Abstract
Objectives
To investigate the association between IL18RAP and body mass index (BMI) and obesity and to verify the effect of a polymorphism in the microRNA136 (MIR136) IL18RAP binding region.
Design
We analysed samples from two Spanish cross-sectional studies, VALCAR (Spanish Mediterranean coast) and Hortega (Spanish centre). These studies aimed at analysing cardiovascular risk and development of cardiovascular disease in the general population. Both populations correspond to regions with different characteristics.
Setting
Five IL18RAP single nucleotide polymorphisms were selected using the SYSNPs web tool and analysed by oligonucleotide ligation assay (SNPlex). For the MIR136 functional study, cells were transfected with plasmids containing different rs7559479 polymorphism alleles and analysed by luciferase reporter assays.
Participants
1970 individuals (Caucasian, both genders): VALCAR (468) and Hortega (1502).
Results
rs2293225, rs2272127 and rs7559479 showed the following associations: rs7559479 G allele correlated with a higher obesity risk (P=0.01; OR=1.82; 95% CI 1.15 to 2.87 for the VALCAR group; P=0.033; OR=1.35; 95% CI 1.03 to 1.79 for the Hortega population) and higher body mass index (BMI) values (P=0.0045; P=0.1 for VALCAR and Hortega, respectively); a significant association with obesity (P=0.0024, OR=1.44, 95% CI 1.14 to 1.82) and increased BMI values (P=0.008) was found when considering both populations together. rs2293225 T allele was associated with lower obesity risk (P=0.036; OR=0.60; 95% CI 0.35 to 0.96) and lower BMI values (P=0.0038; OR=1.41) while the rs2272127 G allele was associated with lower obesity risk (P=0.028; OR=0.66; 95% CI 0.44 to 0.97) only in the VALCAR population. A reporter assay showed that the presence of the A allele in rs7559479 was associated with increased MIR136 binding to IL18RAP.
Conclusions
Our results suggest that polymorphisms in IL18RAP influence susceptibility to obesity. We demonstrated that the A allele in rs7559479 increases MIR136 binding, which regulates IL-18 system activity.
Keywords: obesity, variations, cytokine, genetic polymorphism, microrna, genetics
Strengths and limitations of this study.
Our study was carried out in two Spanish populations from different regions with noticeably distinct characteristics, although the results were similar for both.
The number of individuals included in this study was limited, but its statistical power was sufficient for the reduced number of polymorphisms we analysed.
We did not exhaustively study genetic variability in this gene because this gene region has too many variations, and would have required the study of more polymorphisms.
Our results should be confirmed by other studies, which include analyses of different populations and with larger sample sizes.
Introduction
Obesity is a state in which excess energy accumulates in corporal adipose tissue due to a chronic imbalance between energy intake and energy expenditure1; individuals respond differently to this imbalance because of their differing genetic backgrounds.2 Over the past 20 years there has been an alarming increase in the number of overweight and obese individuals. This is especially worrying because obesity significantly increases the risk of chronic diseases, such as type 2 diabetes, cardiovascular disease, non-alcoholic fatty liver disease, colon cancer and obstructive sleep apnoea.3
Obesity is defined as a low-grade inflammatory state characterised by increased cytokine production.4 Interleukin-18 (IL-18) concentration increases in obesity, suggesting it is involved in obesity and metabolic syndrome by increasing adipogenesis.5 IL-18 is a member of the IL-1 cytokine superfamily. It regulates the immune response and is expressed in cells involved in chronic inflammation, autoimmune diseases and several cancers and infectious diseases: macrophages, dendritic cells, Kupffer cells, keratinocytes, osteoblasts, adrenal cortex cells, intestinal epithelial cells, microglial cells and synovial fibroblasts.6–14 IL-18 signalling is induced by binding of this cytokine to a heterodimeric receptor called IL-18R. IL-18R is formed of two chains, IL-18R1 (with a binding role) and IL18RAP (receptor accessory protein, with a signal-transducing role). The interaction of both IL-18R chains is mandatory to induce IL-18 signalling.15–17
Some polymorphisms can modulate gene activity via different mechanisms, such as modifying the binding or action of specific microRNAs (miRNAs). They are small non-coding RNAs which modulate expression of thousands of genes, and studies performed over the past decade suggested their involvement in disease regulation.18 19 Several miRNAS have been described as biomarkers for cancers, cardiovascular disease and type 2 diabetes.18 20–22 Furthermore, it has been suggested that a failure in miRNA regulation plays a part in obesity.23 These facts support the use of miRNA as biomarkers for the early diagnosis of chronic diseases and as therapeutic targets.24
Many studies have reported on IL-18 and IL18R1, but few specifically discussed IL18RAP; therefore, we investigated, for the first time, the possible association between IL18RAP, body mass index (BMI) and obesity. To do this, we verified the effect of polymorphism rs7559479 located in the MIR136 binding site in the mRNA of IL18RAP and the mRNA levels of this gene.
