Abstract
Targeting of antigens to antigen-presenting cells (APCs) increases CD4+ T cell activation, and this observation can be exploited in the development of new vaccines. We have chosen an antigen-targeting approach in which we make recombinant antibodies (Abs) with T cell epitopes in their constant region and APC-specific variable regions. Three commonly used model epitopes, amino acids 110–120 of hemagglutinin, 323–339 of ovalbumin, and 46–61 of hen egg lysozyme, were introduced as loops in the CH1 domain of human IgG3. For all three epitopes, we show that the recombinant molecules are secreted from transfected cells. The epitopes are presented to specific T cells, and targeting to IgD on B cells in vitro enhances the presentation efficiency by 104 to 105 compared with the free peptide. After i.v. injection, the epitopes targeted to IgD are presented by splenic APCs to activate specific T cells, whereas little or no activation could be detected without targeting, even after the amount of antigen injected was increased 100-fold or more. Because a wide variety of T cell epitopes, in terms of both length and secondary structure, can be tolerated in loops in constant domains of Abs, the Ab constant region seems to have the intrinsic stability that is needed for this fusion molecule strategy. It might thus be possible to load the Ab with several different epitopes in loops in different domains and thereby make a targeted multisubunit vaccine.
Over recent years, considerable efforts have been directed toward the development of efficient vaccines aimed to stimulate T cells. Effector CD4+ T cells are central in this regard because they regulate B cells and are essential for induction of CD8+ CTL responses through both cytokine secretion (1) and dendritic cell sensitization (2). CD4+ T cells mainly recognize peptides derived from the processing of extracellular antigens by antigen-presenting cells (APCs), presented in association with MHC class II molecules. Targeting of antigen to APCs by coupling to APC-specific Abs increases CD4+ T cell activation, probably because of increased uptake of antigen (3, 4).
Several new prospects for preparation of T cell vaccines have been suggested based on the identification and characterization of MHC-associated peptides. Synthetic peptides that correspond to these T cell epitopes may represent ideal subunits for safe vaccines. However, synthetic peptides are poor immunogens because of their small molecular size and very short serum half-life. To circumvent these disadvantages, a variety of recombinant proteins that carry short immunogenic epitopes have been described, and these so-called antigenized molecules represent a new generation of subunit vaccines (5–9). Genetically engineered Ab molecules can function as such delivery systems for T cell peptides (10, 11) and have the advantage of being self-proteins devoid of the side effects sometimes associated with microbial vaccines and, moreover, have a long half-life compared with synthetic peptides.
Abs are processed by APCs, and short peptides are presented on class II molecules to CD4+ T cells (12, 13). A series of papers have been published that describe recombinant Abs in which the variable (V) region complementarity-determining region (CDR) 2, CDR3, or both are replaced with various antigenic peptides recognized by B cells (10, 14–16), CD8+ T cells (17–19), and CD4+ T cells (11, 20, 21). However, when the peptides are introduced in CDRs of V domains, the Abs lose their antigen-binding specificity. Our strategy is to construct recombinant Abs that carry T cell epitopes in loops in their constant (C) domains, thereby allowing the subsequent addition of APC-specific V regions to the Abs. The loops used resemble CDR loops in that they connect β-strands in the β-sheets of the Ig C domain.
We have previously shown that loops in the CH1 domain of human IgG3 can be replaced with the class II-restricted λ2315 T cell epitope, which comprises amino acids 91–101 of the λ2 Ig light (L) chain produced by the MOPC315 plasmacytoma. In the original molecule, this epitope encompasses the V region CDR3 loop. Such Ab-peptide chimeras stimulate specific CD4+ T cells both in vitro and in vivo (22, 23). Targeting of these Abs to B cells increases antigen presentation efficiency in vitro as well as in vivo (23).
One could argue that the use of an epitope derived from a CDR loop may lead to results that cannot be extended to epitopes that are not loop structures or epitopes derived from non-Ab proteins. To demonstrate the versatility of the strategy, we have introduced three different, well characterized T cell epitopes into human IgG. These are amino acids 110–120 of hemagglutinin (HA) of influenza PR8 virus, amino acids 323–339 of ovalbumin (OVA), and amino acids 46–61 of hen egg lysozyme (HEL). All epitopes have been introduced as substitutions of the sixth loop (L6) in the CH1 domain. This 4-aa loop corresponds to the CDR3 loop in the VH domain (22). It is known from x-ray crystallography data that neither the HA, OVA, nor HEL epitope is a loop in its native molecule and, moreover, all are different both in length and secondary structure. Nevertheless, all recombinant Abs are secreted from transfected cells at levels comparable to wild-type (wt) molecules, indicating that the main outline of the C domain framework is maintained. The Abs are internalized and processed by APCs, and the peptides generated stimulate specific T cells. When the Abs are IgD-specific, they are up to 650 times more efficient than nontargeted Abs in activating T cells in vitro, and up to 50,000 times more potent than the corresponding synthetic peptide. In vivo, only the IgD-specific recombinant Abs delivered enough peptide to APCs to activate T cells. Thus, the chimeric Abs stimulate a significantly stronger response than free peptide at equimolar concentrations.
Materials and Methods
Mice.
BALB/cABom (H-2d, IgHa) and C3H/HeNBom (H-2k, IgHa) mice were purchased from Bomholtgaard Breeding and Research Center (Ry, Denmark). T cell receptor (TCR) transgenic (Tg) mice that express an α/β TCR specific for amino acids 110–120 from HA/I-Ed were the gift of Klaus Karjalainen and Sylvie Degermann, Basel Institute for Immunology (24). DO11.10 TCR Tg mice that express an α/β TCR specific for amino acids 323–339 from OVA/I-Ad were originally produced by Murphy et al. (25) and kindly provided by Nils Lycke (University of Gothenburg, Gothenburg, Sweden).
