Abstract
Stony corals (Scleractinia) are marine invertebrates that form the foundation and framework upon which tropical reefs are built. The coral animal associates with a diverse microbiome comprised of dinoflagellate algae and other protists, bacteria, archaea, fungi and viruses. Using a metagenomics approach, we analysed the DNA and RNA viral assemblages of seven coral species from the central Great Barrier Reef (GBR), demonstrating that tailed bacteriophages of the Caudovirales dominate across all species examined, and ssDNA viruses, notably the Microviridae, are also prevalent. Most sequences with matches to eukaryotic viruses were assigned to six viral families, including four Nucleocytoplasmic Large DNA Viruses (NCLDVs) families: Iridoviridae, Phycodnaviridae, Mimiviridae, and Poxviridae, as well as Retroviridae and Polydnaviridae. Contrary to previous findings, Herpesvirales were rare in these GBR corals. Sequences of a ssRNA virus with similarities to the dinornavirus, Heterocapsa circularisquama ssRNA virus of the Alvernaviridae that infects free-living dinoflagellates, were observed in three coral species. We also detected viruses previously undescribed from the coral holobiont, including a virus that targets fungi associated with the coral species Acropora tenuis. Functional analysis of the assembled contigs indicated a high prevalence of latency-associated genes in the coral-associated viral assemblages, several host-derived auxiliary metabolic genes (AMGs) for photosynthesis (psbA, psbD genes encoding the photosystem II D1 and D2 proteins respectively), as well as potential nematocyst toxins and antioxidants (genes encoding green fluorescent-like chromoprotein). This study expands the currently limited knowledge on coral-associated viruses by characterising viral composition and function across seven GBR coral species.
Keywords: Virus, Coral, Symbiodinium, Metagenomics, Holobiont, GBR
Introduction
Reef-building corals are keystone taxa of coral reefs; they are responsible for the deposition of a three-dimensional calcium-carbonate framework that constitutes the reef, and are the ecosystem’s main primary producers (Muscatine, Mccloskey & Marian, 1981). The coral animal lives in close association with a diverse suite of macroscopic and microscopic symbionts, and is thus a complex holobiont (Rohwer et al., 2002). A growing body of evidence supports a critical role for microbial symbionts in the coral holobiont (Blackall, Wilson & Oppen, 2015; Bourne, Morrow & Webster, 2016). Symbiodinium spp. are obligate symbionts and meet most of the nutritional requirements of the coral host animal by translocating photosynthate (fixed carbon) and nitrogen to the coral tissues (Muscatine, Mccloskey & Marian, 1981; Muscatine & Weis, 1992). Bacterial symbionts play a role in nitrogen fixation (Lema, Willis & Bourne, 2012; Lesser et al., 2004), sulphur metabolism (Raina et al., 2009), and coral holobiont immune responses (Krediet et al., 2013), while some bacteria can become opportunistic pathogens under certain conditions (Harvell et al., 2007; Weynberg et al., in press). The diversity and functional roles of other groups of symbionts within these holobiont communities are poorly studied. This is particularly true for viruses, i.e., the eukaryotic viruses, archaeal viruses, and bacteriophages, whose identity and diversity is still largely undescribed for many coral species. Earlier reports of viruses associated with corals were based on transmission electron microscopy observations of virus-like particles within coral tissue or present in the coral surface mucus layer (Davy & Patten, 2007; Wilson et al., 2005). More recently, metagenomics has been used to identify taxonomic groups of viruses present in certain coral species from different geographical locations and exposed to different environmental stressors (Dinsdale et al., 2008; Laffy et al., 2016; Thurber et al., 2008; Weynberg et al., 2014). However, our knowledge of the identity of most viral communities and their function within corals is still scant. Several dsDNA viral families have been reported from stony corals including: Phycodnaviridae; Mimiviridae; Poxvirirdae; Iridoviridae; Herpesviridae; Ascoviridae; with Caudovirales or tailed bacteriophages being the most dominant order (reviewed in Thurber et al., 2017). However, while previous studies have assessed the taxonomic composition of DNA viruses in corals, insights into RNA virus assemblages and viral functional genes are scarce. Here we utilize the recently developed computational pipeline HoloVir (Laffy et al., 2016) to describe the viral taxonomic diversity and gene function based on DNA and RNA viromes of seven scleractinian coral species and their surrounding seawater from the central GBR.
Materials and Methods
Sample collection, processing, and sequencing
Five species of scleractinian coral—Acropora tenuis, Fungia fungites, Goniastrea aspera, Galaxea fascicularis and Pocillopora verrucosa—were collected in early March 2013, under the Great Barrier Reef Marine Park Authority collection permit number G15/37272.1, from sites around Orpheus Island Research Station (OIRS) in the inshore central GBR. Fragments (∼10 cm diameter) were collected from three colonies, or three single-polyped corals in the case of F. fungites, and were returned to the lab’s flow-through seawater tanks until processing (within 3–4 h of collection). Processing was conducted as described in (Weynberg et al., 2014), including tissue removal via air-blasting, mechanical disruption using bead-beating, cesium chloride (CsCl) density fractionation to purify virus fractions, and buffer exchange to remove cesium salts prior to DNA and RNA extraction. This method ensures viral capsids remain intact prior to nucleic acid extraction following nuclease treatment. Amplification of viral DNA and RNA nucleic acids was performed using RP-SISPA, a sequence-independent amplification step (Weynberg et al., 2014), and sequenced using Nextera XT MiSeq 250 bp paired-end sequencing (Illumina, Hayward, CA, USA) at the Ramaciotti Centre, University of New South Wales, Sydney, Australia.
