticola (%) | 0.2 | − | 33 | 7 | 0.82 | 0.83 | 21.6 | 6.8 to 68.4 |
| + | 7 | 32 | ||||||
|
| ||||||||
| T. forsythia (%) | 0.1 | − | 32 | 8 | 0.80 | 0.80 | 15.5 | 5.2 to 46.4 |
| + | 8 | 31 | ||||||
|
| ||||||||
| P. gingivalis (%) | 0.1 | − | 31 | 8 | 0.80 | 0.78 | 13.3 | 4.6 to 39.1 |
| + | 9 | 31 | ||||||
|
| ||||||||
| P. intermedia (%) | 0.4 | − | 29 | 11 | 0.72 | 0.73 | 6.7 | 2.5 to 18 |
| + | 11 | 28 | ||||||
|
| ||||||||
| C. rectus (%) | 0.1 | − | 25 | 15 | 0.62 | 0.63 | 2.7 | 1.1 to 6.6 |
| + | 15 | 24 | ||||||
|
| ||||||||
| F. nucleatum (%) | 2.8 | − | 24 | 16 | 0.59 | 0.60 | 2.2 | 0.9 to 5.3 |
| + | 16 | 23 | ||||||
|
| ||||||||
| E. corrodens (%) | 0.0 | − | 35 | 31 | 0.21 | 0.88 | 1.8 | 0.5 to 6.1 |
| + | 5 | 8 | ||||||
− = no; + = yes.
Table 3 shows the median levels as a percentage of selected red and orange complex organisms for their ability to identify periodontal disease category. A greater diagnostic ability of these organisms was demonstrated compared to the salivary biomarkers. When comparing the healthy/gingivitis group to the periodontitis group, T. denticola, P. gingivalis, T. forsythia, P. intermedia, and C. rectus exhibited significant differences (P <0.001); F. nucleatum and Eikenella corrodens did not. When the diagnostic properties were evaluated for the pathogens demonstrating significant differences between the groups, good sensitivity and specificity for disease category were shown (Table 4). ORs (2.7 to 21.6) were also found to be significant for T. denticola, P. gingivalis, T. forsythia, P. intermedia, and C. rectus (Table 4).
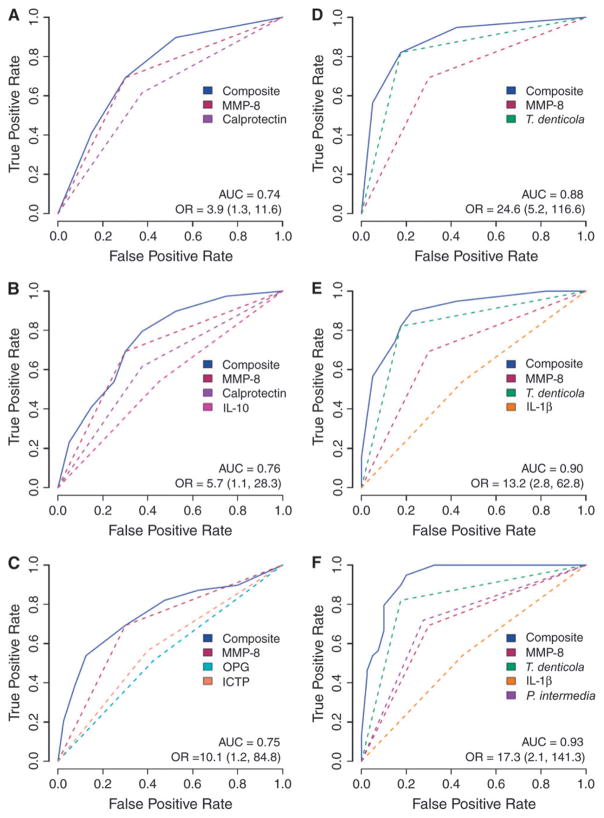
Multianalyte assessments were performed using various combinations of salivary biomarkers and plaque biofilm levels (Fig. 2). For example, when MMP-8 and calprotectin were combined to predict high-risk periodontal status, an AUC of 0.74 was found with a corresponding OR = 3.9 (95% confidence interval [CI]: 1.3 to 11.6). When multiple biomarkers were combined, such as MMP-8, OPG, and ICTP, the AUC increased to 0.75 with OR = 10.1 (95% CI: 1.2 to 84.8; Fig. 2C). When the microbial biofilm was combined with the biomarkers, the predictive values increased markedly. Figure 2D depicts the combination of MMP-8 and T. denticola with a resultant AUC of 0.88 (OR = 24.6; 95% CI: 5.2 to 116.6). Further improvements in the OR were noted when several pathogens were combined. Given the relatively small sample of 99 subjects, the OR could not be determined for many combinations because in all cases, periodontal disease category was correctly identified when comprehensive combinations were chosen and were considered infinite for these permutations (See supplementary table in online Journal of Periodontology). These results suggest that although the study of 99 subjects was able to determine differences in biomarker/biofilm levels to identify disease category, a much larger sample is needed to generate ORs that can be usable given the high level of accuracy demonstrated in this patient cohort.
Figure 2.
A through F) ROC of combinatorial permutations of salivary biomarkers coupled with biofilm subgingival pathogens measured by qPCR. Numbers in parentheses are 95% CIs.
DISCUSSION
