Abstract
The discovery that microorganisms can be etiologic agents of disease has driven clinical, research and public health efforts to reduce exposure to bacteria. However, despite extensive campaigns to eradicate pathogens (via antibiotics, vaccinations, hygiene, sanitation, etc.), the incidence and/or severity of multiple immune-mediated diseases including, paradoxically, infectious disease have increased in recent decades. We now appreciate that most microbes in our environment are not pathogenic, and that many human-associated bacteria are symbiotic or beneficial. Notably, recent examples have emerged revealing that the microbiome augments immune system function. This review will focus on how commensal-derived signals enhance various aspects of the host response against pathogens. We suggest that modern lifestyle advances may be depleting specific microbes that enhance immunity against pathogens. Validation of the notion that absence of beneficial microbes is a risk factor for infectious disease may have broad implications for future medical practices.
Introduction
The discovery of antibiotics in the last century is one of the most significant achievements of modern medicine. Pathogens that once devastated entire civilizations, such as Mycobacterium tuberculosis, could finally be controlled, suggesting a triumph over infectious disease. However, the rampant rise of antibiotic resistance among pathogens, compounded by a drying pipeline of novel antibiotic development by pharmaceutical companies has rendered current therapeutic strategies ineffective. As such, we have entered the post-antibiotic era where pathogens once again reign with limited opposition and a minor scrape may pose the risk of a fatal infection [1,2]. To combat the renewed threat of pathogenic microorganisms, clinical approaches towards eradicating infectious disease must evolve.
The recent increase in the severity and incidence of Clostridium difficile-associated diarrhea (CDAD) is emblematic of medicine’s current failings as well as its possible future. The disruption of intestinal microbiota, most commonly by antibiotics, prompts infection by C. difficile resulting in disease that ranges from mild diarrhea to fulminant colitis [3]. Once fatal, the advent of antibiotics consigned it to a manageable infection. However, the spread of antibiotic-resistant, hypervirulent strains in recent years has created an epidemic that is exceedingly difficult to manage [4]. Currently, 20–25% of patients experience relapsing disease, further reflecting the reduced efficacy of antibiotic therapy [3]. Besieged by an unrelenting pathogen, clinicians began to supplement patients with the fecal contents of healthy donors in an attempt to reestablish the natural resistance afforded by the microbiota against C. difficile. Fecal transplantation embraces the hygiene hypothesis which argues microbial exposure, particularly that of commensal microbes, is beneficial to host health. This approach of administering microbes to combat disease is in shocking contrast to standard medical practices of the last century that, abiding by the principles of germ theory, indiscriminately targets microbes as a means of promoting individual health. Yet, achieving a 91% primary cure rate, the use of fecal transplantation insists upon a reassessment of our clinical strategy towards preventing and treating infectious disease [5].
The commensal microbiota is primarily comprised of indigenous bacteria that colonize the external interfaces of its host. Co-evolution has resulted in microbes with extensive and diverse impacts on multiple aspects of host biology including nutrient acquisition, immune development and neurological function [6–8]. Appropriately, conditions that disrupt the symbiotic host-microbial coexistence significantly alter predisposition to a wide spectrum of disorders. This review will focus on the contribution of commensal microbiota in promoting host resistance against infectious disease. Furthermore, we will discuss how efforts to support the integrity of the microbiota, as through bacteriotherapy or the supplementation with microbial products, may be an effective means of achieving protection against infection.
The intestinal microbiota promotes host resistance against mucosal infection
The development of enteric infection following antibiotic use has long been observed in both clinical practice and animal models of disease [3]. This observation suggests some mechanism by which the commensal microbiota protects against pathogen invasion and dissemination. The utilization of animal models to study the microbiota, including germ-free (GF) mice that lack microbial exposure, has revealed significant insight into the diverse and intricate contribution of the commensal microbes to host resistance against infectious disease.
Commensal microbes directly resist enteric pathogens
The commensal microbiota achieves resistance against opportunistic infection, in part, through niche competition. By competing for sites of colonization and nutrient uptake, commensal microbes are able to limit pathogen expansion at host epithelial surfaces [9]. GF mice are highly susceptible to enteric infection with Citrobacter rodentium, a murine pathogen used to model infection with enterohemorragic and enteropathogenic Escherichia coli [10]. Bacteriotherapy with isolated commensal microbes results in pathogen clearance, in part, due to the enhanced glycan acquisition capabilities of the transferred bacteria. These findings reveal direct competition between commensal microbes and pathogens for nutrients as a means of limiting infection at sites of colonization.