Two additional species of Pocillopora (fragments of approximately 10 cm diameter from three colonies) were included in this study. P. damicornis was collected at Trunk Reef in late November 2012, and P. acuta was collected at Davies Reef in early October 2013, both mid-shelf reefs in the central GBR ∼120 km from Orpheus Island. Pocillopora species were identified in the field based on colony morphology, and species ID was confirmed with a genetic assay (Torda et al., 2013). All species except P. acuta were collected in triplicate (i.e., three colonies) and pooled prior to sequencing. P. acuta samples were not pooled prior to sequencing. A seawater sample (20 L) was collected in the vicinity of the P. acuta samples to compare the holobiont and seawater virus communities. Seawater was transported to the Australian Institute of Marine Science (AIMS) for immediate filtration (0.22 µm Sterivex PES) and FeCl flocculation. In brief, 0.5 mL of FeCl (10 g/L stock) was added to 5 L of seawater, shaken for 1 min (repeated every 20 min), and stored at 25 °C for 1 hr. Flocculated viral particles were collected on a fresh Sterivex (Durapore PDVF), and ∼2 mL 0.2 M ascorbate-0.1 M EDTA-Mg buffer (made day of, pH = 6.0) were added to saturate the filter. The filter unit was capped with Parafilm, shaken vigorously to begin the dissociation of viral particles, and stored at 4 °C until the filter appeared clear. The solution was then purged from the filter, and the recovered viral particles were loaded onto a CsCl gradient and processed as described in (Weynberg et al., 2014).
Sequence analysis
Virome data sets were analysed following the HoloVir protocol (Laffy et al., 2016), an analysis protocol specifically designed for taxonomic and functional analysis of holobiont-associated virus communities. HoloVir utilizes a two-tiered analysis approach, performing taxonomic and marker gene analysis to identify viral community composition, as well as the detection of any potential contamination issues, together with a Swiss-Prot keyword enrichment analysis to characterise the viromic functional profiles.
Quality control, sample dereplication and single read analysis
Sequencing data sets underwent quality trimming using FastQC (version 1.11.5) (Andrews, 2010) overlapping reads were identified and merged using PEAR (Zhang et al., 2014) (Unmerged reads were fused together, separated by 10 padding n residues. Merged reads were dereplicated using cd-hit-est (Li & Godzik, 2006) using a global identity threshold of 99%. Merged and non-merged reads were combined prior to single read BLAST+ analysis for taxonomic assignment, and single read analysis was used to validate assembled gene analysis.
Contig assembly and gene prediction
De novo assembly of viral sequences was performed using CLC Genomics workbench 8.5.1 with subsequent filtering steps for a minimum of 3× coverage and a minimum contig length of 1,000 bp for DNA viromes and 500 bp for RNA viromes. Gene prediction was performed on the resulting contigs using MetaGeneAnnotator (Noguchi, Taniguchi & Itoh, 2008). The resulting predicted genes were subsequently subjected to taxonomic assignment, marker analysis and functional assessment. Since the P. acuta samples were not pooled prior to sequencing, a single sequence run was used to maintain read number in further analyses.
Taxonomic assignment
Comparison of predicted genes to the viral RefSeq database (Brister et al., 2015) was performed via BLAST sequence similarity searches using default parameters (Altschul et al., 1990). Taxonomic assignment was subsequently performed using MEGAN5 Last Common Ancestor (LCA) default parameters, using a minimum support parameter of five matches for taxonomic assignment (Huson et al., 2011). A cellular and viral marker database was generated as described in Laffy et al. (2016) and used in sequence similarity comparisons (via BLAST with default parameters, and MEGAN5 LCA default parameters with a minimum support parameter of one match for taxonomic assignment) to confirm viral RefSeq taxonomic assignments and identify potential cellular contaminants. Taxonomic assignment was defined at the species level, and the 60 most abundant taxonomic assignments were determined across all samples and visualized using ggplot in R.
Functional analysis
The function of predicted genes from virome assemblies was determined by performing BLAST sequence similarity searches to the UniprotKB/Swiss-Prot database (UniProt Consortium, 2015), with an e-value cutoff of 10−10. Swiss-Prot keywords were then assigned to predicted genes based on the best hit. Identified Swiss-Prot keywords were collated for each viral metagenome, and the frequency of each keyword was calculated, relative to the proportion of that keyword within the SwissProt database. The 50 most abundant Swiss-Prot keywords were identified across all samples and visualized using ggplot in R.
