Abstract
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by excess B and T cell activation, the development of autoantibodies against self-antigens including nuclear antigens, and immune complex deposition in target organs which triggers an inflammatory response and tissue damage. The genetic and environmental factors that contribute to development of SLE have been extensively studied in both humans and mouse models of the disease. One of the important genetic contributions to SLE development is an alteration in the expression of the transcription factor Ets1, which regulates the functional differentiation of lymphocytes. Here we review the genetic, biochemical and immunological studies that have linked low levels of Ets1 to aberrant lymphocyte differentiation and to the pathogenesis of SLE.
Keywords: Systemic lupus erythematosus, autoantibodies, Ets1, B cell tolerance, plasma cells, T cell cytokines
INTRODUCTION
Systemic Lupus Erythematosus (SLE) is a potentially fatal chronic and systemic autoimmune disease that affects predominantly females in their childbearing years, with overall prevalence varying depending on geographic location and ethnicity.1 SLE is associated with a loss of immune tolerance to nucleic acid containing antigens, leading to inflammation and organ damage. Multiple tissues and organ systems can be affected, including the skin, the joints, the kidneys, the cardiovascular system, and the central nervous system, leading to significant morbidity and mortality. Treatment for SLE primarily involves non-specific immunosuppression, which has undesirable side effects, and only one new therapy has been approved for SLE in the last 50 years.2,3 Thus, a more thorough understanding of the molecular mechanisms of SLE pathogenesis is important for the development of more targeted therapeutic strategies. Genome-wide association studies have revealed genetic alterations in many genes that are associated with susceptibility to SLE, highlighting potential disease mechanisms.4,5
One such SLE-associated gene encodes the transcription factor Ets1,6–8 which is primarily expressed in lymphocytes9–13 and is present at reduced levels in peripheral blood mononuclear cells (PBMCs) from SLE patients.7,14–17 Consistent with an important role for Ets1 in limiting autoimmune disease, Ets1-deficient mice accumulate plasma cells, produce autoantibodies, and develop several features of lupus-like autoimmune disease.18,19 Here we review the interplay of Ets1 with lupus susceptibility and pathogenesis including (1) the association of Ets1 genetic variants with lupus and other autoimmune and inflammatory diseases, (2) the characterization of autoimmunity in Ets1-deficient mice and the role of Ets1 in B cell tolerance to self-antigens, (3) the signaling pathways that control Ets1 expression in B cells, and (4) functions of Ets1 in B and T cells that likely contribute to the control of autoantibody production, including limiting plasma cell differentiation and skewing T cell subsets and cytokines.
I. Systemic Lupus Erythematosus: Disease Mechanisms
Alterations in three major immune pathways have been shown to contribute to SLE pathogenesis; loss of adaptive immune tolerance, impaired clearance of apoptotic debris, and hyperactivation of the innate immune system, particularly with respect to Toll-like receptor (TLR) signaling and the type I interferon (IFN) pathway (reviewed in20,4,21). These defects synergize to form pro-inflammatory positive feedback loops described briefly below. A breach in adaptive immune tolerance results in an elevated frequency of autoreactive B and T cells, the activation of which is facilitated by the increased availability of nucleic acid-containing self-antigens. These antigens are particularly efficient at activating B cells as they can engage both the B-cell receptor (BCR) and the endosomal, nucleic-acid sensing TLRs such as TLR7 and TLR9. The autoantibodies produced by activated B cells form immune complexes which subsequently stimulate cells of the innate immune system via both Fc receptors and nucleic acid sensing TLRs to produce type I interferons (IFNs) and other proinflammatory cytokines. These cytokines further enhance B cell activation and autoantibody production, resulting in a more robust and sustained inflammatory milieu. In addition, immune complexes deposit in tissues and promote inflammation and subsequent organ damage via both complement and Fc receptor dependent mechanisms. CD4+ T cells also are important for the production of autoantibodies and inflammatory responses in lupus (reviewed in21–24). In particular, T follicular helper (Tfh) cells and Th17 cells are elevated in SLE and can promote the development of autoimmune germinal centers and autoantibodies. Th17 cells also contribute to tissue inflammation and damage. While there are conflicting data regarding the role of Treg defects in lupus, it has been suggested that a shift in the Th17 to Treg balance in favor of Th17 cells plays a role in SLE pathogenesis.25 Alterations in both Th1 and Th2 cells and cytokines have also been reported in SLE, with both IFNγ (a Th1 cytokine)26 and autoreactive IgE (induced by Th2 cytokines)21 suggested to contribute to disease pathogenesis.