Conversely, recent studies show that certain enteric pathogens are able to outcompete commensal microbes by actively triggering host inflammation which favors pathogen invasion and dissemination [11]. C. rodentium, Campylobacter jejuni, and Salmonella enterica serovar Typhimurium (STm) appear to induce inflammation as part of their infectious process, and increasing intestinal inflammation actually promotes disease [12,13]. Further, these reports surprisingly demonstrate that pathogen-induced inflammation adversely affects the microbiota, reducing the numbers of beneficial bacteria, which protect us from infections. Collectively, there is growing evidence for the notion that pathogens and symbiotic bacteria are engaged in an ‘evolutionary combat’, with the host serving as the battlefield.
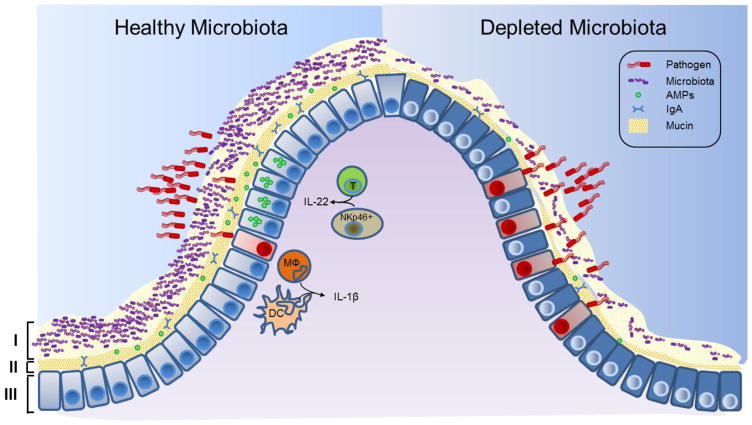
Under conditions in which direct competition is insufficient to limit pathogen invasion, the commensal microbiota promotes resistance to infection by mediating protective host immune responses. Immune modulation by commensal microbes is indispensable in achieving host-microbial symbiotic coexistence and preventing inflammatory disease [7]. We now appreciate that this influence extends into supporting protection against infectious disease by promoting both barrier immunity as well as priming immune defenses against pathogen insult (Fig. 1).
Figure 1. The intestinal microbiota promotes three levels of protection against enteric infection.
I, Saturation of colonization sites and competition for nutrients by the microbiota limit pathogen association with host tissue. II, Commensal microbes prime barrier immunity by driving expression of mucin, immunoglobulin A (IgA) and antimicrobial peptides (AMPs) that further prevents pathogen contact with host mucosa. III, Finally, the microbiota enhances immune responses to invading pathogens. This is achieve by promoting IL-22 expression by T cells and NKp46+ cells, which increases epithelial resistance against infection, as well as priming secretion of IL-1B by intestinal monocytes (MΦ) and dendritic cells (DCs), which promotes recruitment of inflammatory cells into the site of infection. In conditions in which the microbiota is absent, such as following antibiotic treatment, there is reduced competition, barrier resistance and immune defense against pathogen invasion.