Results and Discussion
This study characterised the viral diversity across seven coral species of the central GBR and provided taxonomic and functional information with an additional comparison of viral communities that exist in seawater sampled in the GBR. Analysis of DNA and RNA derived coral viromes revealed a broad taxonomic diversity of viruses associated with central GBR corals. For the coral samples, ∼8.105–3.106 raw reads were obtained for each of the DNA and RNA viromes, and ∼3.105 and ∼7.105 reads were obtained from the water sample (Table 1). The number of assembled predicted genes was highly variable, ranging from ∼100–44,000 per sample (Table 1), with generally lower numbers for RNA compared to the DNA viromes. The viromes contained representatives from 23 dsDNA families (including six of the ten families that collectively form the nucleocytoplasmic large dsDNA viruses, or NCLDVs), six ssDNA families, one dsRNA virus, two ssRNA families and three retrotranscribing viral families. Many viral genes involved in latency and viral host infection, viral replication, propagation and particle assembly were observed, particularly in the DNA viromes. Data sets generated from these samples were submitted to Genbank Sequence Read Archive (see Table 1 for accession numbers).
Table 1. Sequencing statistics of seven coral holobiont viral communities.
Sequencing statistics of DNA and RNA viromes associated with coral holobiont communities of seven Great Barrier Reef coral species.
| Host species | Template | Accession number (Bioproject # PRJNA302344) | #raw reads | #contigs | N50 | Longest contig | #predicted genes |
|---|---|---|---|---|---|---|---|
| Acropora tenuis | DNA | SAMN02709831 | 1,620,989 | 4,268 | 1,530 | 13,669 | 10,679 |
| Fungia fungites | DNA | SAMN04272222 | 1,605,748 | 778 | 1,365 | 7,098 | 1,779 |
| Goniastria aspera | DNA | SAMN04274802 | 1,170,146 | 15,173 | 1,614 | 48,259 | 44,198 |
| Galaxea fascicularis | DNA | SAMN04274801 | 1,869,983 | 989 | 1,501 | 5,386 | 1,512 |
| Pocillopora acuta | DNA | SAMN04277425 | 2,213,465 | 18,029 | 1,615 | 40,341 | 33,726 |
| Pocillopora damicornis | DNA | SAMN02709826 | 2,659,198 | 2,188 | 1,617 | 20,529 | 5,651 |
| Pocillopora verrucosa | DNA | SAMN04277423 | 9,328,233 | 10,351 | 1,594 | 67,301 | 29,381 |
| Seawater | DNA | SAMN06849075 | 320,701 | 1,006 | 1,491 | 18,804 | 3,116 |
| Acropora tenuis | RNA | SAMN02709832 | 853,227 | 1,517 | 731 | 5,386 | 1,499 |
| Fungia fungites | RNA | SAMN04274763 | 1,323,170 | 354 | 693 | 1,597 | 377 |
| Goneastrea aspera | RNA | SAMN04274806 | 1,337,144 | 3,953 | 727 | 5,693 | 5,144 |
| Galaxea fascicularis | RNA | SAMN04277306 | 1,796,476 | 934 | 679 | 5,386 | 921 |
| Pocillopora acuta | RNA | SAMN04277426 | 1,445,938 | 3,761 | 928 | 7,499 | 4,389 |
| Pocillopora verrucosa | RNA | SAMN04277424 | 2,928,367 | 67 | 739 | 1,462 | 106 |
| Seawater | RNA | SAMN06849076 | 667,528 | 60 | 699 | 1,795 | 102 |
Taxonomic assignments
Viruses that target prokaryotes
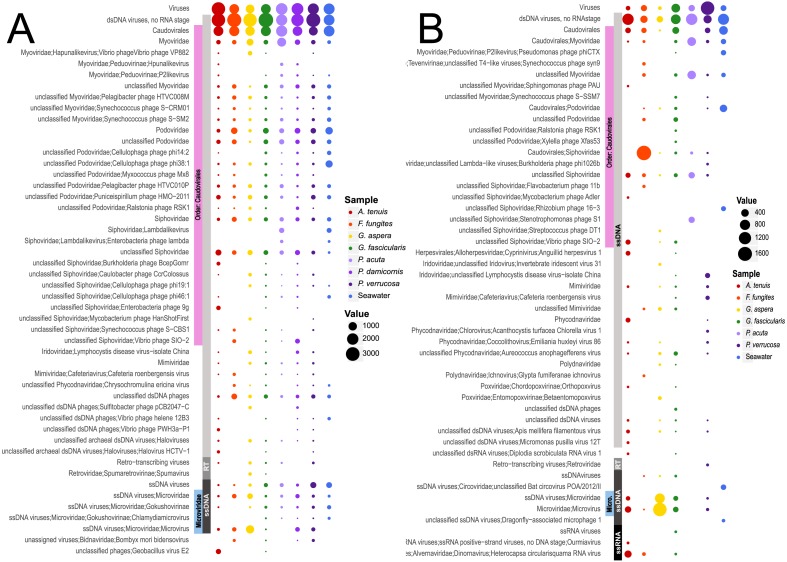
The metavirome sequence data sets were dominated by viruses of prokaryotes, consistent with previous studies in other marine invertebrates (reviewed in Thurber et al., 2017). The most abundant viruses in the DNA viromes from all seven coral species were the dsDNA bacteriophages in the order Caudovirales (Figs. 1A and 1B), reflecting the high abundance and diversity of bacterial hosts within the coral holobiont (∼107 prokaryotic cells/cm2 of coral surface area; 102–104 OTUs per coral colony (Blackall, Wilson & Oppen, 2015)). Previous coral microbial and viral metagenome studies on other species or from other locations also found high abundances of Caudovirales (Correa et al., 2016; Littman, Willis & Bourne, 2011; Soffer, Zaneveld & Vega Thurber, 2014; Wegley et al., 2007). Their prevalence in the RNA viromes was unexpected as viral particles were DNase-treated prior to viral genome isolation. In the DNA viromes, Caudovirales-like sequences were relatively evenly divided across the three families Myoviridae, Siphoviridae and Podoviridae in all seven coral species and the seawater (Fig. 1A). Siphoviridae contain many prophage types (King et al., 2011) (Fig. 1A), indicating a likely prevalence of lysogenic viruses in the coral holobiont. This is consistent with a high proportion of latency genes identified in the functional analyses (see section Functional annotation of coral viromes below).