The central role of autoantibodies in the perpetuation of proinflammatory conditions and the damage to tissues in lupus highlights the importance of understanding how control of B cell development and differentiation is disrupted in this disease. Defects in both B cell tolerance and activation have been observed in SLE patients. A landmark study by Arbuckle et al27 showed that autoantibodies can be detected in SLE patients prior to clinical presentation, suggesting an intrinsic defect in B cell tolerance that contributes to the initiation of disease. Indeed, single cell expression cloning of antibodies from new emigrant (recently emerged from the bone marrow) and mature naïve B cells revealed that unlike healthy controls, SLE patients fail to eliminate autoreactive B cells at this developmental transition.28 Germinal center tolerance checkpoints are also breached; in SLE patients, autoreactive B cells are not excluded from germinal centers,29 and DNA-reactive B cells can emerge from germinal centers as a result of somatic hypermutation.30,31 Studies with mice congenic for various combinations of NZM2410-derived lupus susceptibility alleles have also shown that loss of adaptive immune tolerance conferred by the Sle1 allele is a critical initiating event in the development of autoimmune disease.32
Inappropriate B cell activation and differentiation in SLE patients is reflected by a dramatic increase in circulating plasmablasts, particularly during times of high disease activity.33–36 Repertoire analysis has indicated that these expanded antibody-secreting cells are polyclonal, and many are derived from naïve precursors.36 Interestingly, a partial plasmablast gene expression signature is observed in B cells from a subset of quiescent (inactive) lupus patients, even without a significant difference in peripheral blood B cell subsets in this group as measured by flow cytometry.37 Taken together, these observations suggest that some patients may have intrinsic changes in B cell signaling and resultant gene expression that predispose them to enhanced B cell terminal differentiation.
Genome-wide association studies (GWAS) have identified dozens of genes associated with risk of developing SLE, as reviewed extensively elsewhere.4,5,20,21 These genes fall into several functional categories, including each of the main pathways linked to SLE pathogenesis described above as well as the response of target organs to inflammation. Among these SLE risk alleles are numerous genes involved in B cell activation and differentiation, including the HLA-DR genes, BANK1, BLK, RASGRP3, PTPN22, IL21R, IL10, TLR7, UBE2L3, STAT4, TNFAIP3, FCGR2B, LYN, CSK, PRDM1, and ETS1. Here we review the role of one of these genes, the transcription factor Ets1, in B cell tolerance, plasma cell differentiation, and autoantibody production in lupus.
II. Human ETS1 genetic variants associated with lupus
The transcription factor Ets1 is highly expressed in human and mouse lymphocytes (B cells, T cells and natural killer cells).9–13 The genetic region encoding the human gene ETS1 has been implicated as a susceptibility locus in numerous autoimmune and inflammatory diseases (Table I). As early as 2000, polymorphisms in the 3′ UTR of the human ETS1 gene were associated with particular clinical phenotypes of lupus.38 As described in more detail in the sections below, a few years later, Ets1−/− mice were discovered to develop a lupus-like autoimmune disease,19 supporting a role for Ets1 in regulating immune tolerance to self-antigens. More recently, three independent genome-wide association studies in Chinese and Korean populations have identified genetic variants in and around ETS1 gene as increasing the lupus risk.6–8 These initial genome-wide association studies were later replicated in independent populations of Chinese39 and Malaysian40 origin. As indicated in Table 1, the SNPs associated with lupus in these particular studies all map near the 3′ end of the gene, either in the final intron, in the 3′UTR or downstream of the gene. Exome sequencing in healthy donors and lupus patients has identified a single nucleotide polymorphism (SNP rs34846069) in the final exon of the gene that is associated with lupus, although this SNP does not change the encoded amino acid (Asp440→Asp).41 This SNP may be in linkage disequilibrium with other genetic changes that promote lupus. In addition to lupus, SNPs in or near the ETS1 gene have also been identified as susceptibility alleles in many other autoimmune and inflammatory diseases (Table 1), including rheumatoid arthritis,42–47 psoriasis,48–50 multiple sclerosis,51,52 ankylosing spondylitis,15 uveitis,16 allergy,53 atopic dermatitis,54 and celiac disease.55,56
Table I.