Cell Lines and Abs.
The λ1-expressing murine myeloma cell line J558L was a gift from S. L. Morrison (University of California, Los Angeles). The myeloma cell line NS0 was obtained from the American Type Culture Collection. Antibodies and reagents used for flow cytometry were as follows: anti-hIgG-PE (Southern Biotechnology Associates; PE indicating phycoerythrin), anti-CD4-PE (PharMingen), anti-CD69-PE (PharMingen), anti-BrdUrd-FITC (Becton Dickinson), streptavidin-Cy-Chrome (PharMingen), and an Ab specific for the Tg receptor of the DO11.10 mice, KJ1–26-biotin (26), kindly provided by Nils Lycke.
T Cells and APCs.
The T cell hybridoma 3A9 was a gift from R. Germain, National Institutes of Health (27). These T cells recognize the 46–61 epitope of HEL in the context of I-Ak. The T cell hybridoma DO11.10 was a gift from P. Marrack (National Jewish Medical and Research Center, Denver) (28). These T cells recognize the amino acids 323–339 epitope of OVA in the context of I-Ad (29). The HA amino acids 110–120-specific CD4+ T cell clone Vir-2 was from A. Rolink (Basel Institute for Immunology) (30). In some experiments, lymph node cells from TCR Tg mice were used as a source of T cells. The APCs used in the experiments were spleen cells from either BALB/c or C3H mice.
Antigens.
A synthetic peptide corresponding to amino acid residues 110–120 (SFERFEIFPKE) of HA was a gift from S. Degermann and K. Karjalainen (Basel Institute for Immunology). A synthetic peptide corresponding to amino acid residues 46–61 (NTDGSTDYGILQINSR) of HEL was a gift from R. Germain. OVA protein was purchased from Sigma. A synthetic peptide corresponding to amino acid residues 323–339 (ISQAVHAAHAEINEAGR) of OVA was a gift from B. Fleckenstein (University of Oslo).
Ab Constructs.
OVA, HEL, and HA peptides were expressed on human IgG3 molecules by substituting the L6 loop in the CH1 domain. The loops in the domain are numbered consecutively following the linear amino acid sequence. Because each C domain harbors six loops, L6 is the third “upper” loop in the domain. The resulting recombinant Ab molecules were denoted L6-OVA, L6-HEL, and L6-HA, respectively.
The design of the molecules is illustrated in Fig. 1. In brief, a 0.9-kb HindIII–PstI fragment encoding the CH1 domain was subcloned into M13mp19 and served as template in the in vitro mutagenesis reactions. Site-specific in vitro mutagenesis was performed as described by Kunkel (31). Reagents for mutagenesis reactions were from Bio-Rad. The mutations were confirmed by sequencing. The mutant CH1 fragments were exchanged with the corresponding wt sequence by using HindIII and BglII sites. The resulting mutant Cγ3 genes were subcloned as HindIII–BamHI fragments into the vector pLNOH2 (32). Upstream from the cloning site, this vector contains a cytomegalovirus promoter and originally, the mouse VH gene, VNP. The combination of VNP-containing heavy (H) chains and λ1 L chains creates Abs with specificity for the haptens NP (4-hydroxy-3-nitrophenacetyl) and NIP (4-hydroxy-5-iodo-3-nitrophenacetyl).
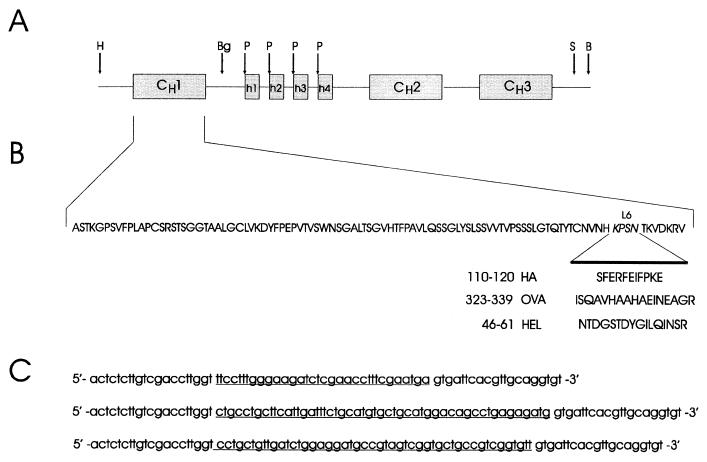
Figure 1.
Strategy for introduction of the HA, OVA, and HEL epitopes into the CH1 domain of human IgG3. (A) Map of the human IgG3 H chain C region gene. H, HindIII; Bg, BglII; P, PstI; S, SphI; B, BamHI. (B) Amino acid sequence of the CH1 domain. The four amino acids of loop 6 (L6) were replaced by those of the HA (110), OVA (323), or HEL (46–61) peptides. (C) Nucleotide sequences of the primers used for the in vitro mutagenesis reactions. The nucleotides encoding the three epitopes are underlined.
The cloning of V region genes that confer specificity for mouse IgD (allotype a) has been described (23). The anti-IgD VH was cloned in pLNOH2, creating a vector encoding the H chain of an IgD-specific IgG3. The wt Cγ3 region gene was subsequently exchanged with the mutant Cγ3 genes on HindIII–BamHI sites. The anti-IgD Vκ was subcloned upstream from the human Cκ gene in the pLNOκ vector as described (23).
