Abstract
Rapid and sensitive point-of-care diagnostics are of paramount importance for early detection of infectious diseases and timely initiation of treatment. Here, we present cellulose paper and flexible plastic chips with printed graphene-modified silver electrodes as universal point-of-care diagnostic tools for the rapid and sensitive detection of microbial pathogens or nucleic acids through utilizing electrical sensing modality and loop-mediated isothermal amplification (LAMP). We evaluated the ability of the developed paper-based assay to detect (i) viruses on cellulose-based paper microchips without implementing amplification in samples with viral loads between 106 and 108 copies per ml, and (ii) amplified HIV-1 nucleic acids in samples with viral loads between 10 fg µl−1 and 108 fg µl−1. The target HIV-1 nucleic acid was amplified using the RT-LAMP technique and detected through the electrical sensing of LAMP amplicons for a broad range of RNA concentrations between 10 fg µl−1 and 108 fg µl−1 after 40 min of amplification time. Our assay may be used for antiretroviral therapy monitoring where it meets the sensitivity requirement of the World Health Organization guidelines. Such a paper microchip assay without the amplification step may also be considered as a simple and inexpensive approach for acute HIV detection where maximum viral replication occurs.
Graphical abstract
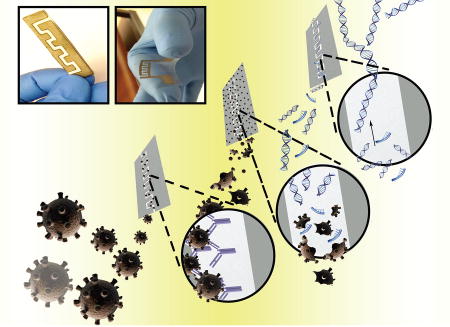
A paper microchip with printed graphene-modified silver nano-composite electrodes was developed for microbial pathogen detection. Human immunodeficiency virus-1 (HIV-1) was captured on cellulose paper substrates functionalized with anti-gp120 antibody and captured viruses were detected through the electrical sensing of viral lysate. Utilizing the loop-mediated isothermal amplification (LAMP) technique to amplify the nucleic acids of the target pathogen enhanced the sensitivity of the paper microchip.
Introduction
Infectious diseases are one of the top three causes of death globally.1 It is estimated that there will be 13 to 15 million deaths annually due to infectious diseases by 2050,2 more than half of which will occur in developing countries.3 The development of effective, rapid, sensitive, and low-cost point-of-care (POC) diagnostics is critical for infectious disease management in developed and developing countries. Recently, various classes of paper- and plastic-based materials have opened a new paradigm in developing a wide range of low-cost and disposable biosensing devices with significant potential for field applications in resource-limited settings.4–9 Such devices are easy-to-fabricate, mass-producible, disposable, and inexpensive and can be integrated with various detection modalities, such as fluorescence,10–12 electrochemical,13–15 photoelectrochemical,16 electrochemiluminescence17, 18 and colorimetric.19–21
Electrical sensing-based modalities are insensitive to light intensity and do not require bulky components usually used in optical-based assays and have been used extensively in developing POC biosensing assays.4, 22–29 Such platforms utilize various conductive electrode materials including gold, carbon, silver or graphene. Of particular interest is silver that is highly conductive, stable, and flexible.30, 31 Similarly, graphene, a single atom layer thick two-dimensional allotrope of carbon, has also an extraordinary electrical double layer capacitance,32 high mechanical strength,33 high carrier electron mobility,34 high surface-to-volume ratio,35, 36 and low signal-to-noise ratio.37 Graphene has been used in the development of optical,38 electrochemical,39 and field effect transistor sensors40 for pathogen detection by leveraging its high carrier electron mobility34 and low signal-to-noise ratio.37 Silver/graphene nano-composites may provide a robust biosensing material for developing diagnostics with an electrical sensing modality,41 as silver/graphene nano-composites showed high electrical and thermal conductivity and flexibility in comparison with graphene alone.42, 43
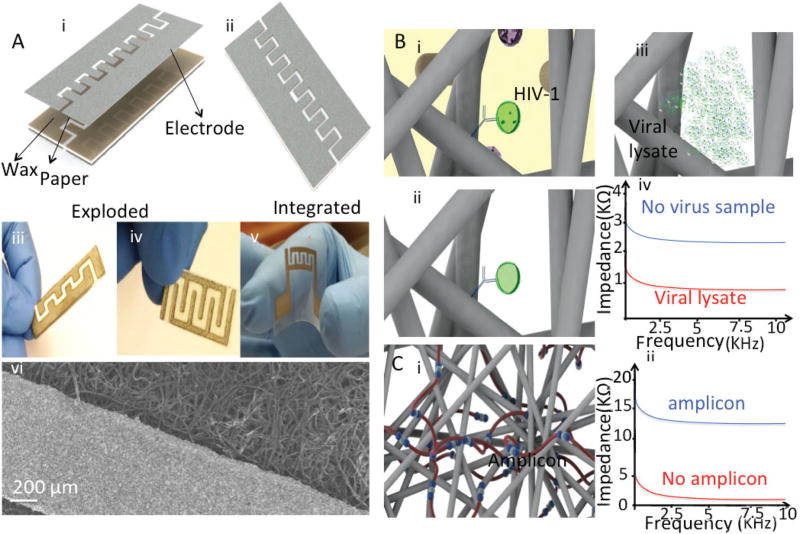
Herein, we developed and tested paper microchips with printed graphene-modified silver electrodes (GSEs) for virus and nucleic acid detection (Fig. 1). The schematic of exploded and integrated paper chip with graphene-modified silver electrodes for virus detection is shown in Fig. 1A(i and ii).
Fig. 1.
(A) Cellulose and plastic paper chips with graphene-modified silver electrodes in the (i) exploded and (ii) integrated modes. (iii) A Maze 4 paper chip, (iv) waxed-modified Maze 4 paper chip, (v) plastic paper chip with printed flexible electrodes, (vi) Scanning Electron Microscopy (SEM) of graphene-modified silver electrodes printed on a cellulose substrate. The scale bar is 200 µm. (B) Detection mechanism without nucleic acid amplification. (i) Intact viruses are captured on the paper chip using anti-gp120 antibody. (ii) The paper is washed with a low electrically conductive solution to remove the electrically conductive background. (iii) Captured viruses are then lysed. (iv) HIV-1 nano-lysate is detected through on-chip impedance measurement. (C) Virus detection mechanism with the LAMP technique and electrical sensing. (i) Target HIV nucleic acids are amplified using the RT-LAMP method. (ii) The LAMP amplicons are then detected through on-chip impedance magnitude measurement.
Electrodes can be printed on cellulose and plastic substrates with different electrode geometries and configurations (Fig. 1A(iii–v)). The scanning electron microscopy (SEM) image of fabricated graphene-modified silver electrodes is shown in Fig. 1A(vi). In the presented detection method, HIV-1 is captured on-chip using an anti-gp120 antibody immobilized on the surface of paper substrates (Fig. 1B(i)) and captured viruses are then washed on-chip to remove the non-target cells, viruses, and molecules (Fig. 1B(ii)). Captured viruses can be detected through the electrical sensing of viral lysate (Fig. 1B(iii and iv)) or LAMP amplicons (Fig. 1C(i and ii)).