Figure 1. Taxonomic analysis of viruses from seven GBR coral species and surrounding seawater.
Taxonomic analysis of viruses from seven coral species and surrounding seawater sampled on the GBR. Relative abundance of the 50 most abundant DNA (A) and RNA (B) viral taxa based on viral Refseq BLASTx comparisons to predicted genes from assigned assembled contigs. Abundances were adjusted based on contig coverage values for each virome community. MEGAN5 default LCA parameters were used to assign taxonomy.
An average 6% and 36% (DNA and RNA viromes, respectively) of all predicted viral genes were most similar to ssDNA viruses, in particular phages in the family Microviridae. Microviridae have previously been reported as an abundant component of coral metaviromes, possibly because the polymerase used in multi-displacement amplification (MDA, phi-29) preferentially amplifies small circular genomes (Correa et al., 2016; Littman, Willis & Bourne, 2011; Soffer, Zaneveld & Vega Thurber, 2014; Wegley et al., 2007). Our SISPA methods are template-independent PCR amplified, validating the abundance of Microviridae in corals. In this study, Microviridae were primarily represented by the genus Microvirus, which commonly infects enterobacteria, and three genera of the subfamily Gokushnovirinae, which are known to infect obligate intracellular parasitic bacteria such as Bdellovibrio and Chlamydia (King et al., 2011). These viruses may infect intracellular parasitic members of the Halobacteriovorax that have been confirmed to be present in the coral microbiome (Welsh et al., 2016), although this requires further experimental validation.
The DNA viromes of all but one coral species (F. fungites) contained sequences affiliated with the archaeal dsDNA virus, Halovirus. Haloviruses target archaea in the genus Haloarcula which are known to associate with corals (Thurber et al., 2009). Archaea are not as widespread as coral-associated bacteria (Wegley et al., 2007), but they can reach densities of >107 cells per cm2 of coral surface area (Wegley et al., 2004). The presence of lemon-shaped virus-like particles, a common morphology of these viruses, in the surface mucus layer of corals (Davy & Patten, 2007) is also consistent with our viral sequence data.
Viruses that target eukaryotes
Of the total number of assembled contigs, between 0.5% (F. fungites) to 18% (P. acuta) of the DNA viromes matched eukaryotic viruses. The majority of predicted dsDNA eukaryotic viruses in the DNA viromes were assigned to five viral families, namely four NCLDV families: Iridoviridae, Phycodnaviridae, Mimiviridae, Poxviridae, as well as Polydnaviridae (Fig. 1). The NCLDVs Ascoviridae and Marseilleviridae were rare, as were other viral families (<0.5%; Fig. 1). The most predominant Iridovirus-like sequences had highest sequence similarity to lymphocystivirus, which commonly infect fish species (King et al., 2011) and have previously been detected in the Caribbean coral Porites sp. (Wegley et al., 2007). Mimivirus-like hits were detected in all coral species but not seawater and these findings are consistent with previous virome, transcriptome and TEM data showing the presence of Mimiviridae in Acropora aspera from the southern GBR (Correa et al., 2016), as well as in Caribbean and Hawaiian corals (Correa, Welsh & Thurber, 2013; Thurber et al., 2008). Phycodnaviridae are also known to be prevalent members of coral viral assemblages (Thurber & Correa, 2011). The presence of phycodnaviruses in corals is likely linked to the dinoflagellate endosymbiont (Symbiodinium spp.) or possibly to endolithic algae present in the coral skeleton (Verbruggen & Tribollet, 2011), as phycodnaviruses are known to infect algae (King et al., 2011). A recent study showed that NCLDV sequences with similarities to Phycodnaviridae and Mimiviridae were abundant in the transcriptomes of GBR Symbiodinium type C1 cultures (Levin et al., 2017), confirming members of these diverse viral families target Symbiodinium. These viral transcripts were regulated during experimental heat stress, suggesting they play a role in the Symbiodinium thermal stress response. Most recently, a transcriptomic study of viruses associated with Symbiodinium microadriaticum clade A1, revealed a prevalence of viruses related to the Potyviridae, and high expression of viral genes under heat shock treatment (Brüwer et al., 2017).
Contrary to previous reports on coral virus assemblages (Correa et al., 2016; Thurber et al., 2008), sequences related to the Herpesviridae were absent or present in very low abundance (<0.1% of all eukaryotic virus reads in five of seven of coral species sampled). Such low representation of herpesviruses may reflect differences in the abundance of these viruses across coral species, geographic locations and coral physiological states, but may also be due to the different computational approaches applied (Wood-Charlson et al., 2015).