Ab Production.
To make anti-NIP Abs, pLNOH2 vectors containing mutant γ3 genes were transfected into the murine plasma cell line J558L by electroporation [BTX (San Diego) apparatus ECM600: ≈1 × 107 cells, 20 μg of DNA, 175 V, 1,300 μF, 129 Ω]. To produce anti-IgD Abs, the pLNOH2 and pLNOκ vectors with anti-IgD V genes and mutant γ3 genes were combined to one vector through a BamHI/BglII cloning step as described (32) and then transfected into NS0 cells by electroporation as explained for J558L cells. Transfected cells were selected in medium containing 800 μg/ml G418 and cloned by limiting dilution. Supernatants from single, G418-resistant colonies were analyzed for Ab secretion after 2–3 wk, by using ELISA as described (33).
Individual clones were cultured in DMEM/5% FCS at high density in the miniPERM bioreactor (Heraeus). Anti-NIP Abs were purified from the cell supernatant by affinity chromatography with the hapten NP coupled to AH-Sepharose (Amersham Pharmacia). Antibodies were eluted from the column with 0.2 mM NIP in PBS/0.02% NaN3. Anti-IgD Abs were affinity purified on protein G (Amersham Pharmacia) or protein L (Actigen) columns. Antibodies were eluted from the columns with 0.1 M glycine⋅HCl (pH 2.7) and rapidly neutralized to avoid aggregation. Eluates were concentrated and dialyzed against PBS and finally against regular growth medium. The purified Abs were analyzed for the presence of aggregates by size exclusion chromatography using Superdex 200 HR 30/10 (Amersham Pharmacia). This gel filtration regularly showed only a trace of aggregates (<1%).
T Cell Activation Assays.
T cell hybridomas as responder cells.
Splenocytes from BALB/c mice or C3H mice were cultured for 20 h in 96-well plates (5 × 105 cells per well) with T cell hybridomas (5 × 104 cells per well) and graded concentrations of antigens. Activation of T cells was assessed by measuring production of IL-2 in the culture supernatant. This was done by [3H]dThd incorporation by using the IL-2-dependent CTLL-2 cells. In brief, diluted culture supernatants (1/3) were incubated with CTLL-2 cells (5 × 103 per well) in 96-well plates for 24 h. Subsequently, 1 μCi (37 kBq) of [3H]dThd was added per well and the culture was continued for an additional 12–14 h. The cells were then harvested onto filters, and [3H]dThd incorporation was counted by using the Matrix96 β-counter (Packard).
Tg lymph node cells as responder cells.
Lymph nodes were isolated from TCR Tg mice. Irradiated (800 rad) splenocytes from BALB/c mice were incubated for 48 h in 96-well plates (5 × 105 cells per well) with Tg lymph node cells (1 × 105 per well) and graded concentrations of antigens. Activation of T cells was assessed by pulsing the cultures with 1 μCi of [3H]dThd for another 16–24 h and then counting the filters as described above.
In Vivo Targeting Assays.
BALB/c (H-2d) mice were injected i.v. in the tail vein with different amounts of recombinant anti-NIP or anti-IgD Ab, synthetic peptide, or native antigenic protein. After 1.5 h, the mice were killed, and spleen cells were isolated. T cell assays were set up with spleen cells (5 × 105 per well) and DO.11.10 (5 × 104 per well) or Vir-2 T cells (2 × 104 per well). After 24 h, supernatant was withdrawn and IL-2 production was measured as described above, whereas IFN-γ production was measured in an ELISA as described (34).
Results
Construction of Antibodies with Epitopes in the CH1 Domain.
The MHC class II-restricted CD4+ T cell epitopes chosen for this work are three different peptides from such proteins as influenza virus HA, chicken OVA, and HEL. The peptides were chosen because they represent well characterized epitopes recognized by MHC class II-restricted T cell hybridomas and because Tg mice with transgenes encoding the TCR chains from such hybridomas exist as a source of specific T cells from lymph nodes.
The peptide corresponding to amino acids 110–120 of influenza virus HA is a helper epitope recognized by CD4+ T cells in association with I-Ed MHC class II molecules (24). The immunogenic peptide from OVA, amino acids 323–339, has been shown to be immunodominant in H-2d mice and is presented by I-Ad (29, 35). The immunodominant epitope from HEL, amino acids 46–61, encompasses the minimal epitope 52–61, which is presented by I-Ak molecules (36, 37).
Three mutated H chains were constructed to include HA, OVA, and HEL peptides, respectively. In all cases, the nucleotides encoding the L6 loop in the IgG3 CH1 gene were deleted and replaced by nucleotide sequences coding for the selected peptide as shown in Fig. 1. We have earlier shown that this loop is permissive for expression of the λ2315 peptide (22, 23). Notably, in previous work, the L6 loop was denoted L3 as its position in the C domain corresponds to that of the CDR3 loop in a V domain. The HA, OVA, and HEL mutants were secreted from transfected cells, indicating that the main tertiary structure was maintained, because misfolded proteins are normally retained intracellularly (38–40). The functional integrity was confirmed in ELISAs in which microtiter plates were coated with antigen (BSA-NIP) or Abs specific for human γ chains. Binding was detected with sheep-anti-human IgG (Fab, Fc, or γ chain specific) or sheep-anti-κ L chains (data not shown). These NIP-specific Abs with HA, OVA, or HEL peptides were denoted αNIP.L6-HA, αNIP.L6-OVA, and αNIP.L6-HEL, respectively.