Commensal microbes promote barrier immunity
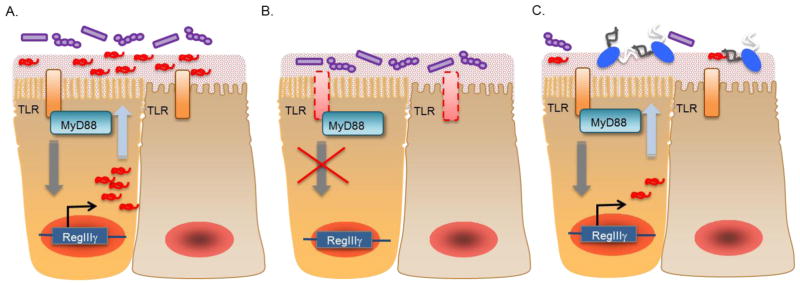
Immune modulation by the microbiota occurs through commensal-derived signals such as microbial associated molecular patterns (MAMPs). Host recognition of MAMPs is achieved by pathogen recognition receptors (PRRs), such as Toll-like receptors (TLRs). At mucosal surfaces, these commensal-derived signals drive epithelial production of mucin, secretion of immunoglobulin A (IgA), and the expression of antimicrobial peptides (AMPs) that limit microbial contact to mucosal tissue [14–16]. One such example is commensal driven expression of RegIIIγ by intestinal epithelial cells (Fig. 2). RegIIIγ is a C-type lectin that possesses antimicrobial activity against Gram-positive microbes [17]. Expression of RegIIIγ requires TLR recognition of commensal MAMPs [18]. As such, disruption of the microbiota, as through antibiotic treatment, reduces production of RegIIIγ resulting in a breakdown of barrier immunity. As a consequence, antibiotic-treated mice are highly susceptible to opportunistic infection with enteric pathogens such as vancomycin-resistant enterococcus (VRE) [19]. Supplementation of antibiotic-treated mice with purified MAMPs is sufficient to prime RegIIIγ expression and achieve resistance against infection. VRE is a common cause of antibiotic-associated diarrhea and, similar to C. difficile, exceedingly difficult to treat. Herein is another example of how current treatment strategies predispose the host to secondary infections and how efforts to maintain the integrity of the microbiota or supplement it during antibiotic treatment may be effective in limiting susceptibly to opportunistic pathogens.
Figure 2. The commensal microbiota primes barrier immunity.
Direct stimulation of epithelial Toll-like receptors (TLRs) by commensal MAMPs primes expression of RegIIIγ (a). Production of RegIIIγ is essential to limit microbial contact with host mucosa. As such, defects in TLR function results in deficient RegIIIγ expression resulting in an increased association of commensal microbes with host tissue as well as a heighten risk of infection with enteric pathogens (b). Additionally, reduced TLR stimulation as a consequence of the depletion of the microbiota is sufficient to reduce RegIIIγ expression and render the host susceptible to infection.
Commensal microbes prime immune resistance to pathogen invasion
Under conditions in which barrier resistance fails, commensal microbes continue to limit pathogen dissemination by enhancing immune clearance mechanisms. One such mechanism by which the microbiota promotes host resistance is through priming interleukin-1 (IL-1) β expression. IL-1β is a proinflammatory cytokine that is expressed in an inactive form (pro IL-1β) that is subsequently cleaved by caspases following inflammasome activation [20]. Intestinal mononuclear phagocytes isolated from specific pathogen-free (SPF) mice express pro-IL-1β, which is deficient in cells isolated from GF mice [21]. Cleavage of pro-IL-1β into its active form occurs after challenge with pathogenic microorganisms, such as STm, but not following exposure to commensal microbes. This would suggest that commensal microbes promote pro-IL-1β expression among intestinal mononuclear cells, which is specifically activated following pathogen insult. Appropriately, commensal-driven pro-IL-1β expression enhances resistance to enteric infection with STm.
Additional mucosal immune responses are driven by the microbiota, including the differentiation of T-helper 17 (Th17) cells and IL-22 expression by intestinal NKp46+ cells [22,23]. While specific details for the role of both cell types in regulating commensal microbes remains to be revealed, both are critical in combating mucosal infection with C. rodentium. It thus appears that the microbiota drives certain immune responses, including the production of pro-IL-1β, with the primary purpose of promoting resistance to pathogenic infection.
Commensal microbes prevent pathogen invasion at colonization sites beyond the gut
While the majority of the studies assessing the contribution of the microbiota to host resistance to infection have focused on the gut, colonization by commensal microbes at other barrier sites also affords pathogen protection. Skin microbes prime local development of Th1, Th17 and IL-17+ gamma-delta T cells [24]. Cutaneous T cell differentiation by commensal microbes is achieved through MAMP-driven IL-1β signaling. This response is independent of the intestinal microbiota as oral antibiotic treatment, which reduced intestinal Th1 and Th17 cells, has no effect on the immune profile within the skin. Furthermore, colonization of GF mice with the prominent skin commensal Staphylococcus epidermidis is sufficient to rescue the defective immune response in GF mice. Priming of these immune responses by skin microbes is instrumental in promoting resistance against cutaneous infection with Leishmania major. Here we see a critical influence, afforded by commensal microbes, in localized host immune development and subsequent protection against infection.