Materials and methods
Population samples
We analysed two general-population-based study samples (a total of 1970 people) from two different regions of Spain: the VALCAR study, with 468 subjects from the Valencian region and the Hortega study, with 1502 subjects from the province of Valladolid. Both the VALCAR and Hortega samples were originally collected in order to study cardiovascular risk and the development of cardiovascular disease in the general population. The Hortega sample comprised subjects from the healthcare authority area representing half the Valladolid province, while the VALCAR patients came from the health authority area covered by the Hospital Clínico Universitario de Valencia. The Hortega study was approved by the ethics committee at the Hospital Río Hortega and the VALCAR study, as well as the genetic studies presented here, were approved by the ethics committee at the Hospital Clínico Universitario de Valencia. All participants gave their signed informed consent. The research was carried out according to the code of ethics of the World Medical Association (Declaration of Helsinki).
Demographic data (age and gender) and anthropometric parameters were collected using standard procedures. The presence of obesity, hypertension (HTN) and type 2 diabetes (as defined by the WHO criteria; http://www.who.int) was recorded and the BMI (kg/m2) was calculated. Briefly, obesity was diagnosed when the BMI was ≥30 kg/m2, HTN was diagnosed when systolic and diastolic blood pressure values were >140 and/or >90 mm Hg, respectively, and diabetes was defined as a fasting plasma glucose level >7.0 mmol/L (126 mg/dL) or a 2-hour plasma glucose level >11.1 mmol/L (200 mg/dL). A previous diagnosis of type 2 diabetes or HTN and detection of the disease at the time of sample collection was also recorded. The general characteristics of the samples analysed are shown in table 1.
Table 1.
General characteristics of the population
| Characteristics | VALCAR | Hortega | VALCAR+Hortega |
| Number of subjects | 468 | 1502 | 1970 |
| Age (years) | 46.4±14.9* | 54.4±19.3 | 52.6±18.7 |
| BMI† (kg/m2) | 28±4.7* | 26.4±4.1 | 27.7±4.4 |
| Waist circumference (cm) | 92.6±12.8* | 89.5±13.0 | 90.2±13.0 |
| Obesity (n, %) | 114 (24.4)* | 356 (23.7) | 470 (23.9) |
| Abdominal obesity (n, %) | 151 (32.3)* | 414 (27.6) | 565 (28.7) |
| Diabetes mellitus (n, %) | 55 (11.8)* | 114 (7.6) | 169 (8.6) |
| Glucose† (mg/dL) | 99.7±24.4* | 92.5±20.4 | 91.9±29.9 |
| Hypertension (n, %) | 129 (27.5)* | 642 (42.7) | 771 (39.1) |
| Systolic blood pressure† (mm Hg) | 126.5±16.1* | 130.7±21.6 | 129.5±20.5 |
| Diastolic blood pressure† (mm Hg) | 77.1±9.9* | 79.1±10.6 | 78.2±10.4 |
| Total cholesterol (mg/dL) | 222.7±61.7* | 201.3±38.1 | 207.6±46.2 |
| HDL cholesterol (mg/dL) | 56.4±15.1* | 51.7±14.2 | 53.9±14.5 |
Values are mean±SD deviation. For qualitative variables, data are expressed as (n, %).
*P<0.0001.
†Number of missing data in VALCAR/Hortega populations: BMI 35/38, systolic blood pressure 53/10, diastolic blood pressure 53/10, glucose 5/2.
BMI, body mass index; HDL, high-density lipoprotein.
The exclusion criteria were the same in both populations: the presence of serious physical conditions which prevented the subject from responding to the survey or donating the samples required, disorders that might influence the collection of reliable information, or any mental or social condition which might complicate or prevent the subject’s participation in the study. The decision to exclude patients was made on an individual basis by the clinician.
SNP selection and genotyping
Venous blood samples were collected in tubes containing EDTA in order to obtain genomic DNA using the Chemagic system (Chemagen, Baesweiler, Germany). DNA was quantified and diluted to a final concentration of 100 ng ⁄µL. The IL18RAP single nuclear polymorphisms (SNPs) were selected for genotyping based on the conjunction of several parameters selected in the SYSNPs web tool25: heterozygosity in a Caucasian population (>10% for the minor allele frequency), position and spacing along the gene and a possible functional effect (http://www.ensembl.org/index.html). Five SNPs in the IL18RAP gene were selected (table 2) and were genotyped using an oligonucleotide ligation assay (SNPlex; Applied Biosystems, Foster City, California, USA), according to the manufacturer’s guidelines. The genomic information about the selected SNPs was obtained from Ensembl release 89 and the single nucleotide polymorphism database (dbSNP) build 149.
Table 2.
Characteristics of selected polymorphisms from the IL18RAP gene (Ensembl ID: ENSG00000115607)
| SNP name | Chromosome position | Location | Reference | % Genotype | HWE | MAF | Allele reference vs minor |
| rs4851581 | 102418289 | Upstream | NM_003853.3:c.-989A>G | 95.2 | 0.196 | 0.053 | A>G |
| rs2293224 | 102419319 | Intron 1 | NM_003853.3:c.-337–242T>C | 96.8 | 0.522 | 0.466 | C>T |
| rs2293225* | 102419429 | Intron 1 | NM_003853.3:c.-337–132C>T | 98.6 | 0.129 | 0.167 | C>T |
| rs2272127 | 102423413 | Intron 3 | NM_003853.3:c.70+66C>G | 99.9 | 0.149 | 0.195 | C>G |
| rs7559479* | 102452327 | 3´-UTR | NM_003853.3:c.*146G>A | 99.7 | 0.42 | 0.277 | A>G |
*Tag-SNP by HapMap in Caucasian patients.