To target the peptides to APCs, the NIP-specific V genes were exchanged with V genes specific for the a allotype of mouse IgD (IgDa) (41). The resulting genes were transfected into the non-Ig-secreting mouse myeloma B cell line NS0. The binding of the chimeric molecules to IgD displayed on the surface of target cells was visualized by flow cytometry analysis and found to be similar to the IgD-specific Ab with the λ2315 epitope (data not shown and ref. 23). These IgD-specific Abs with HA, OVA, or HEL peptides were denoted αIgD.L6-HA, αIgD.L6-OVA, and αIgD.L6-HEL, respectively.
IgG3 with HA Peptide Efficiently Stimulates Tg T Cells.
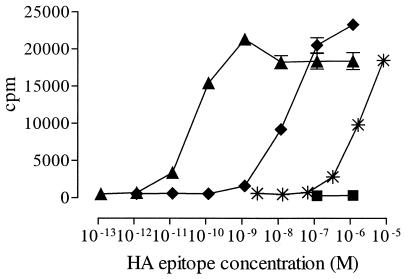
To assess the in vitro antigenicity of the HA-expressing Abs, we used them as antigens in dose–response T cell activation assays. BALB/c spleen cells were used as APCs. The BALB/c strain has the MHC H-2d haplotype and expresses I-Ed MHC class II molecules necessary for presentation of the HA epitope to CD4+ T cells. As responder CD4+ T cells, lymph node cells from mice Tg for a HA-(110–120)-specific TCR were used (24). The results shown in Fig. 2 indicate that the HA epitope was presented 150-fold better as part of αNIP.L6-HA than after addition of free HA peptide.
Figure 2.
Proliferation of HA-specific T cells in response to recombinant Ab with the HA-(110–120) epitope. Lymph node cells from HA-specific TCR Tg mice were cultured together with irradiated BALB/c spleen cells and αIgD.L6-HA (▴), αNIP.L6-HA (♦), αIgD.wt (■), or synthetic HA-(110–120) peptide (✠). Ab and peptide concentrations were as indicated, and proliferation of lymph node cells was measured as incorporation of [3H]dThd.
Targeting IgG.L6-HA to B Cells Further Increases Presentation of the HA Epitope.
We next addressed the targeting property of αIgD.L6-HA. Fig. 2 shows an increase in T cell activation when the chimeric molecules were targeted to B cells. The dose–response curves show that αIgD.L6-HA was ≈300-fold more efficient than its nontargeted counterpart in activating the T cells. Anti-IgD molecules with wt H chain C region, αIgD.wt, did not elicit any response. Compared with synthetic peptide, αIgD.L6-HA was 40,000–50,000 times more efficient than free peptide at equimolar concentrations.
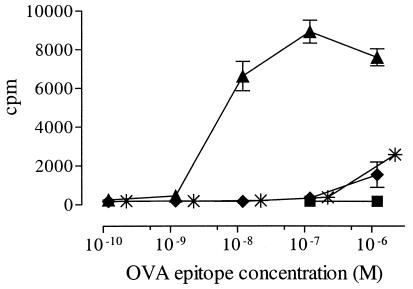
IgG3 with OVA Peptide Efficiently Stimulates Tg Lymph Node T Cells and the T Cell Hybridoma DO11.10.
In a first set of experiments, we compared nontargeted and targeted L6-OVA in an in vitro T cell proliferation assay with BALB/c spleen cells as APCs and lymph node cells from DO11.10 mice as responder cells. As shown in Fig. 3, αIgD.L6-OVA induced a significant increase in the stimulation of DO11.10 cells compared with its nontargeted counterpart. Based on molar ratios, the αIgD.L6-OVA was ≈650 times more effective than αNIP.L6-OVA and OVA protein. Similar results were obtained with the DO11.10 hybridoma as responder cells and measurement of IL-2 in the supernatant (data not shown).
Figure 3.
Proliferation of OVA-specific T cells in response to recombinant Ab with the OVA-(323–339) epitope. αIgD.L6-OVA and controls were titrated into cultures of lymph node cells from DO11.10 TCR Tg mice and irradiated BALB/c spleen cells. αIgD.L6-OVA (▴), αNIP.L6-OVA (♦), αIgD.wt (■), and OVA protein (✠). Tg lymph node cell proliferation was measured as [3H]dThd incorporation.
To investigate more closely the phenotype of proliferating cells, we measured BrdUrd incorporation into DNA of lymph node T cells from DO11.10 TCR Tg mice after coculturing with chimeric Abs and BALB/c spleen cells. Proliferation of specific CD4+ T cells was identified by flow cytometric analysis by use of anti-BrdUrd Abs in combination with anti-CD4 and a clonotypic Ab that recognize the Tg receptor. We also examined the expression of CD69. We found that a considerable fraction of the T cells cocultured with αIgD.L6-OVA was specific, BrdUrd+, CD4+, and CD69+, whereas αNIP.L6-OVA stimulated fewer cells (results not shown).
IgG3 with HEL Peptide Efficiently Stimulates the T Cell Hybridoma 3A9.
Further evidence that Ab molecules can function as carriers for delivery of yet another epitope is presented in Fig. 4. The Abs used carry the 46–61 epitope from HEL. As APCs we used spleen cells from C3H mice, which have the MHC H-2k haplotype and express I-Ak MHC class II molecules necessary for presentation of the HEL epitope to specific 3A9 hybridoma cells. Both the targeted and the nontargeted Ab are superior immunogens compared with free unconjugated peptide, αIgD.L6-HEL being 10,000–15,000 times more efficient than synthetic peptide based on a molar ratio. The experiments also confirm that targeting induces a more vigorous T cell response because αIgD.L6-HEL is 100–150 times more efficient than αNIP.L6-HEL.
