Abstract
IMPORTANCE
Muscle weakness, the most common symptom of neuromuscular disease, may result from muscle dysfunction or may be caused indirectly by neuronal and neuromuscular junction abnormalities. To date, more than 780 monogenic neuromuscular diseases, linked to 417 different genes, have been identified in humans. Genome-editing methods, especially the CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 (CRISPR-associated protein 9) system, hold clinical potential for curing many monogenic disorders, including neuromuscular diseases such as Duchenne muscular dystrophy, spinal muscular atrophy, amyotrophic lateral sclerosis, and myotonic dystrophy type 1.
OBJECTIVES
To provide an overview of genome-editing approaches; to summarize published reports on the feasibility, efficacy, and safety of current genome-editing methods as they relate to the potential correction of monogenic neuromuscular diseases; and to highlight scientific and clinical opportunities and obstacles toward permanent correction of disease-causing mutations responsible for monogenic neuromuscular diseases by genome editing.
EVIDENCE REVIEW
PubMed and Google Scholar were searched for articles published from June 30, 1989, through June 9, 2016, using the following keywords: genome editing, CRISPR-Cas9, neuromuscular disease, Duchenne muscular dystrophy, spinal muscular atrophy, amyotrophic lateral sclerosis, andmyotonic dystrophy type 1. The following sources were reviewed: 341 articles describing different approaches to edit mammalian genomes; 330 articles describing CRISPR-Cas9–mediated genome editing in cell culture lines (in vitro) and animal models (in vivo); 16 websites used to generate single-guide RNA; 4 websites for off-target effects; and 382 articles describing viral and nonviral delivery systems. Articles describing neuromuscular diseases, including Duchenne muscular dystrophy, spinal muscular atrophy, amyotrophic lateral sclerosis, and myotonic dystrophy type 1, were also reviewed.
FINDINGS
Multiple proof-of-concept studies reveal the feasibility and efficacy of genome-editing–meditated correction of monogenic neuromuscular diseases in cultured cells and animal models.
CONCLUSIONS AND RELEVANCE
Genome editing is a rapidly evolving technology with enormous translational potential once efficacy, delivery, and safety issues are addressed. The clinical impact of this technology is that genome editing can permanently correct disease-causing mutations and circumvent the hurdles of traditional gene- and cell-based therapies.
Despite major advances in the identification of monogenic human disease genes, many challenges remain in the amelioration of these disorders. Monogenic disorders are estimated to account for more than 10 000 diagnosed human diseases.1 Although individually relatively rare, together these diseases affect approximately 1 in 100 individuals.1 Neuromuscular diseases, which impair the function of muscles, motor nerves, and/or neuromuscular junctions, are among the most common and severe monogenic disorders. To date, more than 780 monogenic neuromuscular diseases, linked to 417 different genes, have been identified in humans.2 Muscle weakness, spasms, hypertonia, and hypotonia are common, incurable consequences of neuromuscular diseases, including Duchenne muscular dystrophy (DMD), spinal muscular atrophy (SMA), amyotrophic lateral sclerosis (ALS), and myotonic dystrophy type 1 (DM1).
Recently developed genome-editing systems, which allow for precise modification of the genome, are revolutionizing our understanding of the molecular basis of disease and providing the potential to permanently correct the underlying causes of disease. However, this technology is still in its infancy, and many questions and challenges remain to be addressed. This review introduces the most up-to-date genome-editing systems and their potential applications to medicine. We also summarize published proof-of-concept studies on representative neuromuscular diseases and discuss the clinical feasibility of genome editing in the neuromuscular system.
Data Sources and Extraction
We searched PubMed, which comprises more than 26 million citations for biomedical literature from MEDLINE, life science journals, and online books, for articles published from June 30, 1989, through June 9, 2016. We also used Google Scholar, which allows searches across many disciplines and sources, including articles, theses, books, abstracts, and court opinions from academic publishers, professional societies, online repositories, universities, and other websites. Subject headings and indexed text keywords used were genome editing, CRISPR-Cas9, neuromuscular disease, Duchenne muscular dystrophy, spinal muscular atrophy, amyotrophic lateral sclerosis, and myotonic dystrophy type 1. The following sources were reviewed: 341 articles describing different approaches to edit mammalian genomes; 330 articles describing CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 (CRISPR-associated protein 9)–mediated genome editing in cell culture lines (in vitro) and animal models (in vivo); 16 websites used to generate single-guide RNA; 4 websites for off-target effects; and 382 articles describing viral and nonviral delivery systems. Articles describing neuromuscular diseases, including DMD, SMA, ALS, and DM1, were included.