Results
Electrode and paper substrate materials
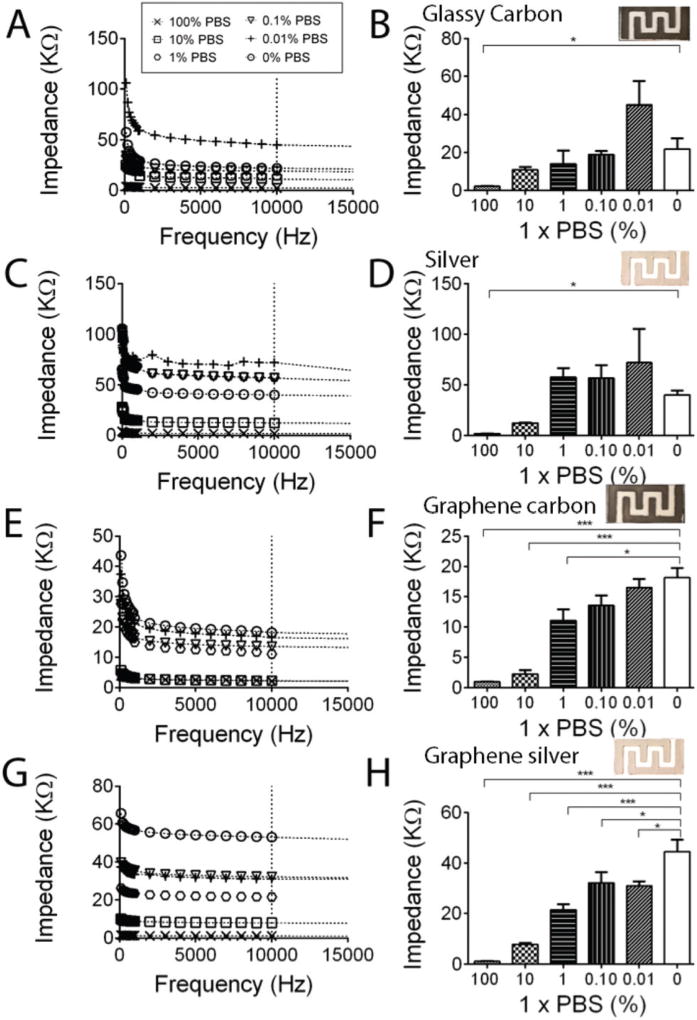
We first analyzed and optimized the effect of the electrode material on the electrical response of the paper chips. We tested 4 different electrode materials, including (i) silver ink, (ii) carbon ink, (iii) 20% (w/w) graphene-modified silver, and (iv) 20% (w/w) graphene-modified carbon electrodes. The electrical responses of the prepared electrode materials were tested using different serial dilutions of 1× PBS (100%, 10%, 1%, 0.1%, 0.01%, 0.001% and 0%) spiked in DI water. We performed impedance spectroscopy of the samples on paper chips with printed electrodes for frequencies between 1 Hz and 1 MHz (Fig. 2). We observed a maximum impedance change in the samples due to the presence of electrically charged biomolecules for frequencies below 10 kHz. We chose 10 kHz for our quantifications and statistical analysis as we observed the lowest signal-to-noise ratio and the highest impedance change at this frequency (Fig. 2). The least diluted detectable PBS sample on paper chips with carbon (Fig. 2A and B, p < 0.05, n = 3) and silver (Fig. 2C and D, p < 0.05, n = 3) electrodes was 100% PBS based on statistical analysis of the impedance magnitude of the samples at 10 kHz. There was no significant difference between the impedance magnitude of 10%, 1%, 0.1%, and 0.01% PBS samples and DI water at 10 kHz (Fig. 2A and B, p > 0.05, n = 3).
Fig. 2.
Electrode material optimization on cellulose paper chips. Impedance spectroscopy (A) and impedance magnitude at 10 kHz (B) of diluted PBS samples on cellulose paper chips with carbon electrodes. 100% PBS showed significantly different impedance magnitudes compared to the DI water sample. Impedance spectroscopy (C) and impedance magnitude at 10 kHz (D) of diluted PBS samples on cellulose paper chips with silver electrodes. The least diluted PBS sample that was detected using chips with silver electrodes was 10% PBS. Impedance spectroscopy (E) and impedance magnitude at 10 kHz (F) of diluted PBS samples on cellulose paper chips with graphene-modified carbon electrodes. The least diluted PBS sample that showed significantly different impedance magnitudes compared to DI water was 1% PBS. Impedance spectroscopy (G) and impedance magnitude at 10 kHz (H) of diluted PBS samples on cellulose paper chips with graphene-modified silver electrodes. The least diluted PBS sample with significantly different impedance magnitudes compared to DI water is 0.01% PBS. All error bars are a standard error of mean (SEM) based on repeating each experiment three times (n = 3).
Fig. 2E and F show the electrical response of the paper chips with graphene-modified carbon electrodes when diluted PBS samples were used. The least diluted PBS sample that was detected using paper chips with graphene-modified carbon electrodes was 1% PBS (Fig. 2E and F, p < 0.0001, n = 3). We observed that paper chips with printed graphene-modified silver electrodes demonstrated the optimum sensitivity as the lowest diluted PBS sample that was detected on these paper chips was 0.01% PBS (Fig. 2G and H, p < 0.05, n = 3).
We also evaluated the effect of the cellulose paper material on the electrical response, stability, and reproducibility of the paper chips with printed electrodes using a cellulose paper pad and Whatman chromatography paper substrates that are usually used in developing paper-based microfluidics.44 The impedance magnitude of the samples on cellulose-based paper substrates can significantly change due to sample evaporation at room temperature. The sample evaporation rate is dependent on the material and thickness of the cellulose-based substrates. Considering the time required for signal readout in our system (1–2 min), we evaluated the effect of sample evaporation at room temperature on the impedance results when Whatman paper and paper pad substrates were used for detection. We observed that samples rapidly evaporate on Whatman paper compared to paper pad substrates which may be attributed to the thickness of these paper substrates. Whatman paper substrates are thinner compared to paper pad substrates. The impedance magnitude of DI water samples (100 µl) at 10 kHz on a cellulose paper pad and Whatman chromatography paper over a period of 10 minute incubation time is shown in Fig. S1A and B,† respectively. The cellulose paper pad showed a consistent impedance magnitude signal for the DI samples during the 10 min incubation time, whereas the impedance magnitude of the samples on Whatman chromatography paper chips changed significantly during the same time period (Fig. S1†). The electrical signal instability on Whatman chromatography paper chips may be due to the rapid sample evaporation on these relatively thinner substrates.