Figure 4.
Proliferation of HEL-specific T cells in response to recombinant Ab with the HEL-(46–61) epitope. 3A9 T hybridoma cells were cultured together with irradiated C3H spleen cells and titrated amounts of recombinant Ab and synthetic HEL peptide. αIgD.L6-HEL (▴), αNIP.L6-HEL (♦), αIgD.wt (■), and synthetic HEL-(46–61) peptide (✠). IL-2 production by 3A9 cells was measured as incorporation of [3H]dThd into CTLL-2 cells.
IgG3 with HA and OVA Peptides Are Targeted to APCs in Vivo and Enhance T Cell Activation.
The recombinant Abs were also tested for their ability to target APCs in vivo. First, the IgD-specific Ab with the HA epitope was compared with its NIP-specific counterpart and with synthetic HA peptide. Preparations of all three molecules were injected i.v. in titrated amounts into BALB/c mice. After 1.5 h, spleen cells were mixed with HA-specific Vir-2 T cells without any further addition of peptide. Spleen cells from mice injected with 10−10 mol (8 μg) or more of αIgD.L6-HA activated HA-specific Vir-2 T cells as measured by IFN-γ production (Fig. 5A), whereas there were no detectable T cell responses after injection of synthetic HA peptide or αNIP.L6-HA, even after increasing the amount of injected material 100-fold. Thus, the targeted Ab was at least 100 times more efficient. In a separate experiment, synthetic HA peptide did not induce a response even when 10−6 mol was injected (data not shown).
Figure 5.
Proliferation of T cells in response to recombinant Ab with the HA (A) and OVA (B) epitopes. BALB/c mice were injected i.v. with recombinant Ab, synthetic peptide, or native antigen. Later (1.5 h), the spleens were removed, and spleen cells were cultured together with HA-specific Vir-2 T cells (A) or OVA-specific DO11.10 T cells (B). After 24 h, supernatant was withdrawn and assayed for cytokine production. Anti-IgD Ab (▴), anti-NIP Ab (♦), synthetic peptide (✠), and complete OVA protein (●) responses are shown. IFN-γ production was measured in an ELISA, whereas IL-2 production by DO11.10 cells was measured as incorporation of [3H]dThd into CTLL-2 cells.
Similar experiments were also performed to compare the efficiency of OVA-recombinant IgD-specific Ab and complete antigenic protein. In these experiments, spleen cells were mixed with DO11.10 T cells and T cell activation was detected as IL-2 production (Fig. 5B). Again, the recombinant IgD-specific Ab was found to be at least 1,000-fold more efficient than complete OVA protein and 100-fold more efficient than synthetic OVA peptide.
Discussion
Engineered Abs are attractive carriers for antigenic peptides because Ab-peptide fusions stimulate potent and specific immunity (8, 9, 23). Complete human Abs are likely to be long-lived molecules devoid of side effects and may be given unique specificities. Our main goal is to construct Abs that contain T cell epitopes integrated in their C parts and have V regions with specificity for APCs.
In earlier work (22, 23), we showed that the λ2315 T cell epitope could replace the CDR-like loops L4 and L6 in the CH1 domain and that the two loop mutants were able to elicit MHC class II-restricted CD4+ T cell responses. In this report, we have extended this concept by inserting a variety of T cell epitopes in the L6 loop in the CH1 domain. The λ2315 epitope is originally derived from a loop structure, the CDR3 region in an L chain, and might for that reason be more easily grafted in loops than other T cell epitopes. It was therefore important to extend the strategy to other T cell epitopes.
The three epitopes used in this work are all different, in terms of both length and secondary structure in the intact proteins. The HAs of influenza virus are divided into three subtypes, H1, H2, and H3, of which the H3 subtype has been crystallized (42). The HA-(110–120) epitope used in this study is derived from the H1 subtype. Despite the low degree of homology with the H3 subtype at the level of amino acid sequence (43), there is a strict conservation between subtypes of many structurally important amino acids. Therefore, it is assumed that the subtypes share many of their structural features as well as a common overall shape (42, 44). The HA-(110–120)-corresponding sequence in the H3 subtype has a secondary structure defined as a bend β-strand γ-turn (43, 45). The OVA-(323–339) epitope is composed mostly of amino acids participating in a β ladder (46), whereas the HEL-(46–61) epitope in its native position in the lysozyme protein has a secondary structure with three turns and two β-strands (47).
Despite these differences in secondary structure motifs, the L6 loop is permissive for all epitopes. Furthermore, the peptides were presented to T cells in a specific manner, indicating that they were all cleaved out of the Abs and bound a diversity of class II molecules, namely I-Ed, I-Ad, and I-Ak. It is known that the flanking residues outside the core sequence of a T cell epitope may influence its processing and presentation (48–52). The Ig flanking regions in L6 did not seem to have any negative effect on the presentation of the HA, OVA, or HEL epitope, which agrees with other observations for T cell peptides expressed as integral parts of unrelated proteins (49, 51). In conclusion, the L6 loop in the CH1 domain of human γ3 chains appears to represent a permissive site for grafting a multitude of foreign epitopes without altering the structural integrity of the Ab molecule or the immunogenicity of the peptides.