To the best of our knowledge, this study demonstrates for the first time the ability to use host-response salivary biomarkers coupled with microbial biofilm DNA to identify individuals with different stages of periodontal disease. The results underscore the robustness of combinatorial measures of disease mediators, such as MMPs with putative periodontal pathogen genes, to more accurately identify a patient’s status. These findings may allow for rapid POC diagnostics to quickly identify and screen at-risk patients in a more time-effective manner compared to extensive clinical examinations.
Our data identified key biomarkers from saliva and biofilm that represent three distinct phases of periodontitis: periodontal tissue inflammation (IL-1 and -6), matrix degradation (MMP-8 and -9), and alveolar bone turnover/resorption (osteoprotegerin and ICTP). Complementing the dataset with anaerobic pathogens (particularly P. gingivalis, T. denticola, and T. forsythia) augments the microbe–host influences on periodontal disease identification to clinical measures of disease status. These results represent an early approach to the identification of disease signatures for periodontitis using rapid diagnostic techniques. Given the multifactorial complexity of periodontitis as a polygenic disease, similar to cardiovascular disease and osteoporosis, the consideration of multiple checkpoints of disease (infection, inflammation, immune dysregulation, and bone resorption) can now be addressed with the use of multiple biomarkers that reflect the distinct stages of periodontitis. The periodontology field has failed to come up with a “silver bullet” or specific biomarker for periodontal disease identification. The results from this study suggest that patient disease status might be able to be determined rapidly using a combined proteomic/microbial genetic approach. The development of such methodologies may have implications for rapid POC diagnostics for oral and other systemic diseases; however, much more information will be gleaned from longitudinal investigation. 27
During the initiation of an inflammatory response in the periodontal connective tissue, numerous cytokines, such as IL-1β and -6 and TNF-α, are released from cells of the junctional epithelia, connective tissue fibroblasts, and macrophages. Additionally, a number of enzymes, such as MMP-8 and -9 and calprotectin, are produced by PMNs and osteoclasts, leading to the degradation of connective tissue collagen and alveolar bone. During connective tissue inflammation and following bone resorption, cytokines and bone resorptive/turnover proteins migrate toward the gingival sulcus or periodontal pocket and further into GCF, where they are released into and contribute to whole saliva. Host cell–derived MMP-8 and -9 are believed to mediate, to a substantial extent, the matrix-destroying events during the stages of periodontal disease. The results from our investigation are in agreement with and extend the overall findings that MMP-8 and -9 seem to be key biomarkers that are elevated in the oral fluids of periodontal patients.28,29
These data support the concept of the development of periodontal signatures or biologic phenotypes for disease classification that consider the host phenotype (response to the microbial insult) and the nature of the invading pathogens that initiate periodontal disease.16 Specific biofilm organisms or exposures may have the capacity to affect the “inflammatory set point” of the local tissues in certain patients via epigenetic mechanisms.30,31 Thus, the use of rapid chairside POC diagnostics that identify disease in the context of the host–microbe interaction will likely lead to more rationally