SNP name: dbSNP build 149. Reference: begins in the first nucleotide of exon 1, build 149 and Ensemble release 89.
HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
Statistical analysis
Before the study, we estimated the statistical power using the lower frequency included in the selection criteria of the selected polymorphisms (minor allele frequency=0.10) for the lower sample size (distributed in cases and controls) by the GAS Power Calculator (http://csg.sph.umich.edu/abecasis/gas_power_calculator/index.html). We considered a P value of 0.01 (based on multiple comparisons). The statistical power obtained for OR=2.5 was 91.9% for one stage study. Applying the same criteria for whole sample the statistical power was 0.975 for OR=1.5.
After genotyping, samples with poor genotyping results (<90%) were removed for all the SNP analyses. All quantitative values are expressed as the mean±SD or as a percentage for qualitative variables. Analysis of variance was used to compare quantitative variables between groups, and χ2 tests were used for categorical variables. Allele and genotype frequencies were calculated for each SNP using SPSS Statistics package version 22.0 and SNPStats (http://bioinfo.iconcologia.net/SNPstats). The Hardy–Weinberg equilibrium (HWE) was tested using a χ2 distribution with one degree of freedom using SNPStat software. HWE was maintained for all the polymorphisms analysed.
The associations between polymorphisms and anthropometric parameters, BMI as a quantitative trait and obesity as a qualitative trait were examined, first using a co-dominant inheritance model in SPSS and SNPStat and then using the remaining models if two genotypes had similar means. Analysis of variance was used to compare the mean differences for continuous variables among genotypes. The association between obesity and each polymorphism and, where appropriate, the haplotype, was tested using logistic regression models. Linkage disequilibrium measurements were calculated using the R2 statistic. Odds ratios were used to evaluate the risk for the presence of obesity for each polymorphism, and age and gender were used as two potential confounding covariates.
MiRNA target prediction
Because the rs7559479 polymorphism is in the 3'-untranslated region (3´-UTR) of the IL18RAP gene, the microRNA.org tool (August 2010 release) was used to predict the microRNA target.
Cell lines and plasmids
HuH-7 cells, an immortal epithelial-like tumorigenic cell line, were used for miRNA assays. Cells were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum, L-glutamine and antibiotics and were grown in a humidified environment at 37°C with 5% CO2. The pEZX-MT05 control reporter, reporters containing the IL18RAP 3´-UTR with either the most frequent rs7559479 A-allele polymorphism or the rs7559479 G-allele polymorphism, as well as the pEZX-MR04 control reporter and the MIR136 reporter, were purchased from GeneCopoeia.
Transfection and luciferase reporter assay
HuH-7 cells were seeded in six-well plates the day before transfection and were 70–90% confluent at the time of use. Transfections were carried out with a calcium phosphate ProFection Mammalian Transfection System kit (Promega, Madison, USA), according to the manufacturer’s instructions. Seventy-two hours after transfection, we collected the media and measured the Gaussia luciferase and secreted alkaline phosphatase activity using a Secrete-Pair Dual Luminescence assay kit (GeneCopoeia, Rockville, USA), according to the manufacturer’s instructions. Luminescence was measured using a Perkin Elmer Wallac 1420 VICTOR2 multilabel counter.
Results
Characteristics of the study population
The association between IL18RAP gene polymorphisms and obesity traits was first identified in the VALCAR population, and similar results were subsequently found in the Hortega study; when we analysed both samples together this association remained. The main characteristics of the two samples are shown in table 1. The VALCAR study included 468 individuals (24.8% obese; 47.2% male) and the Hortega study included 1502 subjects (23.7% obese; 49.5% male).
Association between IL18RAP gene polymorphisms and obesity traits
All the polymorphisms analysed were in HWE. Of the five SNPs analysed in the IL18RAP gene, rs2293225, rs2272127 and rs7559479 were associated with obesity. We focused on rs7559479 because it is the one most strongly associated with obesity. The presence of the G allele was associated with a higher obesity risk (P=0.0024, OR=1.44) and BMI values (P=0.008) in the pooled population (table 3) and was significantly associated with obesity risk in each individual population. We found an association for rs2293225 and rs2272127 only in the VALCAR population: the T allele in rs2293225 was associated with a lower obesity risk (P=0.036; OR=0.60) and with lower BMI values (P=0.0038; OR=1.41) and the G allele in rs2272127 was associated with a lower obesity risk (P=0.028; OR=0.66) but not with BMI values. These associations are summarised in table 4.
Table 3.
Association between obesity and body mass index (BMI) and the rs7559479 polymorphism, adjusted by age and sex in both samples (values for BMI are the mean ±SE; “% Non-obese” indicates the % of non-obese subjects carrying AA or AG-GG genotypes, “% Obese” indicates the % of obese subjects carrying AA or AG-GG genotypes; bold indicates significance)
| Genotype | N | BMI | % Non-obese | % Obese | Obesity, OR (95% CI) |
| AA | 1062 | 26.41 (0.13) | 877 (59.6%) | 185 (50.7%) | 1.00 |
| AG-GG | 774 | 26.96 (0.16) | 594 (40.4%) | 180 (49.3%) | 1.44 (1.14 to 1.82) |
| P value | 0.008 | 0.0024 |
Table 4.