The efficiency of antigen presentation depends, to a large extent, on the concentration of peptide/MHC complexes on the APC surface (53). Thus, by genetically linking the epitopes to APC-specific targeting molecules, the antigen load may be increased, thereby enhancing the antigen uptake, processing, and presentation, and subsequently, increased T cell activation. The use of genetically engineered Abs for delivery of immunogenic peptides has been previously described in an APC-targeting context by Baier and coworkers (9). They achieved immunotargeting by using IgD or MHC class II-specific Fab fragments with T cell epitopes inserted as C-terminal tails. This procedure may have a disadvantage in that peptides attached to the end of a molecule may be more sensitive to proteases (54). By including the epitopes as integral parts of the Ab molecule, we anticipated that the peptides would acquire the same half-life as the Ab molecule itself. We have previously shown that targeting of the λ2315 epitope on complete, bivalent Abs to IgD on B cells increases T cell activation 100–10,000 times in vitro and 20–100 times in vivo (23). The results presented in this study clearly extend this notion because anti-IgD Abs with HA, OVA, or HEL peptides are 150–650 times more efficient than their nontargeted counterparts.
Because a major limitation in the use of synthetic peptide as vaccines is the instability and inherently weak immunogenicity of natural peptides, we show here that targeting of HA, OVA, and HEL epitopes to IgD on the APC dramatically increases the presentation efficiency compared with free peptide, in vitro as well as in vivo. The most striking difference is demonstrated in the experiments with the HA epitope in vitro, in which αIgD.L6-HA is up to 40,000–50,000 times more efficient than free HA peptide. After i.v. injection, the targeted Abs induced T cell activation, whereas control Abs, antigenic protein, as well as synthetic peptide did not. In conclusion, we expect that coupling of peptides to Abs specific for receptors on specialized APCs constitutes several major advantages for vaccination.
The peptide can be targeted to the specific subset of cells expressing the relevant receptors. We have chosen IgD on B cells in our model system. In general, B cells are not optimal targets in vivo. This reservation concerns the question of whether B cells are able to activate T cells that are genuinely naïve (55, 56). However, other candidate molecules may certainly be good targets in vivo. MHC class II or DEC 205 (57), as well as other surface molecules on dendritic cells, are prime candidates, as numerous studies point to dendritic cells as the main APCs in class II-restricted T cell priming in vivo (58, 59) and in vitro (60, 61).
For further in vivo administration, the Fc regions should preferably be made devoid of effector functions (62–65) because Fc receptors expressed on peripheral blood cells play an important role in triggering a variety of cytotoxic, phagocytic, and inflammatory functions. The site for binding of IgGs to Fcγ receptor I (FcγRI) is proposed to be Leu-Leu-Gly-Gly-Pro-Ser (EU numbering 234–239), which lies in the lower hinge region (66). Human IgG4 and mouse IgG2b, which have low or no affinity for FcγRI, contain the sequence Phe-Leu-Gly-Gly-Pro-Ser and Leu-Glu-Gly-Gly-Pro-Ser, respectively, at the FcγRI-binding site. Recent reports (67, 68) have shown that the lower hinge of Igs is likely to be involved in binding to all Fc receptors. Intact Fc parts are indispensable for a long half-life of the Igs in vivo; on the other hand, interaction between the Ig and FcRs may lead to undesired toxicity or side effects. As human IgG3 binds all FcγRs, targeting of the Abs to IgD may lead to crosslinking of IgD and the inhibitory FcγRIIB molecule and inhibition of B cell activity. Moreover, natural killer (NK) cells, a principal cell type involved in Ab-dependent cell-mediated cytotoxicity, do not express the inhibitory FcγRIIB, but express the activating FcγRIII. Interaction between Igs bound to APCs and FcγRIII on NK cells may potentially trigger killing of the APCs in vivo. This possibility underscores the importance of selecting and engineering the Abs to minimize their interactions with FcγRs and make Ab-peptide fusions able to target the appropriate APCs with high efficacy.
The use of C domain loops for epitope insertion may also allow for the introduction of many epitopes into the same Ab molecule. Brumeanu et al. (16) and Xiong et al. (20) have shown that an Ab molecule can contain a T and a B cell epitope simultaneously added to the CDR2 and CDR3 loops of the VH domain. Because each C domain in IgG harbors six loops, there are numerous other candidate positions for insertion of epitopes. Accordingly, our long-term aim is to exchange several C region loops simultaneously to create a multivalent vaccine contained within a single Ab molecule.
Acknowledgments
We are grateful to Randi Sandin at the National Institute of Public Health for her excellent technical assistance and for affinity purifying many of the Abs described in this paper. We especially thank Tone F. Gregers for many valuable discussions. This work was supported by grants from the Norwegian Cancer Society and The Norwegian Research Council.