tailored therapeutic strategies. Offenbacher et al.17 recently described periodontal disease at the biofilm–gingival interface (BGI) and noted from a molecular epidemiologic investigation that patients’ clinical phenotypes are linked to biologic phenotypes based on anti-body response, microbial biofilm levels, and GCF levels of specific proinflammatory cytokines. The identification of these BGI classifications has led to distinct categories that contain elevated antibody titers to P. gingivalis, C. rectus, and T. denticola immunoglobulin G as well as increased GCF concentrations of IL-1 and -6. Our data support and expand these findings using salivary-derived biomarkers for more rapid, easy-to-collect, and more global whole-mouth assessment of inflammatory and matrix-associated markers of periodontal disease. The greatest diagnostic accuracy in disease identification was noted when MMP-8 or -9 was coupled with red-complex periodontal organisms T. denticola, P. gingivalis, or T. forsythia.32 The concept of MMP-8 as a diagnostic has been well described,6,33–36 and the linkage between red-complex bacteria and collagen destruction was reported. 37,38 The red-complex bacteria are known for their potent ability to display trypsin-like enzyme activity that is responsible for destroying collagen matrices. 39 Thus, these data substantiate the combinatorial use of MMP-destroying enzymes and corresponding initiating pathogens, such as T. denticola, for periodontal disease identification. The greatest usefulness of these diagnostic approaches is the development of predictive models for disease that need to be validated in large, longitudinal studies. The patients involved in this investigation were evaluated (Table 5) to determine the ability of these diagnostic approaches to predict progressive periodontal disease.40
Table 5.
Positive and Negative Predictive Values of Specific Thresholds of Selected Salivary Biomarkers and Plaque Biofilm Pathogens
| Biomarker | Threshold | Above Threshold | Periodontitis
|
PPV | NPV | |
|---|---|---|---|---|---|---|
| No (n) | Yes (n) | |||||
|
| ||||||
| MMP-8 (ng/ml) | 87.0 | − | 28 | 12 | 0.69 | 0.70 |
| + | 12 | 27 | ||||
|
| ||||||
| MMP-9 (ng/ml) | 240.0 | − | 28 | 12 | 0.69 | 0.70 |
| + | 12 | 27 | ||||
|
| ||||||
| Calprotectin (ng/ml) | 3.6 | − | 25 | 15 | 0.62 | 0.63 |
| + | 15 | 24 | ||||
|
| ||||||
| IL-6 (pg/ml) | 22.4 | − | 24 | 16 | 0.59 | 0.60 |
| + | 16 | 23 | ||||
|
| ||||||
| IL-1β (pg/ml) | 235.8 | − | 22 | 18 | 0.54 | 0.55 |
| + | 18 | 21 | ||||
|
| ||||||
| IL-10 (pg/ml) | 520.9 | − | 22 | 18 | 0.54 | 0.55 |
| + | 18 | 21 | ||||
|
| ||||||
| OPG (pg/ml) | 2.0 | − | 17 | 22 | 0.43 | 0.44 |
| + | 23 | 17 | ||||
|
| ||||||
| ICTP (ng/ml) | 0.7 | − | 16 | 23 | 0.40 | 0.41 |
| + | 24 | 16 | ||||
|
| ||||||
| T. denticola (%) | 0.2 | − | 33 | 7 | 0.82 | 0.83 |
| + | 7 | 32 | ||||
|
| ||||||
| T. forsythia (%) | 0.1 | − | 32 | 8 | 0.80 | 0.80 |
| + | 8 | 31 | ||||
|
| ||||||
| P. gingivalis (%) | 0.1 | − | 31 | 8 | 0.78 | 0.80 |
| + | 9 | 31 | ||||
|
| ||||||
| P. intermedia (%) | 0.4 | − | 29 | 11 | 0.72 | 0.73 |
| + | 11 | 28 | ||||
|
| ||||||
| C. rectus (%) | 0.1 | − | 25 | 15 | 0.62 | 0.63 |
| + | 15 | 24 | ||||
|
| ||||||
| F. nucleatum (%) | 2.8 | − | 24 | 16 | 0.59 | 0.60 |
| + | 16 | 23 | ||||
|
| ||||||
| E. corrodens (%) | 0.0 | − | 35 | 31 | 0.62 | 0.53 |
| + | 5 | 8 | ||||
PPV = positive predictive value; NPV = negative predictive value; − = no; + = yes.