Results
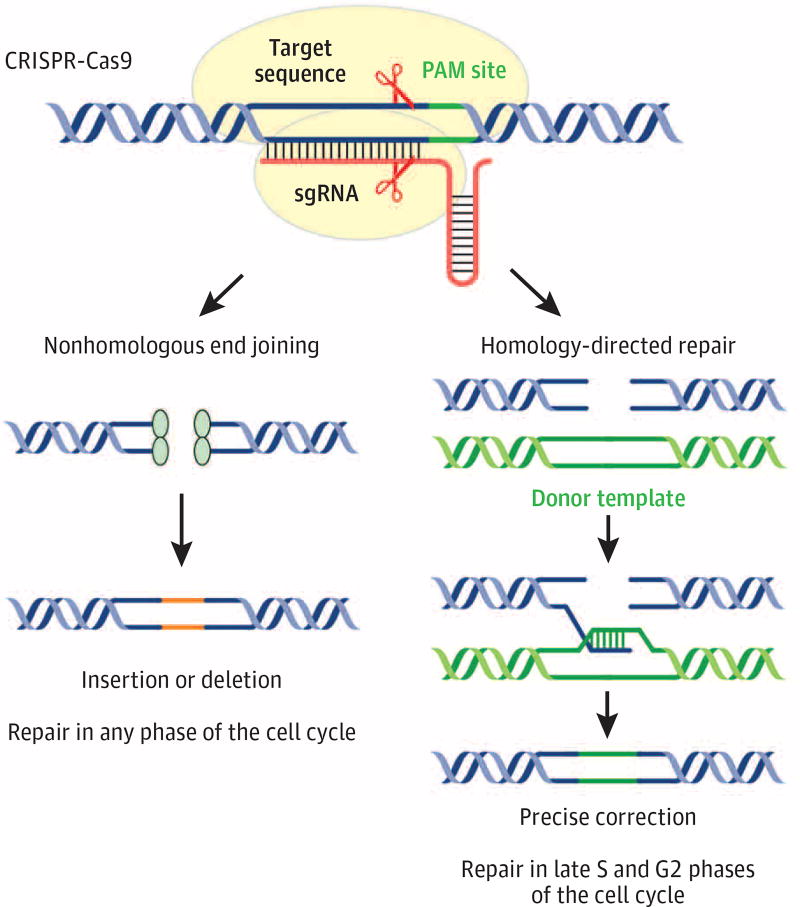
Multiple engineered nucleases, including meganucleases,3 zinc finger nucleases (ZNFs),4,5 transcription activator–like effector nucleases (TALENs),6 and the CRISPR-Cas9 system,7,8 have been developed in recent years to modify the genomes of model organisms and humans. Targeting the cause of monogenic disease represents a promising and rapidly moving aspect of genome-editing technologies. In brief, an engineered nuclease binds to a targeted genomic locus and generates a double-strand break (DSB) (Figure 1). The DSB is then repaired by nonhomologous end joining (NHEJ), which leads to imprecise insertion/deletion (indel) mutations, or homology-directed repair (HDR), which requires an exogenous DNA template and can generate a precise modification at the target locus. The phase of the cell cycle determines the repair pathway choice between NHEJ and HDR. Nonhomologous end joining directly ligates broken DNA ends throughout the cell cycle, whereas HDR is restricted to the S and G2 phases when the sister chromatid is available as a repair template.9 Hence, engineered nucleases can effectively generate NHEJ-mediated mutations in most cell types, whereas HDR-mediated editing generally does not occur in postmitotic cells. Thus, postnatal cells, which exit the cell cycle shortly after birth, such as myofibers, cardiomyocytes, and neurons, are less likely to undergo HDR-mediated genome editing. Engineered nuclease–mediated genome editing represents a potentially powerful technique to correct disease-causing mutations in the genome. This review focuses on the CRISPR-Cas9 system, a relatively rapid and simple editing method.
Figure 1. Schematic Outline of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas9 (CRISPR-Associated Protein 9)–Mediated Genomic Editing.
Cas9 (yellow) guided by single-guide RNA (sgRNA) binds to a target DNA site next to the protospacer adjacent motif (PAM). Scissors indicate the Cas9 cleavage site. Red lines mark the sgRNA scaffold. The Cas9-sgRNA complex generates a double-strand break, which is repaired by nonhomologous end joining or homology-directed repair. Orange lines indicate the insertion or deletion (indel) mutations. The green line marks an exogenous DNA template (donor template).