Abbreviations
- APCs
antigen-presenting cells
- C
constant
- CDR
complementarity-determining region
- FcγR
Fcγ receptor
- H
heavy
- HA
hemagglutinin
- HEL
hen egg lysozyme
- L
light
- NIP
4-hydroxy-5-iodo-3-nitrophenacetyl
- NP
4-hydroxy-3-nitrophenacetyl
- OVA
ovalbumin
- TCR
T cell receptor
- Tg
transgenic
- V
variable
- wt
wild type
References
- 1.Keene J A, Forman J. J Exp Med. 1982;155:768–782. doi: 10.1084/jem.155.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Z, Yuan L, Zhou X, Sotomayor E, Levitsky H I, Pardoll D M. J Exp Med. 2000;191:541–550. doi: 10.1084/jem.191.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carayanniotis G, Barber B H. Nature (London) 1987;327:59–61. doi: 10.1038/327059a0. [DOI] [PubMed] [Google Scholar]
- 4.Casten L A, Pierce S K. J Immunol. 1988;140:404–410. [PubMed] [Google Scholar]
- 5.Leclerc C, Lo-Man R, Charbit A, Martineau P, Clement J M, Hofnung M. Int Rev Immunol. 1994;11:123–132. doi: 10.3109/08830189409061720. [DOI] [PubMed] [Google Scholar]
- 6.Freimuth P, Steinman R M. Res Microbiol. 1990;141:995–1001. doi: 10.1016/0923-2508(90)90139-h. [DOI] [PubMed] [Google Scholar]
- 7.Evans D J, McKeating J, Meredith J M, Burke K L, Katrak K, John A, Ferguson M, Minor P D, Weiss R A, Almond J W. Nature (London) 1989;339:385–388. doi: 10.1038/339385a0. [DOI] [PubMed] [Google Scholar]
- 8.Bona C A, Bot A, Brumeanu T D. Chem Immunol. 1997;65:179–206. [PubMed] [Google Scholar]
- 9.Baier G, Baier-Bitterlich G, Looney D J, Altman A. J Virol. 1995;69:2357–2365. doi: 10.1128/jvi.69.4.2357-2365.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billetta R, Hollingdale M R, Zanetti M. Proc Natl Acad Sci USA. 1991;88:4713–4717. doi: 10.1073/pnas.88.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaghouani H, Steinman R, Nonacs R, Shah H, Gerhard W, Bona C. Science. 1993;259:224–227. doi: 10.1126/science.7678469. [DOI] [PubMed] [Google Scholar]
- 12.Bogen B, Malissen B, Haas W. Eur J Immunol. 1986;16:1373–1378. doi: 10.1002/eji.1830161110. [DOI] [PubMed] [Google Scholar]
- 13.Bikoff E K, Yu H, Eckhardt L A. Eur J Immunol. 1988;18:341–348. doi: 10.1002/eji.1830180304. [DOI] [PubMed] [Google Scholar]
- 14.Cook J, Barber B H. Vaccine. 1995;13:1770–1778. doi: 10.1016/0264-410x(95)00140-v. [DOI] [PubMed] [Google Scholar]
- 15.Zaghouani H, Anderson S A, Sperber K E, Daian C, Kennedy R C, Mayer L, Bona C A. Proc Natl Acad Sci USA. 1995;92:631–635. doi: 10.1073/pnas.92.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brumeanu T D, Bot A, Bona C A, Dehazya P, Wolf I, Zaghouani H. Immunotechnology. 1996;2:85–95. doi: 10.1016/1380-2933(96)85196-7. [DOI] [PubMed] [Google Scholar]
- 17.Zaghouani H, Krystal M, Kuzu H, Moran T, Shah H, Kuzu Y, Schulman J, Bona C. J Immunol. 1992;148:3604–3609. [PubMed] [Google Scholar]
- 18.Kuzu Y, Kuzu H, Zaghouani H, Bona C. Int Immunol. 1993;5:1301–1307. doi: 10.1093/intimm/5.10.1301. [DOI] [PubMed] [Google Scholar]
- 19.Billetta R, Filaci G, Zanetti M. Eur J Immunol. 1995;25:776–783. doi: 10.1002/eji.1830250323. [DOI] [PubMed] [Google Scholar]
- 20.Xiong S, Gerloni M, Zanetti M. Nat Biotechnol. 1997;15:882–886. doi: 10.1038/nbt0997-882. [DOI] [PubMed] [Google Scholar]
- 21.Legge K L, Min B, Cestra A E, Pack C D, Zaghouani H. J Immunol. 1998;161:106–111. [PubMed] [Google Scholar]
- 22.Lunde E, Bogen B, Sandlie I. Mol Immunol. 1997;34:1167–1176. doi: 10.1016/s0161-5890(97)00143-0. [DOI] [PubMed] [Google Scholar]
- 23.Lunde E, Munthe L A, Vabo A, Sandlie I, Bogen B. Nat Biotechnol. 1999;17:670–675. doi: 10.1038/10883. [DOI] [PubMed] [Google Scholar]
- 24.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy K M, Heimberger A B, Loh D Y. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 26.Haskins K, Kubo R, White J, Pigeon M, Kappler J, Marrack P. J Exp Med. 1983;157:1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen P M, Unanue E R. J Immunol. 1984;132:1077–1079. [PubMed] [Google Scholar]
- 28.Walker E, Warner N L, Chesnut R, Kappler J, Marrack P. J Immunol. 1982;128:2164–2169. [PubMed] [Google Scholar]
- 29.Sette A, Buus S, Colon S, Smith J A, Miles C, Grey H M. Nature (London) 1987;328:395–399. doi: 10.1038/328395a0. [DOI] [PubMed] [Google Scholar]
- 30.Hackett C J, Dietzschold B, Gerhard W, Ghrist B, Knorr R, Gillessen D, Melchers F. J Exp Med. 1983;158:294–302. doi: 10.1084/jem.158.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 32.Norderhaug L, Olafsen T, Michaelsen T E, Sandlie I. J Immunol Methods. 1997;204:77–87. doi: 10.1016/s0022-1759(97)00034-3. [DOI] [PubMed] [Google Scholar]
- 33.Michaelsen T E, Aase A, Westby C, Sandlie I. Scand J Immunol. 1990;32:517–528. doi: 10.1111/j.1365-3083.1990.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 34.Lauritzsen G F, Bogen B. Scand J Immunol. 1991;33:647–656. doi: 10.1111/j.1365-3083.1991.tb02537.x. [DOI] [PubMed] [Google Scholar]