CONCLUSIONS
These data support the pairing of microbial and host-response biomarker information for more accurate periodontal diagnoses. Future clinical in-office applications of rapid POC diagnostics that can measure proteins, genes, and biofilm pathogens in saliva should lead to the development of improved disease identification and improved oral health. These studies require the longitudinal validation of these cross-sectional approaches to determine the prediction of disease activity. The patients in this trial are being monitored for the determination of disease progression to better forecast clinical disease outcomes.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Dental and Craniofacial Research (U01-DE014961) and the National Center for Research Resources (M01-RR000042), Bethesda, Maryland, and the Swiss Society of Periodontology, Brig, Switzerland. Dr. Singh is a manager and Dr. Tran is a principal technologist in the Biosystems Research Department at Sandia National Laboratories. Drs. Herr, Shelburne, Braun, Singh, and Giannobile hold intellectual property related to this article. This trial is registered on the www.clinicaltrials.gov database (NCT00277745). The authors appreciate the clinical assistance of Drs. Thiago Morelli, Amy Kim, and Noah Smith, Michigan Center for Oral Health Research.
Footnotes
Schick Technologies, Long Island City, NY.
Emago Advanced, Oral Diagnostic Systems, Amsterdam, The Netherlands.
RNA Protect, Ambion, Austin, TX.
R&D Systems, Minneapolis, MN.
Whatman, Florham Park, NJ.
Immunodiagnostic Systems, Fountain Hills, AZ.
Molecular Devices, Sunnyvale, CA.
GenePix Pro, MDS Analytical Technologies, Toronto, ON.
indicates supplementary slide presentation (with audio) in the online Journal of Periodontology
References
- 1.Taubman MA, Kawai T, Han X. The new concept of periodontal disease pathogenesis requires new and novel therapeutic strategies. J Clin Periodontol. 2007;34:367–369. doi: 10.1111/j.1600-051X.2007.01065.x. [DOI] [PubMed] [Google Scholar]
- 2.Offenbacher S, Barros SP, Beck JD. Rethinking periodontal inflammation. J Periodontol. 2008;79(Suppl 8):1577–1584. doi: 10.1902/jop.2008.080220. [DOI] [PubMed] [Google Scholar]
- 3.Tabak LA. Point-of-care diagnostics enter the mouth. Ann N Y Acad Sci. 2007;1098:7–14. doi: 10.1196/annals.1384.043. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Mauk MG, Wang J, et al. A microfluidic system for saliva-based detection of infectious diseases. Ann N Y Acad Sci. 2007;1098:429–436. doi: 10.1196/annals.1384.024. [DOI] [PubMed] [Google Scholar]
- 5.Mauk MG, Ziober BL, Chen Z, Thompson JA, Bau HH. Lab-on-a-chip technologies for oral-based cancer screening and diagnostics: Capabilities, issues, and prospects. Ann N Y Acad Sci. 2007;1098:467–475. doi: 10.1196/annals.1384.025. [DOI] [PubMed] [Google Scholar]
- 6.Herr AE, Hatch AV, Giannobile WV, et al. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci USA. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 8.Malamud D. Salivary diagnostics: The future is now. J Am Dent Assoc. 2006;137:284, 286. doi: 10.14219/jada.archive.2006.0158. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, St John MA, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann BG, Wong DT. Salivary mRNA targets for cancer diagnostics. Oral Oncol. 2008;44:425–429. doi: 10.1016/j.oraloncology.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaney KP, Branson BM, Uniyal A, et al. Performance of an oral fluid rapid HIV-1/2 test: Experience from four CDC studies. AIDS. 2006;20:1655–1660. doi: 10.1097/01.aids.0000238412.75324.82. [DOI] [PubMed] [Google Scholar]