Introduction of the CRISPR-Cas9 System
The CRISPR-Cas9 system, an RNA-guided, nuclease-mediated form of genome editing, represents a major breakthrough in genomic engineering and offers a revolutionary approach to alter the human genome.7,8,10 The Cas9 and Cpf1 (cas gene of Pasteurella francisella) endonucleases are guided by single-guide RNAs (sgRNAs) to bind a targeted genomic locus next to a protospacer adjacent motif, generating a DSB (Figure 1).11 The DSB can then be repaired by NHEJ or HDR, as described above. CRISPR-Cas9 genome editing can permanently remove the genetic defect, whereas other gene therapy methods add only a functional copy of a gene to the cells but retain the underlying dysfunctional copy of the gene. During the past 3 years, numerous studies have found that the CRISPR-Cas9 system can be used to modify specific sequences in the genome or correct monogenic diseases in tissue culture cells,12 in the mouse or rat germline, and in multiple postnatal organs, such as brain,13 liver,14,15 muscle, and heart.16–18
Correction of DMD Mutations by CRISPR-Cas9 Genome Editing
Duchenne muscular dystrophy is caused by mutations in the DMD (dystrophin) gene (OMIM 300377) on the X chromosome and affects approximately 1 in 5000 boys.19 DMD is the largest gene in the human genome, consisting of 2.6 million base pairs and 79 exons. Dystrophin is a large cytoskeletal protein essential for muscle cell membrane integrity. Without dystrophin, muscles degenerate, causing weakness and myopathy.20 Death of a patient with DMD usually occurs by 25 years of age, typically from breathing complications and cardiomyopathy. Hence, therapy for DMD necessitates sustained rescue of skeletal, respiratory, and cardiac muscle structure and function.
The mdx mouse harbors a premature termination codon in exon 23 of the DMD locus and serves as a useful model for DMD. An initial proof-of-concept study21 found that CRISPR-Cas9 genome editing could correct the premature termination codon in mdx mice by HDR within the germline. However, genome editing within the germline is not feasible in humans, necessitating methods for safe and effective gene correction after birth. A series of articles16–18 published in 2016 reported successful editing of the DMD mutation in mdx mice using recombinant adeno-associated virus (AAV), a harmless virus vector, to systemically deliver Cas9 and sgRNA expression vectors to muscle tissues. In those studies, sgRNAs that flanked exon 23 were used to skip this exon and restore dystrophin expression in cardiac and skeletal muscle cells of postnatal mdx mice (Figure 2A). Similarly, adenovirus-mediated genome editing restores dystrophin expression in specific muscles of mdx mice after intramuscular injection.22 This approach has been validated by CRISPR-Cas9–mediated correction of human DMD mutations using myoblast or induced pluripotent stem cells (iPSCs) derived from patients with DMD that were differentiated into skeletal muscle cells in vitro.23–28
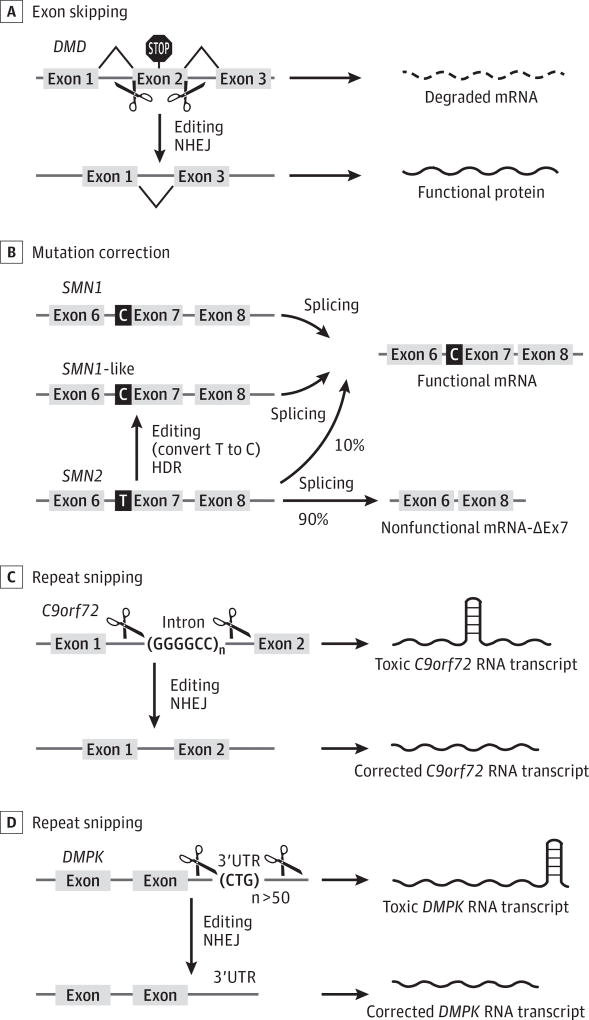
Figure 2. Strategy for Application of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas9 (CRISPR-Associated Protein 9)–Mediated Gene Editing for Monogenic Neuromuscular Diseases.