Immune protection is also achieved by commensal microbes residing within the respiratory mucosa. Antibiotic-treated mice display reduced resistance to influenza infection [25]. Disease susceptibility is characterized by defective IL-1β production as well as reduced dendritic cell recruitment and T cell priming. As a consequence, antibiotic-treated animals display attenuated T cell and B cell responses following viral infection. Interestingly, depletion of the microbiota did not enhance susceptibility to infection with herpes simplex virus type 2 or Legionella pneumophila indicating specificity for pathogens to which the microbiota promotes resistance. Intranasal inoculation with purified MAMPs, such as LPS, is sufficient to restore protective immunity to infection, as is, surprisingly, intrarectal MAMP administration. These findings suggest the imunoprotective properties of commensal microbes are not limited to the sites of colonization, but rather may extend to distal compartments and may even support host resistance against systemic infection.
Commensal microbes promote host resistance to systemic infection
While commensal microbes are physically restricted to external sites of colonization, their influence on host immune responses extends into systemic compartments. This concept was revealed with the finding that GF mice display a diminished splenic CD4+ T cell profile [26]. Monocolonization with a prominent intestinal commensal, Bacteroides fragilis, is sufficient to promote CD4+ T cell development within the spleen. The role of commensal microbes in driving systemic immune maturation suggests that disruption of the microbiota may compromise host immunity and increase susceptibility to systemic infection.
Deliberate depletion of the microbiota reduces resistance to systemic infection with Lymphocytic Choriomeningitis Virus (LCMV) [27]. Antibiotic-treated mice display increased viral burden as a consequence of attenuated anti-viral immune responses following infection. Macrophages isolated from antibiotic-treated mice are deficient in type I and II interferon (IFN) signaling, as well as in controlling viral replication ex vivo. This defect in innate immune resistance contributes to an impaired adaptive immune response, which includes deficient expansion and cytolytic activity of LCMV-specific CD8+ T cells as well as reduced serum titers of anti-LCMV IgG. Furthermore, the defect in anti-viral immunity among microbiota-depleted mice may also reflect altered transcriptional response following infection. Splenic mononuclear cells, isolated from GF mice, are deficient in expressing pro-inflammatory cytokines when stimulated with purified MAMPs [28]. This defective response is associated with reduced transcription of various inflammatory response genes due to chromatin modification of the promoter region. These studies reveal a remarkable role for commensal microbes in programing host systemic defense responses during steady state conditions. Furthermore, as this influence is reversible, temporary depletion of the microbiota is sufficient to compromise systemic immune resistance to pathogen invasion.
In addition to priming anti-viral immune responses during steady state conditions, commensal microbes may also protect against systemic bacteremia. Neutrophils isolated from the bone marrow of antibiotic-treated or GF mice are attenuated in ex vivo killing of extracellular pathogens Staphylococcus aureus and Streptococcus pneumoniae [29]. This defect was reproduced in mice deficient in Nod1, a PRR which recognizes peptidoglycan derived meso-diaminopimelic acid (mesoDAP), but not in mice deficient in other PRRs. Molecules from intestinal microbes are found in the bone marrow neutrophil stores, indicating that direct stimulation by commensal MAMPs primes neutrophil activity. Appropriately, neutrophil antimicrobial activity among antibiotic-treated mice is rescued following stimulation with Nod1 ligand. While it remains to be shown that the absence or disruption of the microbiota actually reduces resistance to bacterial infection, these collective findings suggest immune priming by commensal microbes is critical in promoting host resistance against systemic infections.