- 35.Shimonkevitz R, Colon S, Kappler J W, Marrack P, Grey H M. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- 36.Allen P M, Strydom D J, Unanue E R. Proc Natl Acad Sci USA. 1984;81:2489–2493. doi: 10.1073/pnas.81.8.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latek R R, Unanue E R. Immunol Rev. 1999;172:209–228. doi: 10.1111/j.1600-065x.1999.tb01367.x. [DOI] [PubMed] [Google Scholar]
- 38.Bonifacino J S, Lippincott-Schwartz J. Curr Opin Cell Biol. 1991;3:592–600. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 39.Reddy P S, Corley R B. BioEssays. 1998;20:546–554. doi: 10.1002/(SICI)1521-1878(199807)20:7<546::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 40.Doms R W, Lamb R A, Rose J K, Helenius A. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 41.Oi V T, Herzenberg L A. Mol Immunol. 1979;16:1005–1017. doi: 10.1016/0161-5890(79)90034-8. [DOI] [PubMed] [Google Scholar]
- 42.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 43.Winter G, Fields S, Brownlee G G. Nature (London) 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- 44.Caton A J, Brownlee G G, Yewdell J W, Gerhard W. Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 45.Fleury D, Wharton S A, Skehel J J, Knossow M, Bizebard T. Nat Struct Biol. 1998;5:119–123. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- 46.Stein P E, Leslie A G, Finch J T, Carrell R W. J Mol Biol. 1991;221:941–959. doi: 10.1016/0022-2836(91)80185-w. [DOI] [PubMed] [Google Scholar]
- 47.Maenaka K, Matsushima M, Song H, Sunada F, Watanabe K, Kumagai I. J Mol Biol. 1995;247:281–293. doi: 10.1006/jmbi.1994.0139. [DOI] [PubMed] [Google Scholar]
- 48.Vacchio M S, Berzofsky J A, Krzych U, Smith J A, Hodes R J, Finnegan A. J Immunol. 1989;143:2814–2819. [PubMed] [Google Scholar]
- 49.Lowenadler B, Lycke N, Svanholm C, Svennerholm A M, Krook K, Gidlund M. Mol Immunol. 1992;29:1185–1190. doi: 10.1016/0161-5890(92)90054-2. [DOI] [PubMed] [Google Scholar]
- 50.Eisenlohr L C, Yewdell J W, Bennink J R. J Exp Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leclerc C, Martineau P, Charbit A, Lo-Man R, Deriaud E, Hofnung M. Mol Immunol. 1993;30:1561–1572. doi: 10.1016/0161-5890(93)90447-j. [DOI] [PubMed] [Google Scholar]
- 52.Janssen R, Wauben M, van der Zee R, de Gast M, Tommassen J. Int Immunol. 1994;6:1187–1193. doi: 10.1093/intimm/6.8.1187. [DOI] [PubMed] [Google Scholar]
- 53.Snider D P, Segal D M. J Immunol. 1987;139:1609–1616. [PubMed] [Google Scholar]
- 54.Neuberger M S, Williams G T, Fox R O. Nature (London) 1984;312:604–608. doi: 10.1038/312604a0. [DOI] [PubMed] [Google Scholar]
- 55.Ronchese F, Hausmann B. J Exp Med. 1993;177:679–690. doi: 10.1084/jem.177.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Z, Richter G, Schuler T, Ibe S, Cao X, Blankenstein T. Nat Med. 1998;4:627–630. doi: 10.1038/nm0598-627. [DOI] [PubMed] [Google Scholar]
- 57.Jiang W, Swiggard W J, Heufler C, Peng M, Mirza A, Steinman R M, Nussenzweig M C. Nature (London) 1995;375:151–155. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 58.Crowley M, Inaba K, Steinman R M. J Exp Med. 1990;172:383–386. doi: 10.1084/jem.172.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inaba K, Metlay J P, Crowley M T, Steinman R M. J Exp Med. 1990;172:631–640. doi: 10.1084/jem.172.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inaba K, Steinman R M. J Exp Med. 1984;160:1717–1735. doi: 10.1084/jem.160.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inaba K, Steinman R M. Science. 1985;229:475–479. doi: 10.1126/science.3160115. [DOI] [PubMed] [Google Scholar]
- 62.Clynes R A, Towers T L, Presta L G, Ravetch J V. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 63.Woodle E S, Xu D, Zivin R A, Auger J, Charette J, O'Laughlin R, Peace D, Jollife L K, Haverty T, Bluestone J A, et al. Transplantation. 1999;68:608–616. doi: 10.1097/00007890-199909150-00003. [DOI] [PubMed] [Google Scholar]
- 64.Reddy M P, Kinney C A, Chaikin M A, Payne A, Fishman-Lobell J, Tsui P, Dal Monte P R, Doyle M L, Brigham-Burke M R, Anderson D, et al. J Immunol. 2000;164:1925–1933. doi: 10.4049/jimmunol.164.4.1925. [DOI] [PubMed] [Google Scholar]
- 65.Michaelsen T E, Brekke O H, Aase A, Sandin R H, Bremnes B, Sandlie I. Proc Natl Acad Sci USA. 1994;91:9243–9247. doi: 10.1073/pnas.91.20.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jefferis R, Lund J, Pound J D. Immunol Rev. 1998;163:59–76. doi: 10.1111/j.1600-065x.1998.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 67.Sondermann P, Huber R, Oosthuizen V, Jacob U. Nature (London) 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 68.Garman S C, Wurzburg B A, Tarchevskaya S S, Kinet J P, Jardetzky T S. Nature (London) 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]