A, Duchenne muscular dystrophy (DMD) is shown as an example of the application of exon skipping to skip sections of the gene that harbor mutations, allowing the creation of functional, truncated dystrophin protein. B, Spinal muscular atrophy is shown as an example of the application of a mutation correction strategy in which replacement of 1 nucleotide (T to C) will convert SMN2 (the gene encoding the survival motor neuron protein 2) to a correct copy of the SMN1 gene (termed SMN1-like gene). C and D, Amyotrophic lateral sclerosis and myotonic dystrophy type 1 are shown as examples of the application of a repeat-snipping strategy in which CRISPR-Cas9 editing cuts the nucleotide repeats present in the intron or 3′-untranslated region (3′UTR) of the C9orf72 (chromosome 9 open reading frame 72) or DMPK (dystrophia myotonica-protein kinase) gene, respectively, to restore a functional RNA transcript. HDR indicates homology-directed repair; mRNA, messenger RNA; and NHEJ, nonhomologous end joining.
Exon Skipping as a Strategy to Bypass Mutations in Protein-Coding Genes
CRISPR-Cas9–mediated genome editing of DMD mutations in muscles, which we termed myoediting (Figure 2A), can create internal genomic deletions to correct the open reading frame or disrupt splice sites, thereby allowing splicing between surrounding exons to re-create an in-frame dystrophin protein that lacks the mutations. Whereas DMD, because of loss-of-function mutations in dystrophin, is a fatal disease, Becker muscular dystrophy, caused by in-frame internal deletions of dystrophin, is a comparatively mild muscle disorder such that patients live into their 60s with relatively modest muscle impairment. The functionality of muscle in patients with Becker muscular dystrophy has guided approaches for skipping of mutant exons as an approach to partially restore dystrophin expression in patients with DMD.
Exon skipping is a strategy in which nonessential sections of a gene that harbor mutations are skipped, allowing the creation of partially functional proteins with internal deletions.29 Traditional exon-skipping strategies that involve the use of antisense oligonucleotides to mask splice sites suffer from the inefficiency of tissue uptake of oligonucleotides, the requirement for lifelong delivery of oligonucleotides, and incomplete exon skipping. Genome-editing–mediated exon skipping represents a powerful new approach to permanently eliminate the genetic cause of the disease and restore muscle structure and function in patients with devastating diseases, such as DMD. In principle, CRISPR-Cas9–mediated exon-skipping strategies could be applied to many genes harboring disease-causing mutations, including out-of-frame deletions or insertions, exon duplications, and pseudoexons.
Imprecise deletions, induced by NHEJ, that prevent splicing of exons that harbor mutations are sufficient to restore protein expression by exon skipping. However, this approach is not feasible if the mutation is located in an exon that codes for an essential domain of the protein. For this type of mutation, HDR-mediated precise correction will be required. In this regard, muscle and neural delivery of the genome-editing components, including the HDR DNA template using AAV and in vivo electroporation, was recently reported.30
Gene Correction in Monogenic Neuromuscular Diseases
Spinal muscular atrophy is an autosomal recessive neuromuscular disease characterized by degeneration of α-motor neurons in the spinal cord and brainstem, resulting in progressive proximal muscle weakness, hyposthenia, and paralysis. Spinal muscular atrophy is the leading genetic cause of infant mortality, with an estimated incidence of 1 in 6000 to 1 in 10 000 live births.31 Spinal muscular atrophy is the second most common autosomal recessive lethal disease in white individuals, with a carrier frequency of 1 in 37.32 The disease is caused by mutations in the gene encoding the SMN (survival motor neuron) protein, a protein required for the maintenance of motor neurons, which control muscle movement.33 The common form of the disorder is caused by genetic mutations in the SMN1 gene (OMIM 600354). Prognosis depends on the phenotypic severity, ranging from high mortality within the first year for SMA type 1 to no mortality in later-onset forms. The primary modulator of the clinical phenotype is the paralogue gene SMN2 (OMIM 601627), which differs from SMN1 by a critical single-nucleotide polymorphism in exon 7 (C to T). Although no amino acid substitution is induced by the exonic single-nucleotide polymorphisms, the substitution of a C with a T in exon 7 alters splicing, resulting in exon 7 exclusion in 90% of SMN2 messenger RNA transcripts and the generation of an unstable truncated protein, termed SMNΔ7. Hence, SMN2 cannot produce sufficient functional SMN protein to maintain viability of motor neurons.34
Genome editing might be used to potentially convert SMN2 into an SMN1-like gene by changing a T to a C in exon 7. Corti et al35 generated iPSCs from patients with SMA and used a targeted genome-editing correction approach with single-stranded oligo-nucleotides to convert the SMN2 gene into an SMN1-like gene. CRISPR-Cas9–mediated genome editing could, in principle, generate a fully functional SMN gene that permanently includes exon 7 in motor neurons (Figure 2B). The copy number of SMN2 varies in patients with SMA from 2 to 4, and the SMN2 copy number can modify the clinical phenotypes. Results from SMA animal models and patients with SMA indicate that the severity of SMA inversely correlates with the SMN2 copy number. Interestingly, Ogino et al36 provided evidence that gene conversion from SMN2 to SMN1 occurs in the general population. Hence, genome-editing methods might imitate nature’s way of converting a mutant gene into a functional one (SMN1-like). Similar logic has been applied to oligonucleotide-mediated correction strategies in vitro.35,37 Similar strategies might be applied to correct other point mutations that cause neurodegenerative disorders.