Defects in host-microbial symbiosis may predicate susceptibility to infection
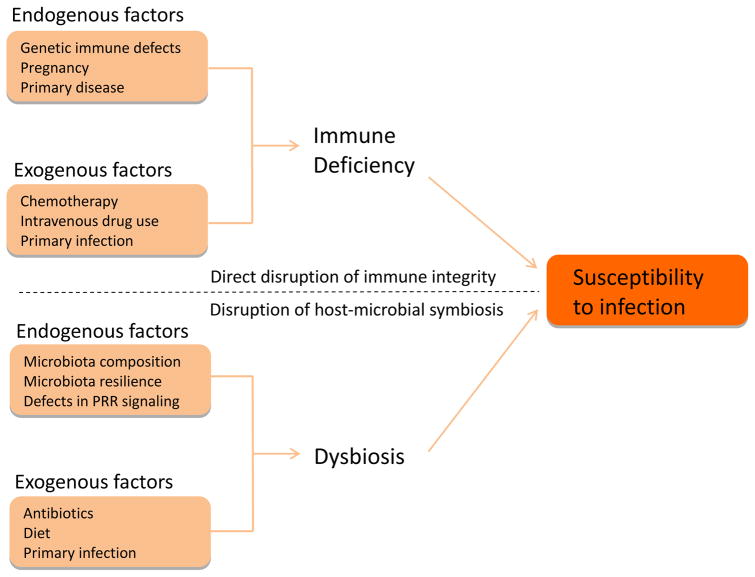
Factors that determine an individual’s susceptibility to infectious disease remain largely unknown. Here we suggest that environmental and genetic influences that disrupt the microbiota or impede host sensing of commensal-derived signals may confer vulnerability to pathogen infection (Fig. 3). As discussed earlier, depletion of the microbiota through antibiotics is sufficient to compromise host immune function and increase the risk of opportunistic infection. Other environmental factors that disrupt the composition of the microbiota, including gastrointestinal infection or diet, may additionally serve as a risk factor for disease [12,30]. Susceptibility to infection may even persist long after exposure to the microbiota-disrupting agent. Tracking the intestinal commensal profile among patients taking oral antibiotics revealed recovery in the composition of the microbiota following cessation of therapy [31]. However, there is a delay of several weeks to months between the final antibiotic administration and recovery of the microbiota to the pre-treatment composition. This delay, in animal models, was associated with increased susceptibility to infection, reflecting persistent consequences of antibiotic therapy [32]. Alternatively, certain individuals display alterations for up to four years after antibiotic treatment, indicating a defect in microbiota resilience [33]. We speculate that such a defect, while asymptomatic, may compromise the protective contribution of the commensal microbiota to host immunity and weaken resistance against pathogenic insult.
Figure 3. Disruption of host-microbial symbiosis as a risk factor for infectious disease.
Exposure to pathogenic microorganisms is often insufficient to cause disease. Rather, susceptibility to infectious disease reflects deficient immune resistance to pathogen challenge. As such, exogenous and endogenous factors that directly compromise individual immune function (including genetic immune defects and chemotherapy) are significant risk factors for infection. We extend this model by proposing that the factors that disrupt the protective benefits of the commensal microbiota similarly compromise individual immune integrity and predispose to infectious disease.
Defects in host sensing of the beneficial influence of commensal microbes may also serve as a risk factor for disease. Nod2 is an intracellular PRR that recognizes muramyl dipeptide, a conserved structural moiety of bacterial peptidoglycan [34]. Nod2 signaling promotes expression of Paneth cell α-defensin, a class of antimicrobial peptides, that, similar to RegIIIγ, limits microbial contact with host tissue [35]. As a consequence of the diminished α-defensin production, Nod2-deficient mice display heightened susceptibility to gastroenteritis by Listeria monocytogenes. Furthermore, as homozygous mutations in this receptor are associated with increased incidence of Crohn’s disease, defects in host sensing of commensal signals may be a risk factor for inflammatory bowel disease (IBD) by reducing clearance of pathogenic bacteria [34]. Indeed, the finding that adhesive and invasive E. coli (AIEC) are tightly associated with the intestinal epithelium among patients with Crohn’s disease may support this notion [36].
Finally, the genetic selection of one’s microbiota composition may reflect individual susceptibility to infection. NIH Swiss (NIH) mice are naturally resistant to gastrointestinal infection with C. rodentium, compared to C3H/HeJ (HeJ) mice which develop lethal disease [37]. Resistance among NIH mice is associated with increased expression of IL-22 and RegIIIβ, relative to HeJ mice. As the microbiota drives the expression of both antimicrobial mediators, susceptibility to infection may be a function of gut bacterial community composition. To test this hypothesis, HeJ mice were depleted of microbiota through antibiotic treatment, and colonized with intestinal microbes from NIH mice. The bacterial community profile of transplanted mice was shown to resemble that of the NIH donor. Remarkably, transfer of commensal microbes from NIH to HeJ mice is sufficient to promote resistance to infection. Protection is associated with increased expression of IL-22 and RegIIIβ, and protection is lost following neutralization of IL-22. Reciprocally, transplantation of HeJ microbiota to NIH mice increased disease burden to C. rodentium. Finally, pups in the subsequent generation inherit the microbiomes transferred to their parents. Offspring remarkably display resistance patterns to C. rodentium infection relative to their microbiota composition, rather than their genetics. These data suggest that familial history of infectious disease may not only reflect the inheritance of susceptibility genes, but possibly the vertical transmission of a microbiota that is less protective against pathogen challenge.