Electrode geometry design
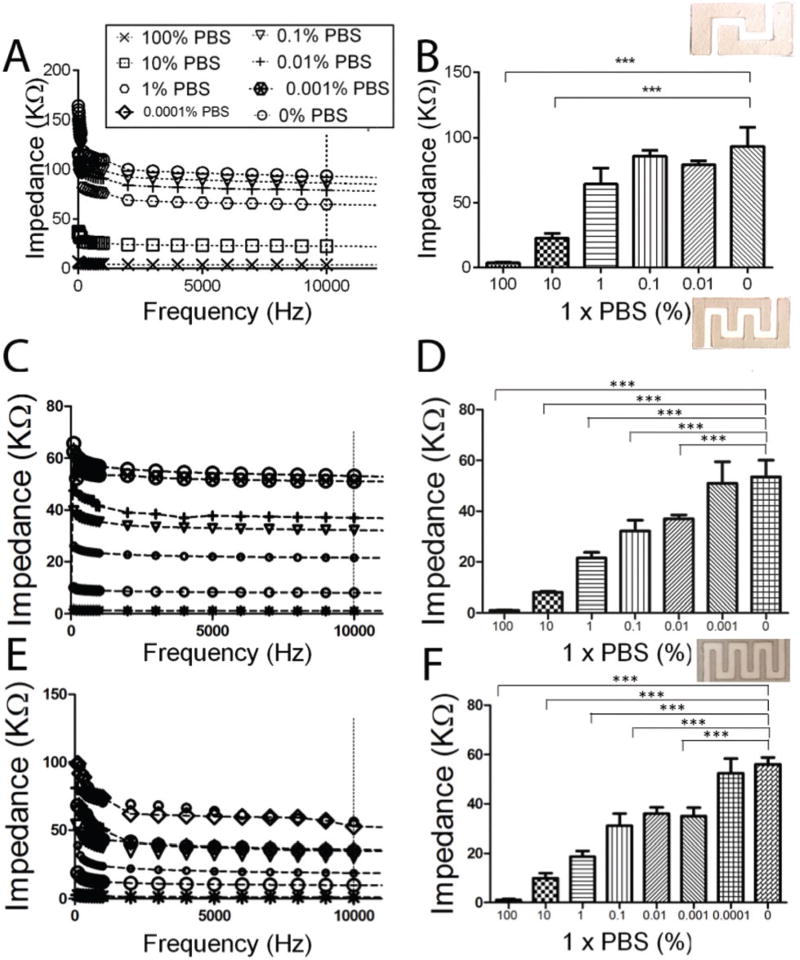
The increase in the electrode surface area is directly correlated to the detection sensitivity of the paper chips.45 We evaluated the effect of the number of interdigitated finger electrodes on the sensitivity of the paper chips through measuring the impedance magnitude of diluted PBS samples on chips with 2, 3, and 4-finger electrode arrays named Maze 2, Maze 3, and Maze 4, respectively. The least possible diluted PBS sample that was detected with Maze 2 chips was 10% PBS (Fig. 3A and B, p < 0.001, n = 3). The sensitivity of Maze 3 chips to detect diluted PBS samples was relatively higher than Maze 2 chips (Fig. 3C and D, 0.01% PBS, p < 0.001, n = 3) with a clear separation between electrical signals for 100%, 10%, 1% and 0.1% PBS samples (Fig. 3E and F, p < 0.001, n = 3). We were able to detect 0.001% PBS using Maze 4 paper chips (Fig. 3E and F, p < 0.001, n = 3). These results suggest that increasing the number of interdigitated finger electrodes could improve the detection limit of the paper chips.
Fig. 3.
The effect of the graphene-modified silver electrode design on the sensitivity of the paper chip. (A, B) Impedance spectroscopy and magnitude at 10 kHz of diluted PBS samples on Maze 2 (A, B), Maze 3 (C, D), and Maze 4 (E, F) chips. The least diluted PBS samples that showed significantly different impedance magnitudes compared to DI on Maze 2, Maze 3, and Maze 4 chips were 10% PBS, 0.01% PBS, and 0.0001% PBS, respectively. All error bars are SEM based on repeating the experiments three times (n = 3).
Intact virus particle detection
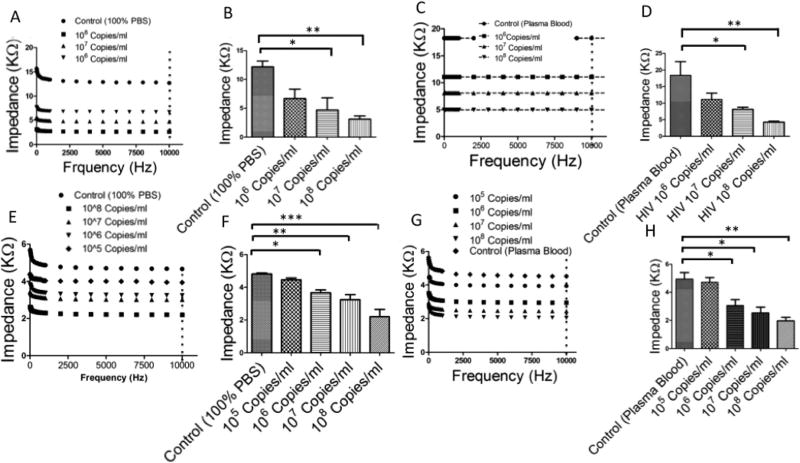
HIV-1 particles spiked in PBS and plasma were initially captured on the surface of Maze 4 chips with graphene-modified silver electrodes using an immobilized monoclonal anti-gp120 antibody. We used two methods for antibody immobilization: (i) the streptavidin–biotin coupling protocol and (ii) the covalent crosslinking of the antibody to cellulose fibers. The substrate surface was then blocked using 5% (v/v) BSA solution in DI water. The captured viruses were washed using DI water to remove electrically conductive solutions from the paper chip (Fig. S2†). The captured viruses were then lysed using 50 µl of 2% Triton X-100 diluted in DI water. As shown in Fig. 4A and B we were able to detect HIV-1 in spiked PBS samples with virus concentrations of 107 copies per ml and 108 copies per ml (p < 0.05, n = 3). The negative control was virus-free 1× PBS samples. The impedance magnitudes for viral lysate samples were significantly lower in comparison with the negative control. We were able to detect HIV-1 in spiked plasma samples with virus concentrations of 107 copies per ml and 108 copies per ml (Fig. 4C and D, p < 0.05, n = 3). We also evaluated the effect of the antibody immobilization mechanism on the sensitivity of the paper chips. The detection limit of the paper chips with a covalently bound antibody was one order of magnitude lower for HIV-spiked plasma samples compared to the detection limit of paper chips with a physically adsorbed antibody (Fig. 4E–H). We were able to detect HIV in spiked PBS and plasma samples with virus concentrations as low as 106 copies per ml (Fig. 4F and H, p < 0.05, n = 3). In addition, these results showed that the covalent-based antibody immobilization provided a more efficient surface chemistry on-chip for repeatable and robust target capture and detection.
Fig. 4.
HIV-1 capture and detection using cellulose paper microchips with graphene-modified silver electrodes. (A–D) Virus capture and detection using on-chip physical antibody immobilization. (A) Impedance spectroscopy of viral lysate for frequencies between 1 Hz and 10 kHz. (B) Impedance magnitudes of viral lysate samples at 10 kHz. We were able to detect HIV-1 in spiked PBS samples with concentrations of 107 copies per ml and 108 copies per ml. (C) Impedance spectroscopy of viral lysate samples in HIV-spiked plasma samples. (D) Impedance magnitude of viral lysate in HIVspiked plasma samples at 10 kHz. (E–H) Virus capture and detection using the on-chip covalent immobilization of anti-gp120 antibody. (E) Impedance spectroscopy of viral lysate samples in HIV-spiked PBS. (F) Impedance magnitude of viral lysate samples in HIV-spiked PBS at 10 kHz. The impedance magnitude of viral lysate samples in HIV-spiked PBS with a viral load of 106 copies per ml was significantly different than virus-free control samples. (G) Impedance spectroscopy of viral lysate for HIV-spiked plasma. (H) Impedance magnitude of viral lysate for HIV-spiked plasma samples at 10 kHz. The impedance magnitude of the viral lysate of the HIV-spiked plasma with a viral load of 106 copies per ml was different than the virus-free control.