Repeat Snipping in Monogenic Neuromuscular Diseases
A major cause of monogenic neuromuscular diseases is the unstable and dynamic transmission of simple, repetitive DNA elements, such as trinucleotide repeat expansion and hexanucleotide repeat expansion. If these disease-causing repeats are localized in a nonessential region of the gene, such as an intron or the 3′-untranslated region, genome editing could potentially expunge the repeats and prevent their expansion (termed repeat snipping) without affecting the normal function of the gene.
Expansion of a hexanucleotide repeat GGGGCC in the first intron of the C9orf72 (chromosome 9 open reading frame 72) gene (OMIM 614260) is the most commonly known genetic abnormality in patients with ALS and in the related disorder, frontotemporal dementia.38 Death of patients with ALS or frontotemporal dementia is attributable to progressive loss of motor neurons in the brain and spinal cord. In a proof-of-concept study, Mutihac et al39 generated iPSC lines from 4 patients with ALS or frontotemporal dementia carrying the C9orf72 repeat expansion. CRISPR-Cas9 genome editing was used to target the expanded (GGGGCC)n repeat (Figure 2C). The corrected ALS-iPSC–derived motor neurons rescued the ALS phenotype by increasing cellular survival and enhancing calcium homeostasis.
A similar strategy can be applied to other nucleotide repeat disorders, such as DM1, a common form of adult muscular dystrophy with a prevalence of 1 in 8000 worldwide.40 Myotonic dystrophy type 1 is an autosomal dominant disease with multisystemic symptoms, including myotonia, muscle wasting, cardiac conduction defects, insulin resistance, cataracts, and cognitive dysfunction.41 It is caused by the progressive expansion of a CTG triplet in the 3′-untranslated region of the dystrophia myotonica-protein kinase (DMPK) gene (OMIM 605377) (Figure 2D). The expanded repeat in the DMPK gene is transcribed into a toxic CUG expansion RNA, which sequesters the muscle blind–like family of splicing factors. Muscle blind–like sequestration causes aberrant splicing of a large number of genes.42 These aberrant splicing events have been proposed to contribute to the multisystem clinical presentation of DM1. Genome editing could potentially be used to eliminate the CTG repeats that cause DM1. A proof-of-concept study37 has been reported using iPSCs derived from patients with DM1. A TALEN genome-editing strategy was used to insert polyA signals upstream of the DMPK CTG repeats, resulting in premature termination of transcription and elimination of the toxic mutant transcripts.43 Genome-editing–mediated repeat snipping represents a potential approach to permanently correct DM1 and other disorders caused by expansion of noncoding trinucleotide repeats,44 such as Friedreich ataxia and spinocerebellar ataxia type 8.
Discussion
Safety and efficacy of CRISPR-Cas9–based gene therapy need to be evaluated and refined before being applied therapeutically to repair mutations in human monogenic diseases. Four key issues, including potential off-target effects, delivery, immunogenicity, and longevity of the benefit of editing in vivo, are addressed below.
Possible Off-Target Effects of Genome Editing
One concern with the use of engineered nucleases and CRISPR-Cas9 to modify the human genome is the specificity of these genome-editing tools. Although significant off-target effects were not observed in an initial study45 of genome editing, it is conceivable that certain sgRNAs will have significant off-target effects. Unexpected cuts and mutations in the genome attributable to promiscuous targeting of nucleases to sequences related to those of the gene to be targeted have the potential to generate adverse effects. Potential off-target effects by ZNFs and TALENs are reduced by the requirement that only heterodimers of the nuclease domains are functional. In addition, the target sequence for ZNFs and TALENs is usually more than 30 base pairs, minimizing the likelihood of homologous sequences at multiple locations in the genome. In contrast, Cas9 from Streptococcus pyogenes only needs a 20-nucleotide sgRNA to bind to its target sequence and make a DSB, and an early study45 found that a few mismatches between the sgRNA and its target were tolerated for Cas9 editing. Recent studies46,47 of genome-wide binding of Cas9 and elucidation of the crystal structure of Cas9, sgRNA, and target DNA have provided a deeper understanding of the molecular basis of target recognition and target specificity of the Cas9 system, which is important for the design of optimized sgRNAs to avoid potential off-target effects in genome editing.