Autoimmune or inflammatory disease-associated polymorphisms in or near the ETS1 gene
| Polymorphisms in or near ETS1 | Autoimmune or Inflammatory Disease | Reference(s) | Location1 |
|---|---|---|---|
| CA repeat polymorphisms | Lupus | Sullivan et al., 2000 | 3′ UTR |
| rs6590330 | Lupus | Han et al., 2009; He et al., 2010; Yang et al., 2010; Zhong et al., 2011; Leng et al., 2013; Wang et al., 2013; Lu et al., 2015 | Downstream of gene |
| rs10893872 | Lupus, Uveitis | Yang et al., 2010; Zhang et al., 2013; Wei et al., 2014 | Downstream of gene |
| rs4937333 | Lupus, Ankylosing spondylitis | Yang et al., 2010, Zhong et al., 2011; Shan et al., 2014 | 3′ UTR |
| rs7932088 | Lupus | Yang et al., 2010 | Downstream of gene |
| rs12223943 | Lupus | Yang et al., 2010 | Proximal promoter |
| rs6590343 | Rheumatoid arthritis | Freudenberg et al, 2011 | Upstream of the gene |
| rs61907765 | Celiac disease | Trynka et al., 2011 | 5′ UTR |
| rs4937362 | Rheumatoid arthritis | Okada et al, 2012 | Upstream of the gene |
| rs11221332 | Rheumatoid arthritis; Celiac disease | Chatzikyriakidou 2012; Dubois et al., 2010 | Intron I |
| rs3802826 | Psoriasis | Tsoi et al., 2012 | Upstream of gene |
| rs34846069 | Lupus | Davis et al, 2013 | Final exon (does not change amino acid sequence) |
| rs970924 | Allergy | Hinds et al., 2013 | Downstream of gene |
| rs1128334 | Lupus, Ankylosing spondylitis | Zhang et al., 2013, Lessard et al., 2015; Shan et al., 2014 | 3′ UTR |
| rs76404385 | Lupus | Molineros et al., 2014 | Intron VII |
| rs4936059 | Rheumatoid arthritis | Kim et al., 2014 | Upstream of the gene |
| rs1128334 | Ankylosing spondylitis | Shan et al., 2014 | 3′ UTR |
| rs73013527 | Rheumatoid arthritis | Okada et al, 2014; Chen et al., 2015 | Upstream of the gene |
| rs12576573 | Lupus | Lessard et al., 2015 | Downstream of gene |
| rs7941765 | Lupus | Bentham et al., 2015; Morris et al., 2016 | Upstream of gene |
| rs3809006 | Multiple sclerosis | Lill et al., 2015 | Upstream of gene |
| rs4520607 | Psoriasis | Stuart et al., 2015 | Upstream of gene |
| rs6590334 | Psoriasis | Yin et al., 2015 | Upstream of gene |
| rs7933433 | Psoriasis | Yin et al., 2015 | Downstream of gene |
| rs4936044 | Psoriasis | Yin et al., 2015 | Downstream of gene |
| rs55974252 | Psoriasis | Yin et al., 2015 | Downstream of gene |
| rs573624 | Psoriasis | Yin et al., 2015 | Downstream of gene |
| rs7127307 | Atopic dermatitis | Paternoster et al., 2015 | Downstream of gene |
| rs61432431 | Lupus | Morris et al., 2016 | Downstream of gene |
Introns are labeled from the first (I) to the last (VII) intron of the major isoform of Ets1
The association of ETS1 with lupus in European populations is less well-replicated than it is in Asian populations. In 2013, a study showed that one of the SNPs in ETS1 (rs6590330) that had been identified in Asian lupus populations was also associated with lupus in people of European ancestry, although it did not reach the statistical threshold of genome wide significance (p<5×10−8).57 Another study with European lupus patients demonstrated that a different SNP in the ETS1 gene (rs7941765, located about 100 kb upstream of the gene) was associated with lupus susceptibility.58 A meta-analysis of GWAS studies of Chinese and European lupus patients confirmed this association of SNP rs7941765 with lupus susceptibility in European populations and the same SNP was also associated weakly with lupus in Asian patients.59 Another SNP (rs61432431) located downstream of ETS1 was associated with lupus susceptibility in both European and Asian cohorts, but the p value was more significant in the Asian cohort.59 Altogether, the data suggest that ETS1 is a lupus susceptibility locus in both European and Asian populations, but the causal variants might well be different.