Members of the Retroviridae were also seen in all coral species sequenced with the exception of F. fungites. Retro-transcribing viruses have previously been reported in corals (Correa et al., 2016; Correa, Welsh & Vega Thurber, 2012; Weynberg et al., 2014; Wood-Charlson et al., 2015) and are known to be abundant in diverse eukaryotes (Koonin, Dolja & Krupovic, 2015). In contrast, prokaryotes are predominantly infected by dsDNA bacteriophages with no reported retroviruses infecting bacteria or archaea. This suggests that eukaryotic members of the coral holobiont are the retroviral targets.
Three of the coral species (A. tenuis, F. fungites and G. fascicularis) contained sequences with similarity to a dinornavirus, Heterocapsa circularisquama ssRNA virus of the Alvernaviridae, that infects free-living dinoflagellates (Tomaru et al., 2004). This virus is likely a new divergent ssRNA virus that targets the coral algal symbiont, Symbiodinium (Levin et al., 2017). Gene transcripts of the major capsid protein were previously shown to be highly expressed in a thermo-sensitive Symbiodinium C1 population at ambient temperature, but not in a conspecific thermo-tolerant population under the same conditions, suggesting it may play a role in thermal susceptibility of this dinoflagellate coral endosymbiont (Levin et al., 2017). The presence of Alvernaviridae sequences in some, but not all, Symbiodinium transcriptome data sets (Correa, Welsh & Thurber, 2013; Levin et al., 2017) suggests not all Symbiodinium types and populations are infected with this virus.
Sequences with similarities to Bombyx mori bidensovirus, a ssDNA virus belonging to the Bidnaviridae family (King et al., 2011; Krupovic & Koonin, 2014) were detected in four of the seven species but not in the surrounding seawater (Fig. 1A). A second viral group previously undescribed from corals, but detected in the RNA viromes of A. tenuis and G. fascicularis, had sequence similarity to the dsRNA mycovirus, Diplodia scrobiculata virus, which is known to infect endophytic fungi in terrestrial plants (King et al., 2011). Fungi are known to associate with the coral skeleton (Amend, Barshis & Oliver, 2012; Bentis, Kaufman & Golubic, 2000), and we hypothesize they may act as a host in corals. As dsRNA does not exist in cells during normal replication processes, this nucleic acid form can be exceptionally vulnerable to attack by nucleases and is less stable than DNA (Mertens, 2004), which may explain the absence of these viruses in previously reported coral viromes.
Functional annotation of coral viromes
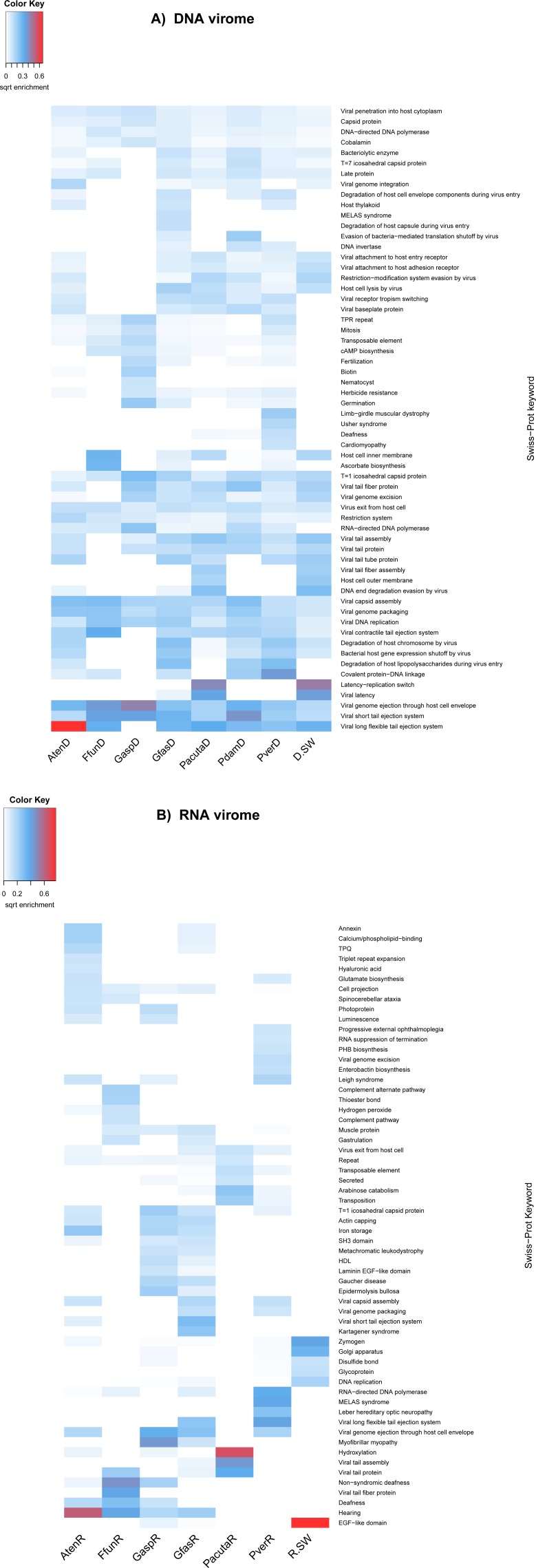
The most abundant genes in the coral and seawater DNA viromes related to viral functions (Fig. 2A). ‘Latency-replication switch’ was the most common Swiss-Prot keyword, and ‘viral latency’ was also highly represented in P.acuta (and seawater), suggesting that prophages are a common feature in the holobiont of this particular coral species. This is consistent with the high prevalence of Siphoviridae, Myoviridae and Podoviridae observed in the taxonomic analyses. It is possible that the high prevalence of latency related genes may have originated from bacteriophages entering a lysogenic stage within their holobiont host genomes. In line with the predominance of bacteriophages in the taxonomic analyses, the second most abundant keyword was ‘viral long flexible tail ejection system’, which represents a viral protein that constitutes the noncontractile ejection system carried by some long-tailed prokaryotic viruses such as the Siphoviridae. Of the top 60 Swiss-Prot keywords, 17 fell in non-viral categories, eleven of which were absent from the seawater sample.