Nucleic acid detection
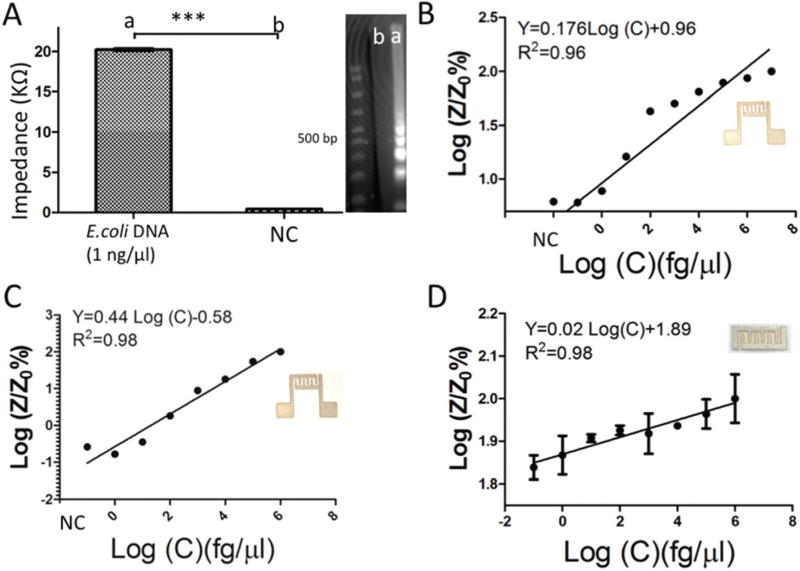
In order to enhance the sensitivity, we integrated LAMP-based nucleic acid amplification with the on-chip electrical sensing method. A set of previously designed primers for amplification of the Tuf gene in E. coli46 and p24 in HIV-147 was used for the LAMP-based amplification of target nucleic acids (Table S1†). The amplification reaction relied on the consumption of primers and deoxynucleoside triphosphates (dNTPs)48 which accumulate non-precipitated magnesium pyrophosphate and protons49 thereby reducing the sample electrical conductivity.50 The reduction in the sample electrical conductivity is correlated to the concentration of the target nucleic acid.49 To optimize the electrical detection of LAMP amplicons, we first evaluated the impedance magnitude change of amplified E. coli DNA (Tuf gene) compared to DNA-free negative control DI samples. We observed that the maximum impedance magnitude change occurs in a range between 1 kHz and 100 kHz and we set 1 kHz for impedance measurement analysis (Fig. S3†).
A sample with 1 ng µl−1 of E. coli DNA concentration in DI water was amplified for 60 min using a benchtop LAMP assay and the impedance magnitude of the amplified target was then measured at 1 kHz on a plastic chip with printed electrodes at room temperature (Fig. 5A).
Fig. 5.
Detection of nucleic acids on plastic- and cellulose-based chips. (A) Impedance magnitude of the E. coli LAMP amplified product after 60 min and negative control (DI water) at 1 kHz. (B) Normalized impedance magnitude of the LAMP E. coli amplified product after 40 min amplification on a plastic-based chip (Maze 4). Z and Z0 are the impedance magnitudes of the sample at 40 min and control, respectively. The sensitivity of this chip was 1 fg µl−1. (C) Normalized impedance magnitude of the HIV-1 LAMP amplified product at 1 kHz using a plastic Maze 4 chip. The sensitivity of this plastic chip for HIV detection was 10 fg µl−1. (D) The electrical response of cellulose paper chips for capturing and detecting HIV. The sensitivity of the cellulose paper chip was 10 fg µl−1.
The impedance magnitude of amplified E. coli (DNA 20 ± 0.03 kΩ) was significantly different than the impedance magnitude of the DNA-free control samples (422.6 ± 7.58 Ω). One of the important factors in such quantitative assays is the amplification time.46, 51–53 We set the amplification time at 40 min which was previously optimized for the quantification of amplified E. coli DNA (Tuf gene)52 and p24 HIV-1 RNA53 using LAMP and RT-LAMP methods. Fig. 5B shows the normalized impedance magnitude of E. coli DNA-spiked samples with concentrations from 0.1 fg µl−1 to 10 ng µl−1 with a sensitivity of 10 fg µl−1 (Fig. 5B, p < 0.05, n = 4). The gel images of E. coli DNA and HIV-1 RNA samples are shown in Fig. S4.† Following the same protocol, we detected different concentrations of HIV-1 RNA through the impedance magnitude measurement of LAMP amplicons using primers targeting p24 on both plastic and cellulose substrates (Fig. 5C and D). There was a linear relationship between the normalized impedance magnitude of the LAMP amplicons and the initial concentration of HIV-1 RNA. The sensitivity of the assay was 10 fg µl−1 for both plastic and cellulose paper Maze 4 chips.
Specificity evaluation
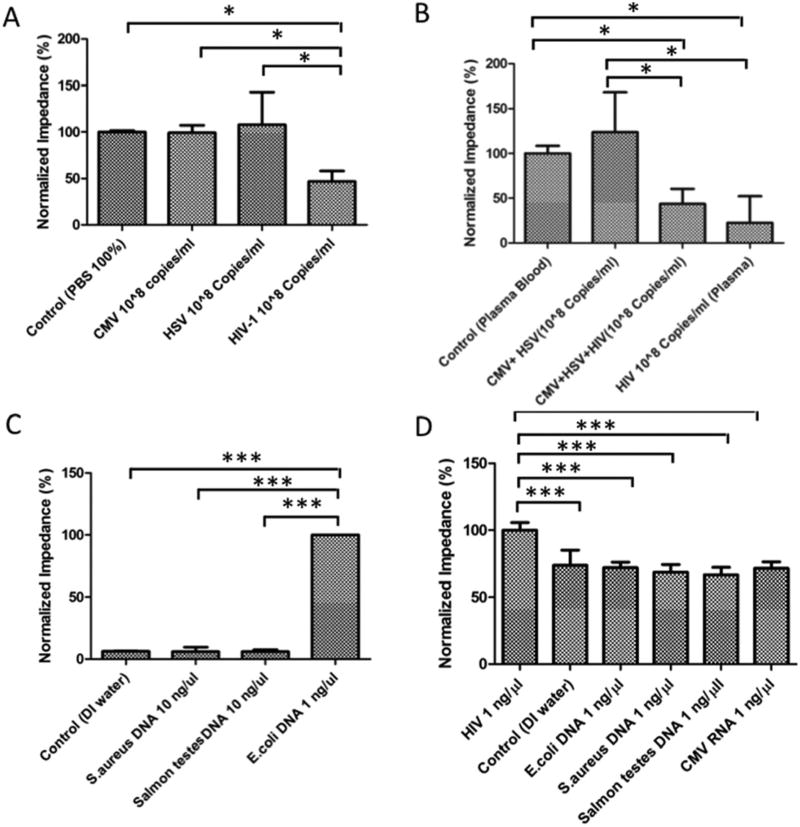
We evaluated the specificity of the assay in detecting target pathogens. The impedance magnitude of the PBS samples spiked with herpes simplex virus (HSV) and cytomegalovirus (CMV) was not significantly different than virus-free control samples (Fig. 6A) (p > 0.05, n = 3). The impedance magnitude of the HIV-spiked PBS samples was significantly different than virus-free control samples (p < 0.05, n = 3). We were also able to specifically detect HIV-1 in spiked plasma samples. Similarly, the impedance magnitude of the mixture of HSV and CMV spiked in plasma samples was not significantly different than virus-free control samples (Fig. 6B, p > 0.05, n = 3), whereas the impedance magnitudes of the HIV-spiked plasma samples as well as the mixture of HIV-1, HSV, and CMV samples were significantly different than virus-free control samples (Fig. 6B, p < 0.05, n = 3). These results showed that HIV-1 was specifically captured and detected on paper chips functionalized with the anti-gp120 antibody.