Genetic variants (including SNPs) in ETS1 have also been associated with disease phenotypes in lupus and other autoimmune diseases. Particular allelic variants of ETS1 have been associated with a variety of clinical phenotypes in lupus, including early age of diagnosis,39, 60 levels of anti-DNA and antinuclear autoantibodies in the serum,17,39 serum IL-17 concentration,61 discoid and malar rash,38,39 photosensitivity,39 arthritis,39 serositis,39 vasculitis,38 hematologic disorders,39 immunologic disorders,39 and renal involvement.39 In rheumatoid arthritis, ETS1 SNPs have also been associated with particular clinical phenotypes including DAS28 (rheumatoid arthritis disease activity score 28) level and serum C-reactive protein level.45 In addition, SNPs in both ETS1 and the IL21 gene form epistatic interactions to cooperatively promote lupus susceptibility.62
Several studies have shown that Ets1 mRNA levels are reduced in PBMCs from autoimmune patients, suggesting that the effects of these genetic variants are to decrease Ets1 expression.7,14–17 Ets1 mRNA is also reduced in regulatory T cells (Tregs) from lupus patients and in bulk CD4+ T cells from multiple sclerosis patients.51,63 Indeed, using pyrosequencing, mRNA levels of Ets1 were measured in patients carrying one copy of a disease-associated allele and one copy of a protective allele in the 3′UTR of Ets1.7 Expression from the allele with the disease-associated SNP (rs1128334) was reduced as compared to the protective allele.
In order to understand how genetic variants in the human ETS1 locus might influence gene transcription, statistical analysis was used to map disease-associated SNPs and identify the most likely causal variants.64 One of these SNPs (rs6590330) showed differential binding in electrophoretic mobility shift assays when comparing the disease-associated allele to the protective allele. Further analysis showed that the disease-associated allele results in enhanced binding of the transcription factor Stat1, which is associated with reduced ETS1 transcription.64 Therefore, interferon signaling, which is prominent in lupus, may decrease the levels of Ets1 in patient PBMCs and promote autoimmunity.
One thing that should be kept in mind when evaluating the role of genetic variants in the ETS1 locus is the ETS1 is located in a head-to-head orientation with another Ets gene family member FLI1. The transcriptional start sites of ETS1 and FLI1 are located about 170 kb apart and some of the SNPs listed in Table I are between the ETS1 and FLI1 genes. Therefore, at least some of the ETS1 SNPs may affect FLI1 expression in addition to or instead of affecting ETS1 expression. In mice, Fli1 has opposite effects as that of Ets1 (i.e., over-expression of Fli1 promotes autoimmunity, while loss of Ets1 promotes autoimmunity).65–67 Accordingly, any changes to FLI1 expression as a result of SNPs in the ETS1/FLI1 locus may affect susceptibility to lupus.
III. Ets1 knockout mice develop autoimmune disease
Analysis of Ets1 knockout mice has demonstrated the critical role of Ets1 in the development and continued proper function of the immune system. Ets1 knockout mice exhibit a variety of lymphocyte abnormalities, including increased B and T cell activation and excessive B cell differentiation to plasma cells (discussed in detail in the section below). These immune abnormalities result in an autoimmune phenotype, reminiscent of human SLE. The phenotype of Ets1 knockout mice shares some similarities with mice deficient for BCR inhibitory signaling components, such as the kinase Lyn or phosphatase SHP1, although the autoimmunity is less severe in Ets1-deficient mice than in Lyn-deficient or SHP1-deficient mice.19,68–73
The phenotype of Ets1 knockout mice was first described by Bories and Muthusamy, using mutant alleles that lacked either the final two exons that encode the Ets DNA binding domain or the second and third exons that encode a conserved region known as the Pointed domain, respectively.74,75 The first allele is a complete null, while the second allele was also originally described as a null allele, but later shown to produce a very small amount of internally deleted Ets1 protein.19 However, both alleles result in similar phenotypic manifestations, including decreased cellularity of the thymus and lymph nodes, reduced T cell populations in the thymus, lymph node and spleen, and markedly increased populations of IgM-secreting plasma cells.74,75 These observations were later confirmed and expanded upon.