Conclusion
The evidence summarized in this review suggests that disruption of the microbiota through environmental influences may compromise immune function, leading to increased susceptibility to infectious disease. In particular, we propose that antibiotic use may paradoxically promote bacterial and viral infections by depleting immune-promoting gut bacteria. For example, antibiotics are routinely administered in the hospital to patients admitted for various non-bacterial illnesses. Not only can this practice select for antibiotic-resistant microbes (an extensively reported phenomenon), but may also lead to nosocomial infections by reducing the ability of the immune system to fight infections. Furthermore, antibiotic use over several generations may reduce gut bacteria diversity in entire populations, a notion proposed by the ‘disappearing microbiota’ hypothesis [38]. In cases where antimicrobial use is justified, we speculate that the administration of commensal-derived products that promote immunity may represent a viable companion therapy to antibiotics. Given the rise of antibiotic resistance among pathogens and the potential loss of beneficial microbes in Western societies, efforts that support microbiome-mediated protection may be an effective approach to achieve resistance to infectious disease in the post-antibiotic era.
Review highlights.
Commensal microbes are critical in promoting host resistance against infectious disease.
Protection by the microbiota from infection can be achieved through direct competition with pathogenic microorganisms for space and/or nutrients.
Priming of immune responses by the microbiota to combat pathogens represents a potentially novel approach to control infectious disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alanis AJ. Resistance to Antibiotics: Are We in the Post-Antibiotic Era? Archives of Medical Research. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Kahrstrom CT. Entering a post-antibiotic era? Nat Rev Micro. 2013;11:146–146. [Google Scholar]
- 3.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 4.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 5.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, et al. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 6.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6:306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid G, Howard J, Gan BS. Can bacterial interference prevent infection? Trends in Microbiology. 2001;9:424–428. doi: 10.1016/s0966-842x(01)02132-1. [DOI] [PubMed] [Google Scholar]
- 10.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007;5:e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, et al. Host-Mediated Inflammation Disrupts the Intestinal Microbiota and Promotes the Overgrowth of Enterobacteriaceae. Cell Host & Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–539. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 17.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, et al. The Antibacterial Lectin RegIIIγ Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. These two studies reveal the critical role of commensal microbes in promoting barrier immunity through driving epithelial expression of RegIIIγ. Furthermore, they show that defects in host detection of commensal MAMPs or depletion of the microbiota through antibiotic treatment compromises RegIIIγ expression and predisposes to infection with VRE, a clinically relevant pathogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathinam VAK, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. This study shows that commensal microbes specifically prime mucosal immune responses against pathogenic microorganisms by promoting expression of pro-IL-1β which is selectively activated following infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 24**.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. The study is one of the first to unveil the contribution of skin commensal microbes in driving cutaneous immune development and resistance to parasitic infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27*.Abt Michael C, Osborne Lisa C, Monticelli Laurel A, Doering Travis A, Alenghat T, et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Ganal Stephanie C, Sanos Stephanie L, Kallfass C, Oberle K, Johner C, et al. Priming of Natural Killer Cells by Nonmucosal Mononuclear Phagocytes Requires Instructive Signals from Commensal Microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. These two studies show commensal microbes promote resistance against systemic viral infection, in part by priming macrophage anti-viral immune responses during steady state conditions. [DOI] [PubMed] [Google Scholar]
- 29.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda S, Hsu L-C, Liu H, Bankston LA, Iimura M, et al. Nod2 Mutation in Crohn’s Disease Potentiates NF-κB Activity and IL-1β Processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-Dependent Regulation of Innate and Adaptive Immunity in the Intestinal Tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 36.Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, et al. Escherichia coli isolated from a Crohn’s disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. Altering host resistance to infections through microbial transplantation. PLoS One. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nature reviews Microbiology. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]