Fig. 6.
Specificity evaluation in a cellulose paper chip. (A) The normalized impedance magnitudes of the viral lysate of CMV- and HSV-spiked samples were not significantly different than the virus-free control (p > 0.05, n = 3), however the normalized impedance magnitude of the viral lysate of HIV-spiked PBS samples was significantly different than the control (p < 0.05, n = 3). (B) The normalized impedance magnitude of the mixture of CMV and HSV in spiked plasma samples was not significantly different than the control (p > 0.05, n = 3), however the normalized impedance magnitude of the viral lysate of HIV-spiked plasma was significantly different than the control (p < 0.05, n = 3). (C) Specificity test for E. coli detection. The normalized impedance magnitude of samples spiked with S. aureus and Salmon testes was not significantly different than the pathogen-free control (p > 0.05, n = 3), but the normalized impedance magnitude of LAMP E. coli products was significantly different than control samples (p < 0.05, n = 3). (D) Specificity test for LAMP HIV-1 detection. The normalized impedance magnitude of HIV-1 LAMP amplicons with an initial concentration of 1 ng µl−1 was significantly different than the control (p < 0.05, n = 3), but the normalized impedance magnitude of LAMP-based amplified HCV RNA, S. aureus DNA, E. coli DNA, and S. aureus DNA was not different than the control (p > 0.05, n = 3).
We also evaluated the paper chip assay in detecting target amplified nucleic acids through the electrical sensing of LAMP amplicons. The normalized impedance magnitude of amplified E. coli DNA samples was significantly different compared to the normalized impedance magnitude of non-target DNA samples including S. aureus DNA, Salmon testes, and HSV DNA on plastic Maze 4 chips (p < 0.001, n = 6, Fig. 6C). The normalized impedance magnitudes of S. aureus DNA, Salmon testes, and HSV DNA samples were not statistically different than pathogen-free control samples (p > 0.05, n = 6, Fig. 6C). The normalized impedance magnitude of amplified HIV-1 RNA samples was significantly different than the normalized impedance magnitude of non-target pathogens including CMV RNA, E. coli DNA, Salmon testes DNA, and S. aureus DNA (p < 0.001, n = 6, Fig. 6D). The normalized impedance magnitudes of CMV RNA, E. coli DNA, Salmon testes DNA, and S. aureus DNA samples were not statistically different than the negative control (p > 0.05, n = 6, Fig. 6D).
Discussion and conclusions
Here, we demonstrated on-chip virus capture and detection using antibody-coated cellulose paper and plastic microchips with printed graphene-modified silver inks. The anti-gp120 antibody was immobilized on the surface of chips through (i) the streptavidin–biotin binding of biotinylated antibodies and streptavidin-coated chips, and (ii) the covalent crosslinking of antibodies on aldehyde-modified cellulose paper chips.
The presented microchip technology has the potential to be used for viral load testing and more specifically for antiretroviral therapy (ART) monitoring. We demonstrated that the developed cellulose paper and plastic chips with printed graphene- modified electrodes can capture and detect intact HIV-1 through the electrical sensing of viral lysate in samples with viral loads between 106 and 108 copies per ml. This simple and inexpensive method can potentially be used for acute HIV-1 detection where virus replication and shedding is at its maximum level.54 We also showed that the sensitivity of the assay could be enhanced through integrating the LAMP technique with the printed-paper chips in order to amplify the target nucleic acid and enhance the electrical signal. We were able to achieve a sensitivity of 10 fg µl−1 (1000 RNA copies per reaction) for HIV-1 detection. The sensitivity of 10 fg µl−1 shows the ability of the presented LAMP-based assay for HIV detection to initiate ART and monitor the treatment (104 to 105 copies per ml)47 when samples with a volume of 25 µl are used. Increasing the sample volume to 100 µl can also increase the sensitivity of our assay to cover a broader range of virus concentrations between 103 and 108 copies per ml.47 Therefore, the LAMP-based paper microchip can potentially be used as a diagnostic tool for HIV-1 detection to initiate ART as well as ART monitoring specifically in developing countries. We were able to specifically detect amplified HIV-1 RNA and E. coli DNA on-chip.
One of the critical aspects in the early detection of infectious diseases and efficient treatment monitoring is rapid and inexpensive viral load testing at the POC, which can potentially enhance treatment expansion in developing countries where there is limited laboratory infrastructure and trained staff.55 Of particular interest is ART monitoring for HIV-infected patients that has shown great promise in suppressing the HIV disease, prolonging life in HIV-infected patients, and reducing the disease transmission rates.56 One of the major challenges in the field is the lack of affordable and rapid diagnostic assays for HIV detection and viral load testing, which causes restricted access to ART in resource-limited settings.57 Among the different HIV diagnosis methods, viral load testing seems to be the most accurate way of effectively monitoring treatment, detecting ART failure, and enabling alternative medication changes based on the World Health Organization (WHO) consolidated guidelines.58, 59 The current nucleic acid-based and rapid HIV diagnostic tools in the developed countries are complex, expensive, time-consuming, and laboratory-based, and cannot be easily transferred to developing countries.4 In addition, the available assays for quantifying p24 and CD4+ cells in the whole blood of patients lack the ability to effectively monitor disease treatment and to detect ART failure.60–64 Thus, there is an urgent and unmet clinical need for the development of accurate, low-cost, and rapid diagnostic tools for routine viral load testing in resource-limited settings.65–69
We developed a diagnostic platform for viral load testing by integrating paper-based microfluidics, the LAMP technique, and electrical sensing. Electrical sensing is an attractive modality that is insensitive to light intensity and can be integrated with miniaturized lab-on-chip platforms without the need for bulky components usually required in optical-based systems for developing portable biosensors.29, 70–73 Paper and plastic substrates are also appropriate materials for developing POC assays being disposable, flexible, inexpensive, and light.74–77 The LAMP technique is a promising mechanism for amplifying target nucleic acids with a great potential to be used in the development of POC molecular diagnostics.48 Compared to other isothermal amplification methods, LAMP provides relatively higher sensitivity and specificity, faster amplification, higher stability, and lower complexity.78–80 It has the ability to specifically amplify target nucleic acids in complex biological samples without the inhibitory issues usually observed in PCR-based methods.81
The material cost per test in our presented method is ~$2 (Tables S2 and S3†). Our LAMP-based assay time can take up to 60 min in a laboratory setting including 10 min sample preparation, 40 min LAMP process, and 10 min electrical signal readout and analysis. The assay time when no nucleic acid amplification was involved, was less than 40 min including 30 min incubation for virus capturing, 5 min for washing and virus lysis, and 5 min for electrical signal readout and analysis.
Compared to the electrochemical sensing modality for nucleic acid detection, electrical sensing does not require any redox molecules or cumbersome redox concentration optimization to perform the assay.49 The current microchip platform has the potential to be integrated with a handheld impedance meter to measure the impedance magnitude at a single frequency, which makes it more suitable for POC pathogen detection and viral load testing. This biosensing platform has the potential to be used for the detection and quantification of other viruses as it has well-described primers for RT-LAMP amplification or capturing antibodies for an immuno-viral test.