Summary table for the associations found in the IL18RAP gene (bold indicates significance)
| SNP name | Gene location | Associated phenotype | Populations where an association has been found | Associated allele |
| rs4851581 | 2:102418289 | No associations | -- | -- |
| rs2293224 | 2:102419319 | No associations | -- | -- |
| rs2293225 | 2:102419429 | Lower obesity, lower BMI | VALCAR | T |
| rs2272127 | 2:102423413 | Lower obesity | VALCAR | G |
| rs7559479 | 2:102452327 | Higher obesity | VALCAR | G |
| Higher obesity | Hortega | |||
| Higher obesity, higher BMI | VALCAR+Hortega |
BMI, body mass index
Haplotype analysis
We performed haplotype analysis for the rs2272127, rs2293225 and rs7559479 variations, which all have a high degree of linkage disequilibrium (D´>0.90). The CCG haplotype (number 2 in table 5) was associated with a higher risk of obesity (P<0.0015) and with BMI (P<0.0088). This haplotype includes the risk genotypes found in each variation and was present in 23% of the individuals in the whole study population.
Table 5.
Haplotype association analysis for rs2272127, rs2293225 and rs7559479 in the IL18RAP gene with the body mass index (BMI) and obesity risk adjusted for age and sex
| No | Haplotype | Frequency | Freq. | OR (95% CI) | P value | BMI | ||
| Non-obese | Obese | Difference (95% CI) | P value | |||||
| 1 | CCA | 0.344 | 0.308 | 0.337 | 1.00 | – | 0.00 | – |
| 2 | CCG | 0.220 | 0.277 | 0.232 | 1.44 (1.15 to 1.80) | 0.0015 | 0.48 (0.12 to 0.85) | 0.0088 |
| 3 | GCA | 0.225 | 0.232 | 0.226 | 1.13 (0.90 to 1.43) | 0.30 | 0.08 (−0.29 to 0.44) | 0.68 |
| 4 | CTA | 0.206 | 0.175 | 0.199 | 0.95 (0.74 to 1.21) | 0.68 | −0.2 (−0.57 to 0.17) | 0.29 |
SNPs used in the haplotype construction: rs2272127, rs2293225 and rs7559479.
Bold indicates significance.
Freq, frequency.
Confirmation of the interaction between MIR136 and the IL18RAP 3'-UTR
Our analyses indicate that the rs4851581, rs2293224, rs2293225 and rs2272127 polymorphisms (http://www.ensembl.org/index.html) do not have a clear functional effect; however, rs7559479 is located in the 3-’UTR of IL18RAP in the MIR136 miRNA-binding region (http://microrna.org). This variation can change the binding of this miRNA and therefore can affect the levels of IL18RAP mRNA. Thus, it may also influence the downstream adipogenic effect of IL-18 and might be involved in BMI regulation.
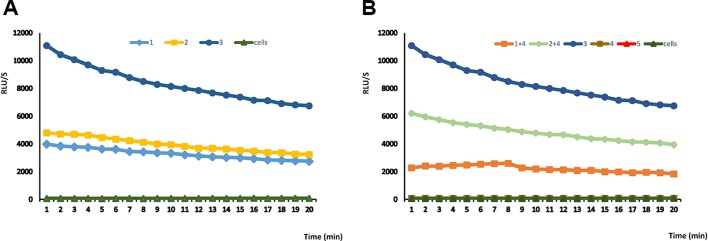
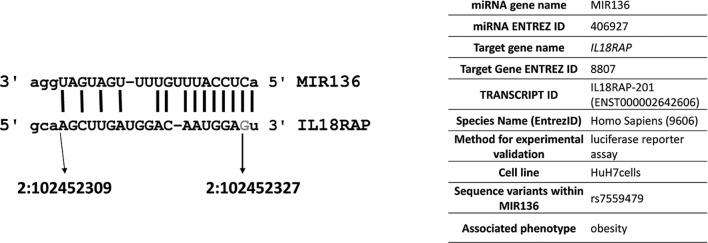
To clarify the functional effect of the rs7559479 SNP on the IL18RAP mRNA MIR136 binding site, we performed an in vitro study in HuH-7 cells. Five plasmids were used: the control reporter pEZX-MT05, a reporter containing the IL18RAP 3´-UTR with either the rs7559479 A- or G-allele polymorphism, the control pEZX-MR04 reporter and the MIR136 reporter, as well as a plasmid that expresses secreted alkaline phosphatase as a transfection control. The expression level was represented as the ratio between the activities of the two enzymes. Figure 1A shows similar luminescence in the reporters containing the IL18RAP 3´-UTR with the rs7559479 A-allele and G-allele polymorphisms in the absence of MIR136. Figure 1B shows that co-transfection of the reporters containing these IL18RAP 3´-UTR rs7559479 polymorphisms alongside the MIR136 reporter produced a significantly higher signal (P<0.05) for the G allele, meaning that miRNA binding was lower for this SNP than for the A-allele polymorphism. Therefore, the A allele (which is associated with a lower risk of obesity) increases MIR136 binding and is thus likely to reduce IL18RAP protein production. Figure 2 shows the characteristics of the interaction between MIR136 and rs7559479 presented according to Piletič and Kunej.26
Figure 1.