Multiple approaches have been developed to evaluate possible Cas9-mediated off-target effects. Multiple similar sequences in the genome can be identified by the Basic Local Alignment Search Tool and tested for off-target cutting. This approach is fast and simple; however, it may not reveal off-target effects on other sites and may not be sensitive enough to identify rare off-target effects in an entire organism. In silico prediction of target sites and testing by deep sequencing have emerged as accepted methods of identification of possible off-target sites. Unbiased whole-genome sequencing would be ideal. However, it is costly and time consuming and therefore unlikely to be widely used for regular analysis. Several new strategies have also been developed to improve the specificity or minimize off-target modification of the CRISPR-Cas9 system, including paired Cas9 nickases,48–50 truncated guide RNA,51 titration of dosage for Cas9-sgRNA,52 and high-fidelity53 or enhanced Cas9.54 Continued advances in this area will undoubtedly minimize possible off-target actions of genome-editing tools.
Current Delivery Strategies
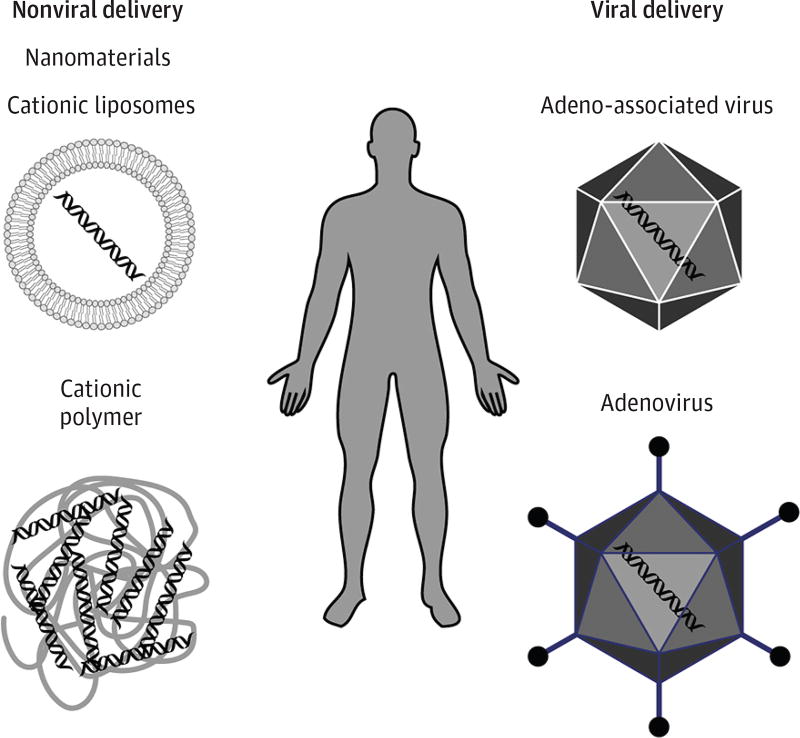
For genome-editing technologies to become clinically viable for a wide range of neuromuscular disorders, the delivery systems need to be efficient and effective. Genome-editing components have been delivered to target cells by nonviral or viral vector–mediated delivery systems (Figure 3). Other delivery approaches that have, thus far, been less effective include hydrodynamic delivery of naked DNA plasmid that contains expression cassettes of Cas9 and sgRNA and nanomaterials, such as cationic liposomes or cationic polymers, which readily associate with negatively charged nucleic acid (DNA and RNA) to form polycationic nanomeric particles. Replicative-defective viruses provide the most efficient method for in vivo delivery of genome-editing components.55 Recombinant adenovirus has been used to deliver Cas9 and sgRNA in mdx mice to effectively edit the Dmd gene. Although recombinant adenovirus efficiently infects postmitotic cells and accepts 37 kilobases of exogenous genetic information, it produces an acute immunologic response, severely limiting clinical applicability.
Figure 3. In Vivo Delivery Strategies for CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas9 (CRISPR-Associated Protein 9) Genomic-Editing Machinery.