Experimental
Reagents
Nuclease-free deionised (DI) water (11-05-01-04) was purchased from Integrated DNA Technologies (IDT, Coralville, IA). Phosphate buffer saline (PBS) was obtained from Life Technologies (Grand Island, NY). Triton X-100, streptavidin from Streptomyces avidinii, bovine serum albumin (BSA), and potassium periodate (KIO4) were purchased from Sigma-Aldrich (St Louis, MO). Biotinylated (ab53937) and non-biotinylated (ab21179) anti-gp120 polyclonal antibodies were purchased from AbCam (Cambridge, MA). Cellulose paper pad (CFSP203000) was obtained from EMD Millipore (Billerica, MA). Whatman chromatography paper (3001-861) was purchased from GE Healthcare (Little Chalfont, UK). Materials for electrode fabrication including silver paste (cl-1001), carbon paste (cl-2001), and graphene paste (UHC-NPD-100 ML) were obtained from Engineered Materials Systems (Delaware, OH), and Graphene Supermarket Inc. (Calverton, NY), respectively.
LAMP reagents
LAMP assays were performed targeting the Tuf gene of E. coli and p24 gene in HIV-1 RNA. For E. coli assay, 20 µl of the master mix contained 2.5 µl of 10× polymerase thermopol buffer (New England Biolabs, Beverly, MA), 0.75 µl of 100 mM MgSO4 (New England Biolabs), 1 µl of 10 mM dNTP, 3.2 µl of 5 M betaine (Sigma-Aldrich), 1 µl of 8000 U ml−1 Bst polymerase, (New England Biolabs), dNTP (Bioshop, Canada), 0.2 µl of 20 µM outer primers (F3, B3) (final concentration of 0.2 µM), 1.8 µl of 20 µM inner primers (FIP, BIP), and 0.8 µl of 20 µM loop primers (LF, LB). All primers were synthesized and purchased from Integrated DNA Technologies (Coralville, IA). The primer sequences were selected based on previously published work.46, 82 Finally, 5 µl of target DNA was added to the reaction mix. LAMP reaction was performed at 65 °C on the benchtop and the amplicon was detected on-chip.
For p24 gene RT-LAMP amplification in HIV-1 RNA previously designed primers47 were used. The whole primer sequences are shown in Table S1.† 25 µl of the reaction mix consists of 2.5 µl of 10× thermopol polymerase buffer, 2.5 µl of 100 mM MgSO4, 1.4 µl of 25 mM dNTP, 0.2 M of 5 M betaine, 1 µl of 8000 U ml−1 Bst polymerase, 0.042 µl of 15 U ml−1 RT AMV enzyme (New England Biolabs), 0.2 µl of 20 µM F3, B3 outer primers (final concentration of 0.2 µM), 1.8 µl of 20 µM inner primers FIP and BIP (final concentration of 1.6 µM), and 0.8 µl of 20 µM loop primers LF, LB (final concentration of 0.8 µM). The remaining reaction sample was filled with HIV-1 target or DI water. LAMP reaction was performed on benchtop assay at 60 °C and the amplicons were detected on-chip using electrical sensing.
HIV culture
HIV-1 samples were cultured from peripheral blood monoclonal cells (PBMCs). PBMCs were isolated from HIV-1 using the Ficoll-Hypaque density gradient cell configuration. The obtained sample was stimulated by phytohemagglutinin (PHA) for a 3 day period and co-cultured with HIV-1 positive PBMCs. Then, samples were incubated at 37 °C in the presence of 5% of CO2. P24 titer measurement was carried out on the supernatant of the co-culture sample with ELISA (Perkin Elmer, NEK050b). When p24 in the sample reached the level of 20 ng ml−1, the co-culture process was stopped. The stock concentration of the sample was 108 copies per ml. The viral load test was performed by using a Roche-COBAS AmpliPrep TaqMan HIV-1 v2.0 system which was located in the microbiology laboratory at Brigham and Women’s Hospital (BWH).4
Chip fabrication
Electrodes were screen-printed on various flexible plastic and cellulose-based paper substrates. An electrode mask was cut into the desirable geometry and sizes on Mask-Ease® (Melissa & Doug, Wilton, CT) using a CO2 laser cutter. The mask sheet was attached on top of the substrate and conductive ink was smeared precisely on the surface of the mask to get electrodes of uniform thickness.83 In fabricating the microchips on plastic flexible substrates, a 2 mm thick poly(methyl methacrylate) PMMA sheet was engraved, with a power, speed and pulse per inch (PPI) of 85, 35 and 500, respectively. For cutting the PMMA, power, speed and PPI were set at 80, 5 and 1000, respectively. The printed electrodes were placed on a hotplate at 90 °C overnight (12 h), the mask was peeled off, and the substrate was then cut into pieces.
For the wax paper chip fabrication, a Xerox Color Qube 8580 wax printer was used. The wax pattern design was printed on both sides of the paper pads. Then the wax paper was placed into an oven at 90 °C for 20 min, the paper was flipped and incubated again for another 20 min to provide a consistent wax barrier to fill across the thickness of the paper pad (Fig. S5†).
Aldehyde-modified cellulose paper
Cellulose paper was modified with KIO4 for covalent crosslinking of the antibody on the surface of the paper.84 3 M KIO4 was prepared in DI water. Paper pads and Whatman chromatography paper were soaked in KIO4 solution at 65 °C for two hours. The substrates were then washed off three times with DI water by inserting the paper into the water container. After the washing step, the paper chip was dried out using a paper towel and was kept in a desiccator for 12 h.
Antibody immobilization and virus detection on cellulose paper
Biotinylated anti-gp120 antibody was non-covalently immobilized on the surface of paper substrates using streptavidin. 50 µl of streptavidin was added onto the paper chip with 5 min of incubation. Then, 50 µl of biotinylated anti-gp120 antibody with the concentration of 225 µg ml−1 was applied to the paper chip followed by 30 min of incubation. 100 µl of the HIV-spiked sample was added to the chip and incubated for 30 min. The chip was washed several times according to the optimized protocol (Fig. S2†) followed by adding 100 µl of 2% Triton X-100 diluted in DI water. Then the impedance of the chip was measured for frequencies between 1 Hz and 1 MHz using a LCR meter (LCR8000G, GW Instek, Taiwan). For the covalent binding of the non-biotinylated anti-gp120 antibody on the aldehyde functioned cellulose paper,84 50 µl of 225 µg ml−1 concentration was applied to the paper chip and the aforementioned procedure for virus detection was repeated accordingly.
LAMP amplified target detection
25 µl of LAMP amplicon was added on both the flexible substrate chip and cellulose paper and the impedance magnitude of the amplicon was detected at 1 kHz.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the support received from the National Institute of Allergy and Infectious Disease (NIAID), the National Institute of Health (NIH) through 1R01AI118502, Brigham and Women’s Hospital (BWH), Harvard Medical School (HMS), through the Bright Futures Prize and Fund to Sustain Research Excellence; the Department of Medicine, Harvard Medical School through the Innovation Evergreen Fund; King Abdulaziz University, Saudi Arabia through Scientific WAQF Fund under grant number 17/1436; and the National Engineering Research Council of Canada (NSERC) through NSERC Postdoctoral fellowship. The authors wish to thank Dr Niloufar Koohestanian and Anish Vasan for their help in performing preliminary experiments and discussions. The authors would like to also thank Professor Donald Coen and Seamus McCarron at Harvard Medical School for providing the team with CMV and HSV samples for specificity testing.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr06417e
Notes and references
- 1.Smartglobalhealth.org Infectious disease: a persistent threat. http://www.smartglobalhealth.org/issues/entry/infectious-diseases.