Luciferase assays for rs7559479 (a MIR136 target) using different reporters and combinations. Reporters used were: rs7559479 polymorphism A allele (1), rs7559479 polymorphism G allele (2), control reporter pEZX-MT05(3), MIR136 (4) and control reporter pEZX-MR04 (5). Data are represented as the mean±SE. Reporters 4, 5 and cells are negative controls.
Figure 2.
Characteristics of the interaction between MIR136 and IL18RAP. MicroRNA targets predicted using the microRNA.org tool are shown; the base in grey corresponds to the position of the rs7559479 polymorphism.
Discussion
In this population-based study we examined five IL18RAP gene polymorphisms related to the IL-18 adipogenic signalling pathway and found that the rs7559479 AG-GG genotypes were associated with higher BMIs and a greater risk of obesity. It is particularly noteworthy that even though the Hortega and VALCAR populations had different characteristics, the findings were replicated in both; furthermore, when we studied the pooled population, the strength of this association with BMI values and obesity became even stronger. As noted in table 4, a haplotype that is associated with a higher obesity risk and which is frequent in the population is congruent with the data obtained in individual polymorphism analyses (haplotype CCG for rs2272127, rs2293225 and rs7559479). These results reinforce the idea that the IL-18 cascade may be involved in BMI regulation and obesity.
These relationships have not been found in previous genome-wide association studies, but this may be the result of different environmental factors, such as nutrition, in the populations studied. Mediterranean populations might specifically differ from other populations, especially because they are usually under-represented in genome-wide association studies, which often do not include any Spanish samples.27–29 Nevertheless, one study analysed 22 obesity-related gene loci in Spanish populations and found that only the FTO gene was clearly associated with BMI.30 Considering the high heritability of obesity, it is very likely that new variants remain to be discovered, especially in the Spanish/Mediterranean population, where polymorphisms do not seem to have the same effects as described elsewhere in the literature.
No previous studies have described the association between increased weight or obesity and the rs7559479 SNP that we have established in this work. However, obesity is strongly associated with inflammation31 and increased IL-18 concentrations are thought to play a pathophysiological role in obesity and metabolic syndromes.32
The association that we describe is important because rs7559479 is in the 3´-UTR region of the IL18RAP gene, which is an MIR136 target. We used a functional assay to demonstrate that the A allele, which is associated with a lower risk of obesity, reduces the mRNA levels of IL18RAP. MIR136 has been widely studied and, in particular, is implicated in several cancers—for example, by modulating the sensitivity of glioma cells to different treatments,33 34 in human non-small cell lung cancer cells35 and in metastasis-associated traits in lung adenocarcinoma cells.36 Although MIR136 is not expressed in adipocytes,37 38 is has been associated with obesity owing to its role in adipogenic differentiation39 and its expression in hypothalamic neurons, which are involved in appetite and whole-body energy-balance control.40 Moreover, to our knowledge, this is the first time the MIR136:IL18RAP target pair has been validated.
The association between inflammatory cytokine genes and obesity has been less studied than for other candidate genes. In humans, circulating IL-18 levels positively correlate with BMI, adiposity and metabolic syndrome disorders.41–45 This finding is consistent with increased adipokine release from adipose tissue in obesity and accordingly, monocyte/macrophage-lineage cells which are resident in fat tissue are major sources of circulating IL-18.46 In addition, adipocytes from obese humans secrete threefold more IL-18 than those from lean donors47 and IL-18 expression in subcutaneous adipose tissue is elevated in obesity and metabolic syndrome disorders.41 44 48 49 Consistent with a metabolic-state signal, circulating IL-18 levels are increased by hyperglycaemia or a high-fat meal,42 50 and intermittent glucose exposure increases IL-18 secretion by adipocytes51; conversely, weight loss and exercise reduce IL-18 levels.41 44 50 Some adipokines, whose levels increase with obesity (eg, leptin), oppose the positive energy-balance via negative feedback.52 53 Interestingly and perhaps reminiscent of obesity-related leptin resistance, leucocytes from patients with obesity or type 2 diabetes are resistant to IL-18.54 Moreover, polymorphisms of the IL-18 gene or its receptor have been associated with obesity55 and metabolic syndrome disorders56–58 in humans. Nevertheless, total loss of IL-1859–61 has the opposite effect (dysregulation of adipogenesis, appetite suppression and energy expenditure) via alternative pathways.
Our data indicate that a reduction in IL18RAP can reduce BMI and obesity; this may occur by an IL18RAP-mediated decrease in IL-18 proinflammatory signalling.62 63 In addition, the effect of these polymorphisms is important in cells during periods of MIR136 expression—for example, because of their potential consequences for cell differentiation and regulation. Therefore, future studies should aim to clarify the pathways by which IL18RAP may mediate this reduction in BMI and obesity risk. As previously mentioned, it has been suggested that IL-18 is an adipogenic cytokine associated with excess adiposity.4 Moreover, IL18RAP enhances IL-18 binding activity via the IL-18 receptor and plays a role in IL-18 signalling. Therefore, although no previous studies relate IL18RAP to obesity, our findings suggest that polymorphisms in this gene might modify individual susceptibility to obesity. Although the number of subjects included in this study is limited, its statistical power is sufficient for the reduced number of polymorphisms we analysed. However, the possible role of IL18RAP SNPs in obesity phenotypes should be confirmed by other studies, including analyses of different populations and with larger sample sizes.