- 12.Yager P, Edwards T, Fu E, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 13.Taba M, Jr, Kinney J, Kim AS, Giannobile WV. Diagnostic biomarkers for oral and periodontal diseases. Dent Clin North Am. 2005;49:551–571. doi: 10.1016/j.cden.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinney JS, Ramseier CA, Giannobile WV. Oral fluid-based biomarkers of alveolar bone loss in periodontitis. Ann N Y Acad Sci. 2007;1098:230–251. doi: 10.1196/annals.1384.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loos BG, Tjoa S. Host-derived diagnostic markers for periodontitis: Do they exist in gingival crevice fluid? Periodontol 2000. 2005;39:53–72. doi: 10.1111/j.1600-0757.2005.00129.x. [DOI] [PubMed] [Google Scholar]
- 16.Casanova JL, Abel L. The human model: A genetic dissection of immunity to infection in natural conditions. Nat Rev Immunol. 2004;4:55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- 17.Offenbacher S, Barros SP, Singer RE, Moss K, Williams RC, Beck JD. Periodontal disease at the biofilm-gingival interface. J Periodontol. 2007;78:1911–1925. doi: 10.1902/jop.2007.060465. [DOI] [PubMed] [Google Scholar]
- 18.Haffajee AD, Socransky SS, Goodson JM. Comparison of different data analyses for detecting changes in attachment level. J Clin Periodontol. 1983;10:298–310. doi: 10.1111/j.1600-051x.1983.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 19.Shelburne CE, Shelburne PS, Dhople VM, et al. Serum antibodies to Porphyromonas gingivalis chaperone HtpG predict health in periodontitis susceptible patients. PLoS ONE. 2008;3:e1984. doi: 10.1371/journal.pone.0001984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullally BH, Dace B, Shelburne CE, Wolff LF, Coulter WA. Prevalence of periodontal pathogens in localized and generalized forms of early-onset periodontitis. J Periodontal Res. 2000;35:232–241. doi: 10.1034/j.1600-0765.2000.035004232.x. [DOI] [PubMed] [Google Scholar]
- 21.Shelburne CE, Prabhu A, Gleason RM, Mullally BH, Coulter WA. Quantitation of Bacteroides forsythus in subgingival plaque comparison of immunoassay and quantitative polymerase chain reaction. J Microbiol Methods. 2000;39:97–107. doi: 10.1016/s0167-7012(99)00106-2. [DOI] [PubMed] [Google Scholar]
- 22.Mandel ID, Wotman S. The salivary secretions in health and disease. Oral Sci Rev. 1976;(8):25–47. [PubMed] [Google Scholar]
- 23.Bamber D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J Math Psychol. 1975;12:387–415. [Google Scholar]
- 24.Breiman L. Randomforests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 25.Borrell LN, Papapanou PN. Analytical epidemiology of periodontitis. J Clin Periodontol. 2005;32(Suppl 6):132–158. doi: 10.1111/j.1600-051X.2005.00799.x. [DOI] [PubMed] [Google Scholar]