Marginal zone B cells are lacking in Ets1 knockout mice,19,76 while follicular B cells have an activated cell phenotype, with increased surface staining of MHC II, CD23, CD80 and CD86,19 and undergo increased differentiation to IgM- and IgG-secreting plasma cells.74,77 The dramatic expansion of the plasma cell population in Ets1 knockout mice correlates with increased serum levels of secreted immunoglobulin, even in the absence of any overt stimulus.18,74,78,79 The titer of circulating IgM in Ets1 deficient mice has been observed to be 4–10 times that of wild type counterparts and levels of IgG1 and IgE have also been shown to be elevated.18,79 Conversely, these mice have a significant decrease in circulating IgG2a, due to the role of Ets1 in positively regulating expression of T-bet, a transcription factor integral in class switching to the IgG2a isotype.79
Of particular interest are the antigen specificities of the circulating immunoglobulin. The serum of Ets1 knockout mice has high titers of autoantibodies of both IgM and IgG isotypes that bind to double-stranded DNA and histones.18,19 Notably, anti-nuclear autoantibodies (including those that bind to DNA and histones) are considered hallmarks of human SLE and correlate with disease activity and severity in SLE patients.80,81 Antibodies against cardiolipin, another autoantigen frequently associated with SLE, are also increased in Ets1 knockout mice.19 Furthermore, these mice also have high titers of rheumatoid factor autoantibodies targeting IgG,19 which are found in about 10% of lupus patients,82 but are more commonly associated with rheumatoid arthritis. Ets1-deficient mice also develop autoantibodies against myelin basic protein,19 an antigen that is frequently targeted in multiple sclerosis,83,84 but is not characteristic of human lupus. The similarities of the serological autoantibody compositions of human autoimmune disease patients and Ets1 knockout mice suggest that the mice are a suitable model for understanding aspects of human autoimmune disease pathogenesis.
Enhanced B cell activation in Ets1−/− mice and excessive secretion of antibodies could be a defect intrinsic to B cells, but could alternatively be reflective of defects in the T cell compartment. Adoptive transfer of Ets1-deficient B cells to wild-type recipients or wild-type B cells to Ets1 knockout recipients demonstrated that the activated B cell phenotype is heavily influenced by its environment.19 However, mixed bone marrow chimeras generated with allotype-labeled wildtype and Ets1 deficient fetal liver cells showed that the Ets1-deficient B cells give rise to increased numbers of plasma cells as compared with wild-type B cells developing in the same host.85 Therefore, there are likely both B cell-intrinsic and B cell-extrinsic functions of Ets1 that together promote B cell activation and autoantibody secretion. These will be described in detail below.
Mice deficient for Ets1 also display autoimmune-related organ pathology. The spleens are often enlarged in these mice compared to wild-type counterparts.18 There are infiltrates of lymphocytes in the lung, liver and other tissues and these infiltrates worsen as the mice age (19 and unpublished data). In the kidneys, there is extensive deposition of IgG and IgM immune complexes,19 reflective of a classic renal pathology associated with SLE. Although these immune complexes effectively activate complement and C3 is deposited in the glomeruli, Ets1−/− mice display weak proteinuria, suggesting that kidney function is largely preserved.19 Despite the weak proteinuria, Ets1−/− mice have a shortened lifespan (median 18 months) in comparison with wild-type littermates (median 28 months) (unpublished data), presumably due to organ damage caused by autoimmune disease. Overall, the autoimmune disease that develops in Ets1−/− mice shares many features with human lupus (activated B cells and T cells, increased numbers of plasma cells and high titers of autoantibodies, deposition of immune complexes in the kidney, infiltration of lymphocytes into target organs and a shortened lifespan). As described above, reductions in Ets1 levels are found in human lupus leukocytes and are associated with disease pathogenesis. Below we review studies of Ets1 function in B and T cells with an emphasis on their relationship to autoimmune disease development and progression.
IV. Ets1 maintains B cell quiescence and preserves peripheral tolerance
Ets1 levels are high in quiescent mouse B cells, but are downregulated upon antigen stimulation at both the mRNA and protein levels prior to differentiation.85 In mice lacking Ets1, B cells are hyper-activated as shown by increased expression of activation markers,19 increased isotype-switching to IgG1 and IgE18,79 and increased propensity to terminally differentiate into plasma cells secreting IgM and IgG.19,74,77,78 These observations suggest that Ets1 may play a role in retaining B cells in a resting state.