Supplementary Material
Footnotes
Contributors: All authors participated in protocol and experiment design. VM-B, GdM, VG-A, CP-S and VA-F performed the experimental procedures. VM-B and FJC analysed the data. SM-H, GR, JFA, JTR, RC, JCM-E and ABG-G revised the results and contributed intellectually to discussion and interpretation of the data. The manuscript was written by VM-B, FJC and ABG-G and reviewed by all the authors.
Funding: This work was supported by funds for health science research from the Carlos III Health Institute (PI07/0497, PI11/00726, PI14/00874), by the Centro de Investigación Biomédica en Red Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM) an initiative by Carlos III Health Institute in Madrid and the Spanish Health Ministry, by PROMETEO/2009/029 (AP-091/11, ACOMP/2013/039) from the Valencian Government, and by the GRS 279/a/08 research project from the Junta De Castilla y Leon. V. Martínez-Barquero was also awarded a grant from the Social Politics and Sports Program “Formación del Profesorado Universitario” (AP2010-4754) from the Spanish Ministry of Education.
Disclaimer: None of the funding bodies played a role in the study design or data analysis or interpretation.
Competing interests: None declared.
Ethics approval: The VALCAR and Hortega studies were approved by the ethics committees at the Hospital Clínico Universitario de Valencia and the Hospital Rio Hortega (Valladolid), respectively. All patients gave their written permission to participate in the study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No unpublished data are included in this manuscript.
References
- 1.Cummings DE, Schwartz MW. Genetics and pathophysiology of human obesity. Annu Rev Med 2003;54:453–71. 10.1146/annurev.med.54.101601.152403 [DOI] [PubMed] [Google Scholar]
- 2.Tan LJ, Zhu H, He H, et al. . Replication of 6 obesity genes in a meta-analysis of genome-wide association studies from diverse ancestries. PLoS One 2014;9:e96149 10.1371/journal.pone.0096149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malnick SD, Knobler H. The medical complications of obesity. QJM 2006;99:565–79. 10.1093/qjmed/hcl085 [DOI] [PubMed] [Google Scholar]
- 4.Hung J, McQuillan BM, Chapman CM, et al. . Elevated interleukin-18 levels are associated with the metabolic syndrome independent of obesity and insulin resistance. Arterioscler Thromb Vasc Biol 2005;25:1268–73. 10.1161/01.ATV.0000163843.70369.12 [DOI] [PubMed] [Google Scholar]
- 5.Wood IS, Wang B, Jenkins JR, et al. . The pro-inflammatory cytokine IL-18 is expressed in human adipose tissue and strongly upregulated by TNFalpha in human adipocytes. Biochem Biophys Res Commun 2005;337:422–9. 10.1016/j.bbrc.2005.09.068 [DOI] [PubMed] [Google Scholar]
- 6.Nakamura K, Okamura H, Wada M, et al. . Endotoxin-induced serum factor that stimulates gamma interferon production. Infect Immun 1989;57:590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui K, Yoshimoto T, Tsutsui H, et al. . Propionibacterium acnes treatment diminishes CD4+ NK1.1+ T cells but induces type I T cells in the liver by induction of IL-12 and IL-18 production from Kupffer cells. J Immunol 1997;159:97–106. [PubMed] [Google Scholar]
- 8.Stoll S, Jonuleit H, Schmitt E, et al. . Production of functional IL-18 by different subtypes of murine and human dendritic cells (DC): DC-derived IL-18 enhances IL-12-dependent Th1 development. Eur J Immunol 1998;28:3231–9. [DOI] [PubMed] [Google Scholar]
- 9.Stoll S, Müller G, Kurimoto M, et al. . Production of IL-18 (IFN-gamma-inducing factor) messenger RNA and functional protein by murine keratinocytes. J Immunol 1997;159:298–302. [PubMed] [Google Scholar]
- 10.Udagawa N, Horwood NJ, Elliott J, et al. . Interleukin-18 (interferon-gamma-inducing factor) is produced by osteoblasts and acts via granulocyte/macrophage colony-stimulating factor and not via interferon-gamma to inhibit osteoclast formation. J Exp Med 1997;185:1005–12. 10.1084/jem.185.6.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti B, Jahng JW, Tinti C, et al. . Induction of interferon-gamma inducing factor in the adrenal cortex. J Biol Chem 1997;272:2035–7. 10.1074/jbc.272.4.2035 [DOI] [PubMed] [Google Scholar]
- 12.Pizarro TT, Michie MH, Bentz M, et al. . IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J Immunol 1999;162:6829–35. [PubMed] [Google Scholar]
- 13.Prinz M, Hanisch UK. Murine microglial cells produce and respond to interleukin-18. J Neurochem 1999;72:2215–8. 10.1046/j.1471-4159.1999.0722215.x [DOI] [PubMed] [Google Scholar]
- 14.Gracie JA, Forsey RJ, Chan WL, et al. . A proinflammatory role for IL-18 in rheumatoid arthritis. J Clin Invest 1999;104:1393–401. 10.1172/JCI7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinarello CA. Interleukin-18. Methods 1999;19:121–32. 10.1006/meth.1999.0837 [DOI] [PubMed] [Google Scholar]