- 26.Tonetti MS, Claffey N. Advances in the progression of periodontitis and proposal of definitions of a periodontitis case and disease progression for use in risk factor research. Group C consensus report of the 5th European Workshop in Periodontology. J Clin Periodontol. 2005;32(Suppl 6):210–213. doi: 10.1111/j.1600-051X.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- 27.Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 28.Beklen A, Tuter G, Sorsa T, et al. Gingival tissue and crevicular fluid co-operation in adult periodontitis. J Dent Res. 2006;85:59–63. doi: 10.1177/154405910608500110. [DOI] [PubMed] [Google Scholar]
- 29.Söder B, Airila Månsson S, Söder PO, Kari K, Meurman J. Levels of matrix metalloproteinases-8 and -9 with simultaneous presence of periodontal pathogens in gingival crevicular fluid as well as matrix metalloproteinase-9 and cholesterol in blood. J Periodontal Res. 2006;41:411–417. doi: 10.1111/j.1600-0765.2006.00888.x. [DOI] [PubMed] [Google Scholar]
- 30.Bobetsis YA, Barros SP, Lin DM, et al. Bacterial infection promotes DNA hypermethylation. J Dent Res. 2007;86:169–174. doi: 10.1177/154405910708600212. [DOI] [PubMed] [Google Scholar]
- 31.Kornman K, Duff G, Reilly P. Re: A critical assessment of interleukin-1 (IL-1) genotyping when used in a genetic susceptibility test for severe chronic periodontitis. Greenstein G, Hart TC (2002;73:231–247) J Periodontol. 2002;73:1553–1556. doi: 10.1902/jop.2002.73.12.1553. [DOI] [PubMed] [Google Scholar]
- 32.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 33.Christodoulides N, Floriano PN, Miller CS, et al. Labon-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann N Y Acad Sci. 2007;1098:411–428. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]
- 34.Golub LM, Lee HM, Greenwald RA, et al. A matrix metalloproteinase inhibitor reduces bone-type collagen degradation fragments and specific collagenases in gingival crevicular fluid during adult periodontitis. Inflamm Res. 1997;46:310–319. doi: 10.1007/s000110050193. [DOI] [PubMed] [Google Scholar]
- 35.Kinane DF, Darby IB, Said S, et al. Changes in gingival crevicular fluid matrix metalloproteinase-8 levels during periodontal treatment and maintenance. J Periodontal Res. 2003;38:400–404. doi: 10.1034/j.1600-0765.2003.00663.x. [DOI] [PubMed] [Google Scholar]
- 36.Prescher N, Maier K, Munjal SK, et al. Rapid quantitative chairside test for active MMP-8 in gingival crevicular fluid: First clinical data. Ann N Y Acad Sci. 2007;1098:493–495. doi: 10.1196/annals.1384.019. [DOI] [PubMed] [Google Scholar]
- 37.Oringer RJ, Palys MD, Iranmanesh A, et al. C-telopeptide pyridinoline cross-links (ICTP) and periodontal pathogens associated with endosseous oral implants. Clin Oral Implants Res. 1998;9:365–373. doi: 10.1034/j.1600-0501.1996.090602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palys MD, Haffajee AD, Socransky SS, Giannobile WV. Relationship between C-telopeptide pyridinoline cross-links (ICTP) and putative periodontal pathogens in periodontitis. J Clin Periodontol. 1998;25:865–871. doi: 10.1111/j.1600-051x.1998.tb02383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loesche WJ, Syed SA, Stoll J. Trypsin-like activity in subgingival plaque. A diagnostic marker for spirochetes and periodontal disease? J Periodontol. 1987;58:266–273. doi: 10.1902/jop.1987.58.4.266. [DOI] [PubMed] [Google Scholar]
- 40.Ligtenberg AJ, de Soet JJ, Veerman EC, Amerongen AV. Oral diseases: From detection to diagnostics. Ann N Y Acad Sci. 2007;1098:200–203. doi: 10.1196/annals.1384.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.