The in vitro behavior of Ets1-deficient B cells reflects the more activated phenotype observed in vivo. Even when left unstimulated, B cells isolated from Ets1 knockouts undergo a low level of differentiation into plasma cells.19 Purified Ets1−/− B cells also undergo increased differentiation to plasma cells when cultured with TLR ligands such as oligonucleotides containing unmethylated CpG sequences (TLR9 ligand)19 or bacterial lipopolysaccharide (TLR4 ligand).85 This suggests that Ets1 may be particularly important in regulating TLR responses. Nguyen et al. also noted a significant increase in the secretion of IgM and IgG2b by Ets1−/− B cells in response to LPS, consistent with increased IgM and IgG-secreting plasma cells in these cultures.79 The enhanced in vitro differentiation and Ig secretion of Ets1−/− B cells can be suppressed by enforced expression of Ets1 via a retroviral construct.74,77,85,86 Taken together, it is clear that Ets1 expression is a critical block to plasma cell differentiation and it has B cell-intrinsic roles in regulating this process.
Appropriately maintaining the resting state of B cells is crucial, particularly for the autoreactive B cells that escape central tolerance checkpoints to persist in the periphery. Using BCR transgenic mice, we have demonstrated that peripheral B cell tolerance is compromised in the absence of Ets1, while central B cell tolerance mediated by clonal deletion is intact. In transgenic mice (Ets1 wild-type) that express a hen egg lysozyme (HEL)-specific BCR and membrane-bound HEL antigen, immature B cells undergo clonal deletion in the bone marrow in response to strong BCR crosslinking.87,88 B cells from Ets1−/− transgenic mice carrying the same HEL-specific BCR and membrane-bound HEL antigen also undergo clonal deletion.89 On the other hand, when mice carrying the HEL-specific BCR are crossed to mice carrying a soluble HEL transgene, B cells are not deleted, but rather move to the periphery and become anergic in response to constant antigen binding.90–92 In this transgenic background, Ets1−/− B cells develop certain properties of anergic B cells in that they express the expected surface marker phenotype and display expected defects in BCR signaling.89 However, these mice have elevated levels of HEL-specific antibody in circulation and increased number of Ig-secreting cells in their spleens89, indicating that despite the apparent anergic state of the B cell population, there is still a breach in self-tolerance and differentiation of B cells to antibody-secreting plasma cells.
Some B cells that recognize their ligands only with low affinity do not become anergic and instead mature similar to normal non-self-reactive B cells, a process termed clonal ignorance.93,94 Using BCR transgenic strains, we have also shown that clonal ignorance is compromised in Ets1-deficient B cells. Transgenic expression of a rheumatoid factor BCR (AM14) that has low affinity for its ligand (IgG2a of the “a” allotype) results in clonal ignorance in a wild-type background expressing autoantigen.93 However, when the same AM14 BCR transgene is crossed onto an Ets1-deficient background, the B cells are not tolerized, but rather become activated and differentiate into plasma cells.89 This evidence bolsters the idea that Ets1 prevents inappropriate B cell activation and differentiation. In its absence, peripheral tolerance is breached, and B cells spontaneously undergo terminal differentiation to Ig-secreting cells, accounting for the expanded plasma cell population and elevation in circulating autoantibodies in Ets1 knockout mice.
V. Mechanisms by which Ets1 limits plasma cell differentiation
The accumulation of autoantibodies and plasma cells in the absence of Ets1 suggests that it has an important function in limiting B cell terminal differentiation under normal circumstances. Indeed, Ets1 is expressed in resting B cells where it has several activities that could contribute to such a role. Despite the importance of Ets1 in B cell biology, relatively few target genes of Ets1 in B cells are known. To address this, we recently performed chromatin immunoprecipitation-sequencing (ChIP-seq) for Ets1 in primary mouse B cells and correlated it with gene expression changes found in Ets1−/− B cells as compared to wild-type B cells.95 Ets1 binds to ~10,000 sites in the B cell genome (representing ~9,000 target genes), including the promoters of many genes involved in B cell differentiation, function and activation.