- 16.Sims JE. IL-1 and IL-18 receptors, and their extended family. Curr Opin Immunol 2002;14:117–22. 10.1016/S0952-7915(01)00306-5 [DOI] [PubMed] [Google Scholar]
- 17.Kato Z, Jee J, Shikano H, et al. . The structure and binding mode of interleukin-18. Nat Struct Biol 2003;10:966–71. 10.1038/nsb993 [DOI] [PubMed] [Google Scholar]
- 18.Couzin J. MicroRNAs make big impression in disease after disease. Science 2008;319:1782–4. 10.1126/science.319.5871.1782 [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. . Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–8. 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 20.D’Alessandra Y, Devanna P, Limana F, et al. . Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J 2010;31:2765–73. 10.1093/eurheartj/ehq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, et al. . MicroRNA expression profiles classify human cancers. Nature 2005;435:834–8. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 22.Zampetaki A, Kiechl S, Drozdov I, et al. . Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–7. 10.1161/CIRCRESAHA.110.226357 [DOI] [PubMed] [Google Scholar]
- 23.Martinelli R, Nardelli C, Pilone V, et al. . miR-519d overexpression is associated with human obesity. Obesity 2010;18:2170–6. 10.1038/oby.2009.474 [DOI] [PubMed] [Google Scholar]
- 24.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med 2009;60:167–79. 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 25.Lorente-Galdos B, Medina I, Morcillo-Suarez C, et al. . Select your SNPs (SYSNPs): a web tool for automatic and massive selection of SNPs. Int J Data Min Bioinform 2012;6:324–34. 10.1504/IJDMB.2012.049249 [DOI] [PubMed] [Google Scholar]
- 26.Piletič K, Kunej T. Minimal standards for reporting microRNA: target interactions. OMICS 2017;21:197–206. 10.1089/omi.2017.0023 [DOI] [PubMed] [Google Scholar]
- 27.Speliotes EK, Willer CJ, Berndt SI, et al. . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–48. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loos RJ. Recent progress in the genetics of common obesity. Br J Clin Pharmacol 2009;68:811–29. 10.1111/j.1365-2125.2009.03523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berndt SI, Gustafsson S, Mägi R, et al. . Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet 2013;45:501–12. 10.1038/ng.2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-García F, Mansego ML, Rojo-Martínez G, et al. . Impact of obesity-related genes in Spanish population. BMC Genet 2013;14:111 10.1186/1471-2156-14-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilherme A, Virbasius JV, Puri V, et al. . Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol 2008;9:367–77. 10.1038/nrm2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HL, Cho SO, Kim SY, et al. . Association of interleukin-18 gene polymorphism with body mass index in women. Reprod Biol Endocrinol 2012;10:31 10.1186/1477-7827-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, Liu Q, Cai T, et al. . MiR-136 modulates glioma cell sensitivity to temozolomide by targeting astrocyte elevated gene-1. Diagn Pathol 2014;9:173 10.1186/s13000-014-0173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W, Yang Y, Chen B, et al. . MiR-136 targets E2F1 to reverse cisplatin chemosensitivity in glioma cells. J Neurooncol 2014;120:43–53. 10.1007/s11060-014-1535-x [DOI] [PubMed] [Google Scholar]
- 35.Shen S, Yue H, Li Y, et al. . Upregulation of miR-136 in human non-small cell lung cancer cells promotes Erk1/2 activation by targeting PPP2R2A. Tumour Biol 2014;35:631–40. 10.1007/s13277-013-1087-2 [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Liu L, Cai J, et al. . Targeting Smad2 and Smad3 by miR-136 suppresses metastasis-associated traits of lung adenocarcinoma cells. Oncol Res 2013;21:345–52. 10.3727/096504014X14024160459285 [DOI] [PubMed] [Google Scholar]
- 37.Ortega FJ, Moreno-Navarrete JM, Pardo G, et al. . MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One 2010;5:e9022 10.1371/journal.pone.0009022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller P, Gburcik V, Petrovic N, et al. . Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 2011;11:7 10.1186/1472-6823-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaragosi LE, Wdziekonski B, Brigand KL, et al. . Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol 2011;12:R64 10.1186/gb-2011-12-7-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneeberger M, Gomez-Valadés AG, Ramirez S, et al. . Hypothalamic miRNAs: emerging roles in energy balance control. Front Neurosci 2015;9:41 10.3389/fnins.2015.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruun JM, Stallknecht B, Helge JW, et al. . Interleukin-18 in plasma and adipose tissue: effects of obesity, insulin resistance, and weight loss. Eur J Endocrinol 2007;157:465–71. 10.1530/EJE-07-0206 [DOI] [PubMed] [Google Scholar]