Figure 2. Predicted gene functional analysis of DNA (A) and RNA (B) viromes in seven GBR coral species and surrounding seawater.
Functional enrichment based on the 60 most enriched Swissprot keywords from SwissProt BLASTx comparisons to assembled predicted genes. Keyword counts were adjusted based on contig coverage values for each virome community.
Several viral genes with host auxiliary function that may be beneficial to the host were detected. Viruses associated with three of the seven coral species examined (A. tenuis, G. fascicularis and P. verrucosa) had genes encoding proteins associated with the Swiss-Prot keyword ‘host thylakoid’. The thylakoid is the compartment within the chloroplast where light-dependent reactions of photosynthesis occur. The genes in Swiss-Prot associated with this keyword encode the photosystem II (PSII) D2 protein (psbD), a protein that, together with the D1 protein, forms the photochemically active reaction center of PSII (Barber, Chapman & Telfer, 1987). All seven coral species harboured genes of the key word ‘herbicide resistance’, which is represented as psbA encoding the PSII D1 protein. Some of these sequences matched the psbA gene from other dinoflagellates. Host genes encoding the D1 and D2 proteins have previously been observed in cyanophages that infect marine Synechococcus and Prochlorococcus (Lindell et al., 2004; Mann et al., 2003; Weigele et al., 2007), including those from coral atolls in the Line Islands (Dinsdale et al., 2008; Sharon et al., 2009). Impairment of PSII results in an increase in reactive oxygen species in the cell and may lead to coral bleaching (Warner, Fitt & Schmidt, 1996; Warner, Fitt & Schmidt, 1999). Therefore, viruses carrying PSII genes may alleviate and/or delay some of the damage to the Symbiodinium PSII from high seawater temperature, herbicides or other stressors, thereby providing energy and more time for their own replication.
The keywords ‘photoprotein’ and ‘luminescence’ were present in the A. tenuis and G. aspera RNA viromes. Both are represented by genes encoding green fluorescent-like protein (GFP) chromoproteins (CPs). GFP-like fluorescent proteins are responsible for the bright coloration many corals display, and are encoded by a large numbers of genes (Alieva et al., 2008). While the function of many fluorescent proteins remains ambiguous (Roth, 2014), there is some evidence for an antioxidant role of CPs (Palmer, Modi & Mydlarz, 2009). As viral infections are known to cause an increase in ROS in the infected cells (Bidle & Vardi, 2011), it is possible that these virally encoded genes may prolong the life of the coral host cells allowing the virus to produce more offspring than it would have without providing antioxidants to its host, although further experimental work would be needed to validate this mechanism.
Genes encoding proteins classified under the key word ‘nematocyst’ were highly abundant in one of the coral species sampled, P. verrucosa, and also present in G. aspera. These sequences match Delta-thalatoxin genes known from anemones (Oshiro et al., 2004). We speculate that these toxin genes acquired by coral-associated viruses may provide a benefit to the coral host by assisting in prey acquisition or repelling predators.
Conclusions
Metagenomic analyses of viral assemblages from seven GBR coral species have revealed considerable viral diversity in corals. The abundance and diversity of bacteriophages suggests they target and control bacterial populations associated with corals. The presence of viral genes associated with functions such as nematocyst toxins and auxiliary metabolic processes indicate a potential benefit of viral infection for hosts within the holobiont, providing a foundation for further experimental investigation and validation.
Supplemental Information
Funding Statement
This research was supported by the Australian Research Council (ARC) through a Super Science Fellowship #FS110200034 (Karen D. Weynberg), two Future Fellowships (#FT100100088 Madeleine J.H. van Oppen; #FT120100480 Nicole Webster) and additional funding from AIMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Thomas Rattei is an Academic Editor for PeerJ.
Author Contributions
Karen D. Weynberg conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Patrick W. Laffy analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Elisha M. Wood-Charlson conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, reviewed drafts of the paper and prepared figures/tables.
Dmitrij Turaev analyzed the data, contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Thomas Rattei contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Nicole S. Webster analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Madeleine J.H. van Oppen conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
Field permit G15/37272.1 was issued by the Great Barrier Reef Marine Park Authority.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The DNA and RNA viromes described here are accessible via Genbank under Bioproject # PRJNA302344 and accession numbers SAMN02709831, SAMN04272222, SAMN04274802, SAMN04274801, SAMN04277425, SAMN02709826, SAMN04277423, SAMN06849075, SAMN02709832, SAMN04274763, SAMN04274806, SAMN04277306, SAMN04277426, SAMN04277424, and SAMN06849076.