- 2.Dye C. Philos. Trans. R. Soc. London, Ser. B. 2014;369:20130426. doi: 10.1098/rstb.2013.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foudeh A, Fatanat Didar T, Veres T, Tabrizian M. Lab Chip. 2012;12:3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- 4.Shafiee H, Kanakasabapathy MK, Juillard F, Keser M, Sadasivam M, Yuksekkaya M, Hanhauser E, Henrich TJ, Kuritzkes DR, Kaye KM, Demirci U. Sci. Rep. 2015;5 doi: 10.1038/srep09919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobjörk D, Österbacka R. Adv. Mater. 2011;23:1935–1961. doi: 10.1002/adma.201004692. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, Xu F. Biosens. Bioelectron. 2014;54:585–597. doi: 10.1016/j.bios.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 7.Olkkonen J, Lehtinen K, Erho T. Anal. Chem. 2010;82:10246–20250. doi: 10.1021/ac1027066. [DOI] [PubMed] [Google Scholar]
- 8.Wong WS, Salleo A. Flexible electronics: materials and applications. Springer; New York, NY: 2009. [Google Scholar]
- 9.Shafiee H, Asghar W, Inci F, Yuksekkaya M, Jahangir M, Zhang MH, Durmus NG, Gurkan UA, Kuritzkes DR, Demirci U. Sci. Rep. 2015;5:8719. doi: 10.1038/srep08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Crooks RM. J. Am. Chem. Soc. 2011;133:17564–17566. doi: 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- 11.Connelly JT, Rolland JP, Whitesides GM. Anal. Chem. 2015;87:7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 12.Horning MP, Delahunt CB, Singh SR, Garing SH, Nichols KP. Lab Chip. 2014;14:2040–2046. doi: 10.1039/c4lc00293h. [DOI] [PubMed] [Google Scholar]
- 13.Dungchai W, Chailapakul O, Henry CS. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 14.Lan W-J, Maxwell EJ, Parolo C, Bwambok DK, Subramaniam AB, Whitesides GM. Lab Chip. 2013;13:4103–4108. doi: 10.1039/c3lc50771h. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Scida K, Crooks RM. Anal. Chem. 2015;87:9009–9015. doi: 10.1021/acs.analchem.5b02210. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Ge L, Wang P, Yan M, Ge S, Li N, Yu J, Huanga J. Lab Chip. 2013;13:3945–3955. doi: 10.1039/c3lc50430a. [DOI] [PubMed] [Google Scholar]
- 17.Mani V, Kadimisetty K, Malla S, Joshi AA, Rusling JF. Environ. Sci. Technol. 2013;47:1937–1944. doi: 10.1021/es304426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S, Wei S, Ying J, Safavieh M, Ahmed M. Biosens. Bioelectron. 2016;86:346–352. doi: 10.1016/j.bios.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 19.Martinez AW, Phillips ST, Whitesides GM. Anal. Chem. 2009;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 20.Dungchai W, Chailapakul O, Henry CS. Anal. Chim. Acta. 2010;674:227–233. doi: 10.1016/j.aca.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Safavieh M, Ahmed M, Sokullu E, Ng A, Braescu L, Zourob M. Analyst. 2014;139:482–487. doi: 10.1039/c3an01859h. [DOI] [PubMed] [Google Scholar]
- 22.Varshneya M, Li Y. Biosens. Bioelectron. 2011;24:2951–2960. doi: 10.1016/j.bios.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Yanga L, Bashir R. Biotechnol. Adv. 2008;26:135–150. doi: 10.1016/j.biotechadv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Mannoor MS, Tao H, Clayton JD, Sengupta A, Kaplan DL, Naik RR, Verma N, Omenetto FG, McAlpine MC. Nat. Commun. 2012;3:763. doi: 10.1038/ncomms1767. [DOI] [PubMed] [Google Scholar]
- 25.Mannoor M, Zhang S, Link A, McAlpine M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19207–19212. doi: 10.1073/pnas.1008768107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varshneya M, Li Y. Biosens. Bioelectron. 2009;24:2951–2960. doi: 10.1016/j.bios.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Ghafar-Zadeh E, Sawan M, Chodavarapu VP, Hosseini-Nia T. IEEE Trans. Biomed. Circuits Syst. 2010;4:232–238. doi: 10.1109/TBCAS.2010.2048430. [DOI] [PubMed] [Google Scholar]
- 28.Yang L. Talanta. 2008;74:1621–1629. doi: 10.1016/j.talanta.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Shafiee H, Jahangir M, Inci F, Wang S, Willenbrecht RBM, Giguel FF, Tsibris AMN, Kuritzkes DR, Demirci U. Small. 2013;9:2553–2563. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shankar R, Groven L, Amert A, Whites KW, Kellar JJ. J. Mater. Chem. 2011;21:10871–10877. [Google Scholar]
- 31.Huang G-W, Xiao H-M, Fu S-Y. Nanoscale. 2014;6:8495–8502. doi: 10.1039/c4nr00846d. [DOI] [PubMed] [Google Scholar]
- 32.Miller JR, Outlaw RA, Holloway BC. Science. 2010;329:1637–1639. doi: 10.1126/science.1194372. [DOI] [PubMed] [Google Scholar]
- 33.Lee C, Wei XD, Kysar JW, Hone J. Science. 2008;321:385–388. doi: 10.1126/science.1157996. [DOI] [PubMed] [Google Scholar]
- 34.Bolotin KI, Sikes KJ, Jiang Z, Klima M, Fudenberg G, Hone J, Kim P, Stormer HL. Solid State Commun. 2008;146:351–355. [Google Scholar]
- 35.Pumera M. Mater. Today. 2011;14:308–315. [Google Scholar]
- 36.Choi W, Lahiri I, Seelaboyina R, Kang YS. Crit. Rev. Solid State Mater. Sci. 2010;35:52–71. [Google Scholar]
- 37.Liu Y, Dong X, Chen P. Chem. Soc. Rev. 2012;41:2283–2307. doi: 10.1039/c1cs15270j. [DOI] [PubMed] [Google Scholar]
- 38.Li C-Y, Cao D, Kang Y-F, Lin Y, Cui R, Pang D-W, Tang H-W. Anal. Chem. 2016;88:4432–4439. doi: 10.1021/acs.analchem.6b00065. [DOI] [PubMed] [Google Scholar]
- 39.Liu F, Choi KS, Park TJ, Lee SY, Seo TS. BioChip J. 2011;5:123–128. [Google Scholar]
- 40.Liu F, Kim YH, Cheon DS, Seo TS. Sensors. 2013;186:252–257. [Google Scholar]
- 41.Kim J, Lee M-S, Jeon S, Kim M, Kim S, Kim K, Bien F, Hong SY, Park J-U. Adv. Mater. 2015;27:3292–3297. doi: 10.1002/adma.201500710. [DOI] [PubMed] [Google Scholar]
- 42.Marinho B, Ghislandia M, Tkalya E, Koning CE, With Gd. Powder Technol. 2012;221:351–358. [Google Scholar]
- 43.Pasricha R, Gupta S, Srivastava A. Small. 2009;5:2253–2259. doi: 10.1002/smll.200900726. [DOI] [PubMed] [Google Scholar]
- 44.Yetisen AK, Akram MS, Lowe CR. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 45.Stulík K, Amatore C, Holub K, Marecek V, Kutner W. Pure Appl. Chem. 2000;72:1483–1492. [Google Scholar]
- 46.Safavieh M, Ahmed M, Ng A, Zourob M. Biosens. Bioelectron. 2014;58:101–106. doi: 10.1016/j.bios.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Curtis KA, Rudolph DL, Owen SM. J. Virol. Methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Safavieh M, Kanakasabapathy MK, Tarlan F, Ahmed MU, Zourob M, Asghar W, Shafiee H. ACS Biomater. Sci. Eng. 2016;2:278–294. doi: 10.1021/acsbiomaterials.5b00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Li Q, Jin X, Jiang C, Lu Y, Tavallaie R, Gooding JJ. Sci. Rep. 2015;5:12539. doi: 10.1038/srep12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Liu W, Lu X, Gooding J, Li Q, Qu K. Anal. Biochem. 2014;466:16–18. doi: 10.1016/j.ab.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Safavieh M, Ahmed M, Tolba M, Zourob M. Biosens. Bioelectron. 2012;31:523–528. doi: 10.1016/j.bios.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 52.Tlili C, Sokullu E, Safavieh M, Tolba M, Ahmed M, Zourob M. Anal. Chem. 2013;85:4893–4901. doi: 10.1021/ac302699x. [DOI] [PubMed] [Google Scholar]
- 53.Liu C, Sadik MM, Mauk MG, Edelstein PH, Bushman FD, Gross R, Bau HH. Sci. Rep. 2014;4:7335. doi: 10.1038/srep07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.WHO. Global update on the health sector response to HIV. http://apps.who.int/iris/bitstream/10665/128494/1/9789241507585_eng.pdf?ua=1.