- 42.Esposito K, Pontillo A, Ciotola M, et al. . Weight loss reduces interleukin-18 levels in obese women. J Clin Endocrinol Metab 2002;87:3864–6. 10.1210/jcem.87.8.8781 [DOI] [PubMed] [Google Scholar]
- 43.Hung J, McQuillan BM, Chapman CM, et al. . Elevated interleukin-18 levels are associated with the metabolic syndrome independent of obesity and insulin resistance. Arterioscler Thromb Vasc Biol 2005;25:1268–73. 10.1161/01.ATV.0000163843.70369.12 [DOI] [PubMed] [Google Scholar]
- 44.Leick L, Lindegaard B, Stensvold D, et al. . Adipose tissue interleukin-18 mRNA and plasma interleukin-18: effect of obesity and exercise. Obesity 2007;15:356–63. 10.1038/oby.2007.528 [DOI] [PubMed] [Google Scholar]
- 45.Sun L, Hu FB, Yu Z, et al. . Lean body mass, interleukin 18, and metabolic syndrome in apparently healthy Chinese. PLoS One 2011;6:e18104 10.1371/journal.pone.0018104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fain JN. Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam Horm 2006;74:443–77. 10.1016/S0083-6729(06)74018-3 [DOI] [PubMed] [Google Scholar]
- 47.Skurk T, Kolb H, Müller-Scholze S, et al. . The proatherogenic cytokine interleukin-18 is secreted by human adipocytes. Eur J Endocrinol 2005;152:863–8. 10.1530/eje.1.01897 [DOI] [PubMed] [Google Scholar]
- 48.Membrez M, Ammon-Zufferey C, Philippe D, et al. . Interleukin-18 protein level is upregulated in adipose tissue of obese mice. Obesity 2009;17:393–5. 10.1038/oby.2008.535 [DOI] [PubMed] [Google Scholar]
- 49.Weiss TW, Arnesen H, Trøseid M, et al. . Adipose tissue expression of interleukin-18 mRNA is elevated in subjects with metabolic syndrome and independently associated with fasting glucose. Wien Klin Wochenschr 2011;123:650–4. 10.1007/s00508-011-0028-6 [DOI] [PubMed] [Google Scholar]
- 50.Esposito K, Pontillo A, Di Palo C, et al. . Effect of weight loss and lifestyle changes on vascular inflammatory markers in obese women: a randomized trial. JAMA 2003;289:1799–804. 10.1001/jama.289.14.1799 [DOI] [PubMed] [Google Scholar]
- 51.Sun J, Xu Y, Dai Z, et al. . Intermittent high glucose stimulate MCP-l, IL-18, and PAI-1, but inhibit adiponectin expression and secretion in adipocytes dependent of ROS. Cell Biochem Biophys 2009;55:173–80. 10.1007/s12013-009-9066-3 [DOI] [PubMed] [Google Scholar]
- 52.Plata-Salamán CR. Cytokines and feeding. Int J Obes Relat Metab Disord 2001;25(Suppl 5):S48–S52. 10.1038/sj.ijo.0801911 [DOI] [PubMed] [Google Scholar]
- 53.Wong S, Pinkney J. Role of cytokines in regulating feeding behaviour. Curr Drug Targets 2004;5:251–63. 10.2174/1389450043490532 [DOI] [PubMed] [Google Scholar]
- 54.Zilverschoon GR, Tack CJ, Joosten LA, et al. . Interleukin-18 resistance in patients with obesity and type 2 diabetes mellitus. Int J Obes 2008;32:1407–14. 10.1038/ijo.2008.109 [DOI] [PubMed] [Google Scholar]
- 55.Melén E, Himes BE, Brehm JM, et al. . Analyses of shared genetic factors between asthma and obesity in children. J Allergy Clin Immunol 2010;126:631–7. 10.1016/j.jaci.2010.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans J, Collins M, Jennings C, et al. . The association of interleukin-18 genotype and serum levels with metabolic risk factors for cardiovascular disease. Eur J Endocrinol 2007;157:633–40. 10.1530/EJE-07-0463 [DOI] [PubMed] [Google Scholar]
- 57.Koch W, Wolferstetter H, Schatke A, et al. . Interleukin 18 gene variation and risk of acute myocardial infarction. Cytokine 2011;56:786–91. 10.1016/j.cyto.2011.09.006 [DOI] [PubMed] [Google Scholar]
- 58.Presta I, Andreozzi F, Succurro E, et al. . IL-18 gene polymorphism and metabolic syndrome. Nutr Metab Cardiovasc Dis 2009;19:e5–e6. 10.1016/j.numecd.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 59.Netea MG, Joosten LA, Lewis E, et al. . Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat Med 2006;12:650–6. 10.1038/nm1415 [DOI] [PubMed] [Google Scholar]
- 60.Zorrilla EP, Sanchez-Alavez M, Sugama S, et al. . Interleukin-18 controls energy homeostasis by suppressing appetite and feed efficiency. Proc Natl Acad Sci USA 2007;104:11097–102. 10.1073/pnas.0611523104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zorrilla EP, Conti B. Interleukin-18 null mutation increases weight and food intake and reduces energy expenditure and lipid substrate utilization in high-fat diet fed mice. Brain Behav Immun 2014;37:45–53. 10.1016/j.bbi.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Born TL, Thomassen E, Bird TA, et al. . Cloning of a novel receptor subunit, AcPL, required for interleukin-18 signaling. J Biol Chem 1998;273:29445–50. 10.1074/jbc.273.45.29445 [DOI] [PubMed] [Google Scholar]
- 63.Hedl M, Zheng S, Abraham C. The IL18RAP region disease polymorphism decreases IL-18RAP/IL-18R1/IL-1R1 expression and signaling through innate receptor-initiated pathways. J Immunol 2014;192:5924–32. 10.4049/jimmunol.1302727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.