Recombinant AAV (rAAV) has a variety of serotypes, offering variation in tissue specificity and infectivity. In addition, rAAVs do not integrate into the genome and are not associated with human disease, making them attractive delivery systems for gene therapy. The major disadvantage of these vectors is the limited transgene capacity of 4.7 to 4.9 kilobases, thereby limiting the size of the genome-editing components that can be delivered. Owing to high tropism for specific tissues and nonpathogenicity in humans, rAAV gene therapy has emerged as one of the most promising delivery approaches. Recent studies16–18 have successfully used this delivery method to correct dystrophin expression in mdx mice using CRISPR-Cas9.
Immunogenicity
Potential immunologic complications of CRISPR-Cas9 therapies include the immune response to (1) the rescued protein product; (2) genome-editing components, such as Cas9 protein; and (3) delivery particles, such as viral proteins. In patients with monogenic disorders in which the product of the mutated gene is completely absent or in an abnormal form, introduction of previously unseen epitopes may trigger an immune response. However, in the case of DMD, approximately 50% of patients have 0.2% to 4% dystrophin-positive revertant muscle fibers,56 attributable to spontaneous exon skipping, which may mitigate possible immune responses. With regard to Cas9 immunogenicity, high-level expression of Cas9 in various tissues of transgenic mice evoked no overt abnormalities.57,58 However, in humans, the potential immunogenicity of Cas9 remains to be determined.
Recombinant adenoviruses elicit the strongest immune responses among current viral delivery systems. However, new strategies have been developed to limit immunologic responses to recombinant adenoviruses, such as preemptive administration of immunosuppressants to prevent innate immune responses to the vector and the development of adenoviruses that lack viral genes. Although rAAV gene therapy is the favored viral delivery system, a substantial fraction of the population harbors preexisting humoral immunity to AAV serotypes, rendering them resistant to AAV delivery approaches. Current clinical trials circumvent this issue by excluding research participants with preexisting anti-AAV antibodies and providing prophylactic pretreatment with anti-inflammatory drugs. Efforts are also underway to generate rAAV capsids that are less immunogenic and more efficient by coating rAAV particles, modifying the capsid of rAAV to reduce the immune response, and creating novel chimeric vectors.
Efficiency and Longevity of the Benefit of Editing In Vivo
Genome editing of monogenic neuromuscular diseases is clearly a goal worth pursuing. Although multiple proof-of-concept studies have revealed the potential of genome editing to cure disease in animal models, long-term benefits and effects need to be examined in large animal models and humans. Skeletal muscle is well suited for genome-editing therapies because correcting a small subset of skeletal muscle cells leads to progressive improvement in muscle function at least in part attributable to the multinucleation of this tissue. In addition, genome editing of adult skeletal muscle stem cells (termed satellite cells), in principle, provides the possibility of a renewable population of corrected cells for continuous regeneration of diseased muscle. Because the heart and nervous system lack endogenous regenerative potential, more efficient genome-editing strategies of these tissues are needed for clinical use. On the other hand, because cardiomyocytes and neurons have a very low turnover rate, once these cell types are corrected, they can confer long-lasting clinical benefits. Indeed, a recent study16 revealed the progressive rescue of cardiomyocytes in mdx mice.
Conclusions
Monogenic neuromuscular diseases impair the function of muscles, motor nerves, and/or neuromuscular junctions. These debilitating diseases are commonly noticed during early childhood, but as the disease progresses, there is no effective treatment. Most therapies focus on alleviating symptoms and are ineffective at ameliorating the disease. New genome-editing technologies target the genetic cause of monogenic disease. In particular, CRISPR-Cas9–mediated genome editing is revolutionizing our understanding of the molecular basis of numerous monogenic neuromuscular diseases and is providing a path toward potential cures of these devastating diseases in patients.
Key Points.
Question
What is the clinical potential of genome-editing technology for treating neuromuscular monogenic disorders?
Findings
This review describes recent advances in genome-editing technology, the latest applications of the method, and potential clinical applications by highlighting 4 monogenic neuromuscular disorders as examples. Opportunities and obstacles in the path toward efficient and permanent correction of the genetic cause of these disorders are discussed.
Meaning
Genome editing is a powerful, revolutionary approach to permanently eliminate or correct the genetic cause of monogenic diseases and has broad translational potential when efficacy, delivery, and safety issues are addressed.
Acknowledgments
Funding/Support: This work was supported in part by grant U54 HD 087351 from the Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center, the Hamon Center for Regenerative Science and Medicine, and grant 1-0025 from the Robert A. Welch Foundation (Dr Olson).
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Long and Amoasii had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Concept and design: All authors.
Acquisition, analysis, or interpretation of data: Long, Amoasii.
Drafting of the manuscript: Long, Amoasii.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Long.
Obtained funding: Long, Bassel-Duby, Olson.