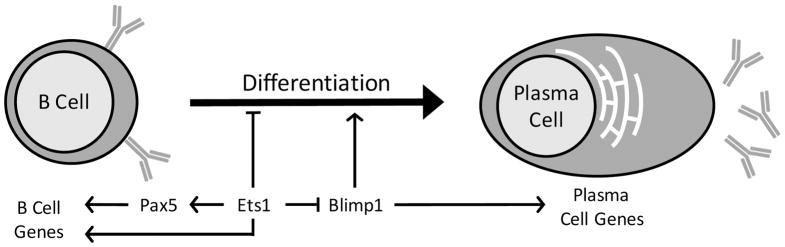
Pax-5, a key B cell identity factor (Figure 1), is among the genes harboring Ets1 binding sites (in both its promoter and the intron 5 B cell-specific enhancer). We had previously shown that Ets1 can transactivate the Pax5 promoter in transient transfection assays.86 Furthermore, retroviral transduction of Ets1 into primary B cells cultured with the TLR9 ligand CpG DNA was able to maintain Pax5 expression, whereas it would normally be downregulated during the differentiation process triggered by TLR signaling.77,86 Conserved arginine residues (R391 and R394) in the Ets domain of Ets1 are required for both Ets1 DNA binding and its ability to upregulate Pax5.77 Therefore, Ets1 might be an important transcription factor regulating Pax5 expression in B cells. However, RNA-sequencing shows that Pax5 mRNA is not decreased in Ets1−/− B cells.95 Together, these observations suggest a model in which Ets1 has the capability of stimulating Pax5 expression, but is not uniquely required for this process, at least in resting B cells. Even under conditions where Ets1 does not directly regulate Pax5 expression, it may stimulate the ability of Pax5 to function as a transcriptional regulator. ChIP-seq data for Ets1 and Pax5 show ~45% of the target sites of each factor are also bound by the other transcription factor. Ets1 and Pax5 are known to co-regulate the CD79a (mb-1) gene, which encodes Igα by forming a cooperative DNA binding complex.96 Ets1 and Pax-5 may interact similarly at the regulatory regions of other B cell-specific target genes to control their expression.
FIGURE 1. Transcriptional pathways by which Ets1 regulates B cell differentiation to plasma cells.
Ets1 functions to stimulate and/or maintain the expression of the transcription factor Pax5, which is crucial for regulating B cell identity genes. Pax5 and Ets1 may also cooperate to regulate B cell genes. Ets1 also represses both the expression and the function of the transcription factor Blimp1, which is necessary to promote the plasma cell fate. Ets1 may also promote the expression of additional target genes that regulate other aspects of B cell function, like formation of germinal centers, isotype-switching and memory B cell formation.
RNA-sequencing assays show that only about 500 genes are expressed differentially in B cells the absence of Ets1.95 Of these genes with altered expression, approximately half have a nearby Ets1 binding motif, indicating that they may be direct targets of Ets1. About two dozen of the potential direct Ets1 targets are genes that have been implicated in autoimmune disease susceptibility by genome-wide association studies, including Stat4 and Ptpn22.95 To determine whether Stat4 and Ptpn22 contribute to the role of Ets1 in regulating development of plasma cells, we restored their expression in Ets1−/− B cells using retroviral constructs.95 Unexpectedly, both Stat4 and Ptpn22 promoted the development of plasma cells, while Ets1 blocked this process. This indicates that Stat4 and Ptpn22 are not key target genes of Ets1 that modulate plasma cell formation. However, Ets1-dependent regulation of Stat4 and Ptpn22 might influence other aspects of B cell responses.
Another gene whose expression is altered in the absence of Ets1 is the Prdm1 gene which encodes Blimp1, a key transcription factor involved in promoting the plasma cell fate (Figure 1). Blimp1 is over-expressed by ~2-fold in sorted follicular B cells from Ets1−/− mice95 and is also over-expressed in Th1 cells that were cultured from Ets1−/− CD4+ T cell precursors.97 Furthermore, Ets1 binds to potential regulatory sequences localized in and near the Prdm1 gene.95,97 Because Blimp1 is crucial for B cell differentiation to plasma cells, it would be reasonable to think that over-expression of Blimp1 in Ets1−/− B cells could drive plasma cell formation. However, crossing Ets1 knockout mice to mice heterozygous for a null allele of Blimp1 (which reduces Blimp1 levels in B cells to those found in wild-type B cells) does not reverse the excess production of plasma cells caused by loss of Ets1.95 This observation indicates that the excess plasma cell phenotype of Ets1 knockout mice cannot be simply explained by over-expression of Blimp1 in the B cells.