References
- Alieva et al. (2008).Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV. Diversity and evolution of coral fluorescent proteins. PLOS ONE. 2008;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amend, Barshis & Oliver (2012).Amend AS, Barshis DJ, Oliver TA. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME Journal. 2012;6:1291–1301. doi: 10.1038/ismej.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews (2010).Andrews S. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc. 2010.
- Barber, Chapman & Telfer (1987).Barber J, Chapman DJ, Telfer A. Characterisation of a photosystem II reaction centre isolated from chloroplasts of Pisum sativum. FEBS Letters. 1987;220:67–73. doi: 10.1016/0014-5793(87)80877-3. [DOI] [Google Scholar]
- Bentis, Kaufman & Golubic (2000).Bentis CJ, Kaufman L, Golubic S. Endolithic fungi in reef-building corals (Order: Scleractinia) are common, cosmopolitan, and potentially pathogenic. Biological Bulletin. 2000;198:254–260. doi: 10.2307/1542528. [DOI] [PubMed] [Google Scholar]
- Bidle & Vardi (2011).Bidle KD, Vardi A. A chemical arms race at sea mediates algal host-virus interactions. Current Opinion in Microbiology. 2011;14:449–457. doi: 10.1016/j.mib.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Blackall, Wilson & Oppen (2015).Blackall LL, Wilson B, Oppen MJ. Coral–the world’s most diverse symbiotic ecosystem. Molecular Ecology. 2015;24:5330–5347. doi: 10.1111/mec.13400. [DOI] [PubMed] [Google Scholar]
- Bourne, Morrow & Webster (2016).Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annual Review of Microbiology. 2016;70:314–340. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- Brister et al. (2015).Brister JR, Ako-Adjei D, Bao Y, Blinkova O. NCBI viral genomes resource. Nucleic Acids Research. 2015;43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüwer et al. (2017).Brüwer JD, Agrawal S, Liew YJ, Aranda M, Voolstra CR. Association of coral algal symbionts with a diverse viral community responsive to heat shock. BMC Microbiology. 2017;17:174. doi: 10.1186/s12866-017-1084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa et al. (2016).Correa AMS, Ainsworth TD, Rosales SM, Thurber AR, Butler CR, Vega Thurber RL. Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00127. Article 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, Welsh & Thurber (2013).Correa AMS, Welsh RM, Thurber RLV. Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME Journal. 2013;7:13–27. doi: 10.1038/ismej.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, Welsh & Vega Thurber (2012).Correa AMS, Welsh RM, Vega Thurber RL. Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME Journal. 2012;7:13–27. doi: 10.1038/ismej.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy & Patten (2007).Davy JE, Patten NL. Morphological diversity of virus-like particles within the surface microlayer of scleractinian corals. Aquatic Microbial Ecology. 2007;47:37–44. doi: 10.3354/ame047037. [DOI] [Google Scholar]
- Dinsdale et al. (2008).Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F. Microbial ecology of four coral atolls in the northern line Islands. PLOS ONE. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell et al. (2007).Harvell D, Jordan-Dahlgren E, Merkel S, Rosenberg E, Raymundo L, Smith G, Weil E, Willis B, Global Envrionm Facility C Coral disease, environmental drivers and the balance between coral and microbial associates. Oceanography. 2007;20:172–195. [Google Scholar]
- Huson et al. (2011).Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Research. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King et al. (2011).King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press; San Diego: 2011. [Google Scholar]
- Koonin, Dolja & Krupovic (2015).Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet et al. (2013).Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proceedings of the Royal Society of London B: Biological Sciences. 2013;280 doi: 10.1098/rspb.2012.2328. Article 20122328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic & Koonin (2014).Krupovic M, Koonin EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Scientific Reports. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffy et al. (2016).Laffy PW, Wood-Charlson EM, Turaev D, Weynberg KD, Botté ES, Van Oppen MJH, Webster NS, Rattei T. HoloVir: a workflow for investigating the diversity and function of viruses in invertebrate holobionts. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00822. Article 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, Willis & Bourne (2012).Lema KA, Willis BL, Bourne DG. Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Applied and Environmental Microbiology. 2012;78:3136–3144. doi: 10.1128/aem.07800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser et al. (2004).Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- Levin et al. (2017).Levin RA, Voolstra CR, Weynberg KD, Van Oppen MJH. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. The ISME Journal. 2017;11:808–812. doi: 10.1038/ismej.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Godzik (2006).Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lindell et al. (2004).Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proceedings of the National Academy of Science of the United States of America. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman, Willis & Bourne (2011).Littman R, Willis BL, Bourne DG. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environmental Microbiology Reports. 2011;3:651–660. doi: 10.1111/j.1758-2229.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- Mann et al. (2003).Mann NH, Cook A, Millard A, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- Mertens (2004).Mertens P. The dsRNA viruses. Virus Research. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Muscatine, Mccloskey & Marian (1981).Muscatine L, Mccloskey LR, Marian RE. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnology and Oceanography. 1981;26:601–611. doi: 10.4319/lo.1981.26.4.0601. [DOI] [Google Scholar]