- 55.Shafiee H, Wang SQ, Inci F, Toy M, Henrich TJ, Kuritzkes DR, Demirci U. Annu. Rev. Med. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. [DOI] [PubMed] [Google Scholar]
- 56.The International AIDS Society Scientific Working Group on HIV Cure. Nat. Rev. Immunol. 2012;12:8. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. Antiretroviral therapy in low- and middle-income countries by region. Available at: http://www.who.int/hiv/topics/treatment/data/en/2011.
- 58.Marconi VC, Grandits G, Okulicz JF, Wortmann G, Ganesan A, Crum-Cianflone N, Polis M, Landrum M, Dolan MJ, Ahuja SK, Agan B, Kulkarni H H.I.V.W.G. Infectious Disease Clinical Research Program. PLoS One. 2011;6:e17956. doi: 10.1371/journal.pone.0017956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.World Health Organization. March 2014 supplement to the 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. http://www.who.int/hiv/pub/guidelines/arv2013/arvs2013upplement_march2014/2014. [PubMed]
- 60.Stevens G, Rekhviashvili N, Scott LE, Gonin R, Stevens W. J. Clin. Microbiol. 2005;43:857–861. doi: 10.1128/JCM.43.2.857-861.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schupbach J. Int. Arch. Allergy Immunol. 2003;132:196–209. doi: 10.1159/000074552. [DOI] [PubMed] [Google Scholar]
- 62.Labib M, Shipman PO, Martic S, Kraatz HB. Analyst. 2011;136:708–715. doi: 10.1039/c0an00741b. [DOI] [PubMed] [Google Scholar]
- 63.van Oosterhout JJG, Brown L, Weigel R, Kumwenda JJ, Mzinganjira D, Saukila N, Mhango B, Hartung T, Phiri S, Hosseinipour MC. Trop. Med. Int. Health. 2009;14:856–861. doi: 10.1111/j.1365-3156.2009.02309.x. [DOI] [PubMed] [Google Scholar]
- 64.Mee P, Fielding KL, Charalambous S, Churchyard GJ, Grant AD. AIDS. 2008;22:1971–1977. doi: 10.1097/QAD.0b013e32830e4cd8. [DOI] [PubMed] [Google Scholar]
- 65.Calmy A, Ford N, Hirschel B, Reynolds SJ, Lynen L, Goemaere E, Garcia de la Vega F, Perrin L, Rodriguez W. Clin. Infect. Dis. 2007;44:128–134. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- 66.Stevens WS, Scott LE, Crowe SM. J. Infect. Dis. 2010;201(Suppl 1):S16–S26. doi: 10.1086/650392. [DOI] [PubMed] [Google Scholar]
- 67.Tanriverdi S, Chen L, Chen S. J. Infect. Dis. 2010;201(Suppl 1):S52–S58. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- 68.Usdin M, Guillerm M, Calmy A. J. Infect. Dis. 2010;201(Suppl 1):S73–S77. doi: 10.1086/650384. [DOI] [PubMed] [Google Scholar]
- 69.World Health Organization. 2007 available at: http://whqlibdoc.who.int/publications/2007/9789241595568_eng.pdf.
- 70.Salmanzadeh A, Romero L, Shafiee H, Gallo-Villanueva RC, Stremler MA, Cramer SD, Davalos RV. Lab Chip. 2012;12:182–189. doi: 10.1039/c1lc20701f. [DOI] [PubMed] [Google Scholar]
- 71.Salmanzadeh A, Sano MB, Shafiee H, Stremler MA, Davalos RV. IEEE EMBS. 2012:590–593. doi: 10.1109/EMBC.2012.6346000. [DOI] [PubMed] [Google Scholar]
- 72.Shafiee H, Caldwell JL, Sano MB, Davalos RV. Biomed. Microdevices. 2009;11:997–1006. doi: 10.1007/s10544-009-9317-5. [DOI] [PubMed] [Google Scholar]
- 73.Shafiee H, Sano MB, Henslee EA, Caldwell JL, Davalos RV. Lab Chip. 2010;10:438–445. doi: 10.1039/b920590j. [DOI] [PubMed] [Google Scholar]
- 74.Daar A, Thorsteinsdóttir H, Martin D, Smith A, Nast S, Singer P. Nat. Genet. 2002;32:3. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 75.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 76.Solomon AW, Engels D, Bailey RL, Blake IM, Brooker S, Chen JX, Chen JH, Churcher TS, Drakeley CJ, Edwards T, Fenwick A, French M, Gabrielli AF, Grassly NC, Harding-Esch EM, Holland MJ, Koukounari A, Lammie PJ, Leslie J, Mabey DC, Rhajaoui M, Secor WE, Stothard JR, Wei H, Willingham AL, Zhou XN, Peeling RW. PLoS Neglected Trop. Dis. 2012;6:e1746. doi: 10.1371/journal.pntd.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 78.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Notomi T, Mori Y, Tomita N, Kanda H. J. Microbiol. 2015;53:1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 80.Mori Y, Notomi T. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Safavieh M, Nahar S, Zourob M, Ahmed MU. Microfluidic biosensors for high-throughput screening of pathogens in food. In: Bhunia AK, Taitt CR, Kim MS, editors. High Throughput Screening for Food Safety Assessment: Biosensor Technologies, Hyperspectral Imaging and Practical Applications. Woodhead Publishing Limited; 2015. pp. 327–357. [Google Scholar]
- 82.Ahmed M, Nahar S, Safavieh M, Zourob M. Analyst. 2013;138:907–915. doi: 10.1039/c2an36153a. [DOI] [PubMed] [Google Scholar]
- 83.Ahmed MU, Hossain MM, Safavieh M, Wong YL, Abd Rahman I, Zourob M, Tamiya E. Crit. Rev. Biotechnol. 2016;36:495–505. doi: 10.3109/07388551.2014.992387. [DOI] [PubMed] [Google Scholar]
- 84.Badu-Tawiah AK, Lathwal S, Kaastrup K, Al-Sayah M, Christodouleas DC, Smith BS, Whitesides GM, Sikes HD. Lab Chip. 2015;15:655–659. doi: 10.1039/c4lc01239a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.