Administrative, technical, or material support: Long.
Study supervision: Long, Olson.
Conflict of Interest Disclosures: None reported.
References
- 1.World Health Organization. [Accessed August 19, 2016];Genes and human disease. http://www.who.int/genomics/public/geneticdiseases/en/index2.html.
- 2.Kaplan JC, Hamroun D. The 2016 version of the gene table of monogenic neuromuscular disorders (nuclear genome) Neuromuscul Disord. 2015;25(12):991–1020. doi: 10.1016/j.nmd.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27(22):4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7(2):91–92. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 10.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class-2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Wang L, Bell P, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabebordbar M, Zhu K, Cheng JK, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendell JR, Shilling C, Leslie ND, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71(3):304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 20.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14(6):373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 21.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Park KH, Zhao L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24(3):564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li HL, Fujimoto N, Sasakawa N, et al. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4(1):143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojtal D, Kemaladewi DU, Malam Z, et al. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet. 2016;98(1):90–101. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maggio I, Stefanucci L, Janssen JM, et al. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44(3):1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyombe-Engembe JP, Ouellet DL, Barbeau X, et al. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol Ther Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young CS, Hicks MR, Ermolova NV, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18(4):533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aartsma-Rus A. Overview on DMD exon skipping. Methods Mol Biol. 2012;867:97–116. doi: 10.1007/978-1-61779-767-5_7. [DOI] [PubMed] [Google Scholar]
- 30.Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. 2016;165(7):1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Human Genome Research Institute. Learning about spinal muscular atrophy. [Accessed August 19, 2016]; https://www.genome.gov/20519681/learning-about-spinal-muscular-atrophy/#top. Updated February 19, 2012.
- 32.Hendrickson BC, Donohoe C, Akmaev VR, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet. 2009;46(9):641–644. doi: 10.1136/jmg.2009.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercuri E, Bertini E, Iannaccone ST. Childhood spinal muscular atrophy: controversies and challenges. Lancet Neurol. 2012;11(5):443–452. doi: 10.1016/S1474-4422(12)70061-3. [DOI] [PubMed] [Google Scholar]
- 34.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corti S, Nizzardo M, Simone C, et al. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4(165):165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S, Gao S, Leonard DG, Paessler M, Wilson RB. Inverse correlation between SMN1 and SMN2 copy numbers: evidence for gene conversion from SMN2 to SMN1. Eur J Hum Genet. 2003;11(9):723. doi: 10.1038/sj.ejhg.5201032. [DOI] [PubMed] [Google Scholar]
- 37.DiMatteo D, Callahan S, Kmiec EB. Genetic conversion of an SMN2 gene to SMN1: a novel approach to the treatment of spinal muscular atrophy. Exp Cell Res. 2008;314(4):878–886. doi: 10.1016/j.yexcr.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 38.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mutihac R, Ababneh N, Scaber J, Wade-Martins R, Cowley S, Talbot K. Modelling amyotrophic lateral sclerosis (ALS) using mutant and CAS9/CRISPR-corrected motor neurons from patients with C9ORF72 mutations reveals disease-specific cellular phenotypes. J Neurol Sci. 2015;357(suppl 1):e48. [Google Scholar]
- 40.National Human Genome Research Institute. Learning more about myotonic dystrophy. [Accessed August 19, 2016]; https://www.genome.gov/25521207/learning-about-myotonic-dystrophy/. Updated June 4, 2012.
- 41.Meola G, Cardani R. Myotonic dystrophies: an update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim Biophys Acta. 2015;1852(4):594–606. doi: 10.1016/j.bbadis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37(pt 6):1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Y, Guo X, Santostefano K, et al. Genome therapy of myotonic dystrophy type 1 iPS cells for development of autologous stem cell therapy [published online June 28, 2016] Mol Ther. doi: 10.1038/mt.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum Mol Genet. 2000;9(6):909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 45.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang F, Taylor DW, Chen JS, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351(6275):867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen B, Zhang W, Zhang J, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 50.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maggio I, Chen X, Gonçalves MA. The emerging role of viral vectors as vehicles for DMD gene editing. Genome Med. 2016;8(1):59. doi: 10.1186/s13073-016-0316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burrow KL, Coovert DD, Klein CJ, et al. CIDD Study Group. Dystrophin expression and somatic reversion in prednisone-treated and untreated Duchenne dystrophy. Neurology. 1991;41(5):661–666. doi: 10.1212/wnl.41.5.661. [DOI] [PubMed] [Google Scholar]
- 57.Carroll KJ, Makarewich CA, McAnally J, et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–343. doi: 10.1073/pnas.1523918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dow LE, Fisher J, O’Rourke KP, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33(4):390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]