- Muscatine & Weis (1992).Muscatine L, Weis VM. Productivity of zooxanthellae and biogeochemical cycles. In: Falkowski PG, Woodhead AE, editors. Primary productivity in the sea. Plenum Press; New York: 1992. pp. 257–272. [Google Scholar]
- Noguchi, Taniguchi & Itoh (2008).Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Research. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro et al. (2004).Oshiro N, Kobayashi C, Iwanaga S, Nozaki M, Namikoshi M, Spring J, Nagai H. A new membrane-attack complex/perforin (MACPF) domain lethal toxin from the nematocyst venom of the Okinawan sea anemone Actineria villosa. Toxicon. 2004;43:225–228. doi: 10.1016/j.toxicon.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Palmer, Modi & Mydlarz (2009).Palmer CV, Modi CK, Mydlarz LD. Coral fluorescent proteins as antioxidants. PLOS ONE. 2009;4:e7298. doi: 10.1371/journal.pone.0007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina et al. (2009).Raina J-B, Tapiolas D, Willis BL, Bourne DG. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Applied and Environmental Microbiology. 2009;75:3492–3501. doi: 10.1128/aem.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer et al. (2002).Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral- associated bacteria. Marine Ecology Progress Series. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- Roth (2014).Roth MS. The engine of the reef: photobiology of the coral-algal symbiosis. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00422. Article 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon et al. (2009).Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, Pinter RY, Partensky F, Koonin EV, Wolf YI, Nelson N, Beja O. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer, Zaneveld & Vega Thurber (2014).Soffer N, Zaneveld J, Vega Thurber R. Phage–bacteria network analysis and its implication for the understanding of coral disease. Environmental Microbiology. 2014;17(4):1203–1218. doi: 10.1111/1462-2920.12553. [DOI] [PubMed] [Google Scholar]
- Thurber et al. (2008).Thurber RLV, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, Edwards RA, Haynes M, Angly FE, Wegley L, Rohwer FL. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber & Correa (2011).Thurber RLV, Correa AMS. Viruses of reef-building scleractinian corals. Journal of Experimental Marine Biology and Ecology. 2011;408:102–113. doi: 10.1016/j.jembe.2011.07.030. [DOI] [Google Scholar]
- Thurber et al. (2017).Thurber RLV, Payet JP, Thurber AR, Correa AMS. Virus-host interactions and their roles in coral reef health and disease. Nature Reviews Microbiology. 2017;15:205–216. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- Thurber et al. (2009).Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. Metagenomic analysis of stressed coral holobionts. Environmental Microbiology. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Tomaru et al. (2004).Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, Nagasaki K. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquatic Microbial Ecology. 2004;34:207–218. doi: 10.3354/ame034207. [DOI] [Google Scholar]
- Torda et al. (2013).Torda G, Schmidt-Roach S, Peplow LM, Lundgren P, Van Oppen MJH. A rapid genetic assay for the identification of the most common Pocillopora damicornis genetic lineages on the great barrier reef. PLOS ONE. 2013;8:e58447. doi: 10.1371/journal.pone.0058447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2015).UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen & Tribollet (2011).Verbruggen H, Tribollet A. Boring algae. Current Biology. 2011;21:R876–R877. doi: 10.1016/j.cub.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Warner, Fitt & Schmidt (1996).Warner ME, Fitt WK, Schmidt GW. The effects of elevated temperature on the photosynthetic efficiency of zooxanthellae in hospite from four different species of reef coral: a novel approach. Plant, Cell and Environment. 1996;19:291–299. doi: 10.1111/j.1365-3040.1996.tb00251.x. [DOI] [Google Scholar]
- Warner, Fitt & Schmidt (1999).Warner ME, Fitt WK, Schmidt GW. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8007–8012. doi: 10.1073/pnas.96.14.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley et al. (2007).Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environmental Microbiology. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wegley et al. (2004).Wegley L, Yu YN, Breitbart M, Casas V, Kline DI, Rohwer F. Coral-associated archaea. Marine Ecology-Progress Series. 2004;273:89–96. doi: 10.3354/meps273089. [DOI] [Google Scholar]
- Weigele et al. (2007).Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environmental Microbiology. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- Welsh et al. (2016).Welsh RM, Zaneveld JR, Rosales SM, Payet JP, Burkepile DE, Thurber RV. Bacterial predation in a marine host-associated microbiome. ISME Journal. 2016;10:1540–1544. doi: 10.1038/ismej.2015.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weynberg et al. (2015).Weynberg KD, Voolstra CR, Neave MJ, Buerger P, Van Oppen MJH. From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Scientific Reports. 2015:17889. doi: 10.1038/srep17889. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weynberg et al. (2014).Weynberg KD, Wood-Charslon EM, Suttle C, Van Oppen MJ. Generating viral metagenomes from the coral holobiont. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00206. Article 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson et al. (2005).Wilson WH, Dale AL, Davy JE, Davy SK. An enemy within? Observations of virus-like particles in reef corals. Coral Reefs. 2005;24:145–148. doi: 10.1007/s00338-004-0448-0. [DOI] [Google Scholar]
- Wood-Charlson et al. (2015).Wood-Charlson EM, Weynberg KD, Suttle CA, Roux S, Van Oppen MJH. Metagenomic characterization of viral communities in corals: mining biological signal from methodological noise. Environmental Microbiology. 2015;17(10):3440–3449. doi: 10.1111/1462-2920.12803. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.