We have found that Ets1 also functions to regulate B cell differentiation in a non-DNA-dependent fashion. In addition to regulating the Prdm1 gene, Ets1 also binds to and inhibits the function of the product of this gene, Blimp1.77,86 This is dependent on a direct protein/protein interaction between Ets1 and Blimp1, which prevents Blimp1 from binding to its DNA target sequences and thereby inhibits its activity.77,86 Normally during plasma cell formation, Ets1 is downregulated, allowing Blimp1 to function properly and promote the plasma cell phenotype. This downregulation of Ets1 is important, since retrovirally-driven forced expression of Ets1 leads to a block in plasma cell generation in response to TLR stimulation.77,85,86 We have mapped the domains of Ets1 required for interaction with Blimp1 and found that optimal interaction of Ets1 with Blimp1 requires a large fraction of the Ets1 protein, including the Ets DNA binding domain, the N terminus, the acidic transactivation domain, and the Pointed domain,77,86 suggesting the overall 3-dimensional structure of the Ets1 protein may be important for this process. Mutants of Ets1 that fail to interact with Blimp1 are also ineffective in blocking plasma cell formation in the retroviral assay.77,86
In summary then, we suggest a model in which Ets1-dependent control of Blimp1 activity via a protein/protein interaction is the most important influence in regulating plasma cell formation (Figure 1). Ets1-dependent regulation of a cohort of target genes such as Stat4 and Ptpn22 might influence other aspects of B cell activation and functional competence and the combined activities of these factors are important for Ets1’s role in preventing autoimmune disease.
VI. Positive and negative signaling events control expression of Ets1 in B cells
Consistent with its critical role in maintaining B cell tolerance and preventing autoimmunity, Ets1 expression in B cells is under tight control. In the absence of strong activating signals, Ets1 expression is maintained in B cells by inhibitory signaling pathways.85 A series of immunoreceptor tyrosine-based inhibitory motif (ITIM)-containing cell surface inhibitory receptors serve to keep B cell activation in check. These include FcγRIIb, PIR-B, CD72, CD22, and SiglecG. Phosphorylation of ITIM motifs in these receptors by the tyrosine kinase Lyn results in the recruitment and activation of the inhibitory phosphatases SHIP and SHP-1 (reviewed in98,99). To varying degrees, mice deficient in any of these inhibitory signaling molecules accumulate plasma cells, produce autoantibodies and develop lupus-like autoimmune disease; in many cases these effects have been shown to occur in a B cell-intrinsic manner. Loss of Lyn, SHP-1, or SHIP has a more profound effect than deficiency of individual receptors, suggesting that the receptors may be partially redundant for maintaining B cell tolerance.70,72,100–105
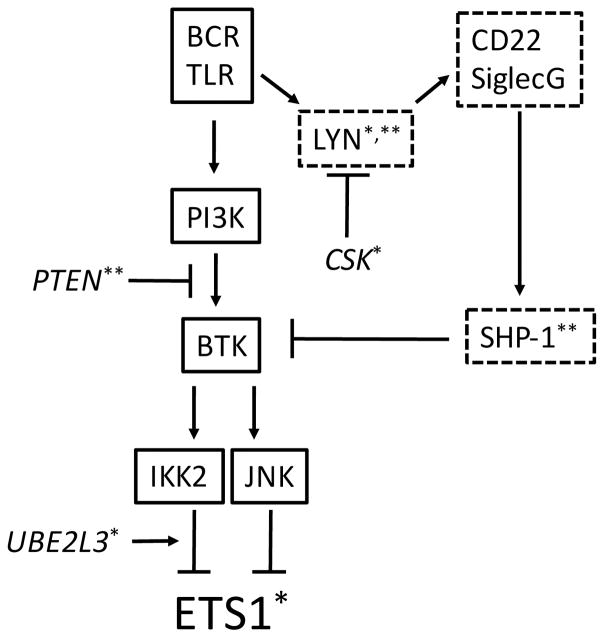
B cells from Lyn−/− mice demonstrate a dramatic reduction in the expression of both Ets1 mRNA and protein.85 This occurs prior to the accumulation of autoantibodies and the development of autoimmune disease manifestations85 and is independent of the inflammatory cytokine IL-6,106 strongly suggesting that a Lyn-dependent signaling pathway directly controls Ets1 expression in B cells. Indeed, a similar loss of Ets1 expression was observed in B cells with mutations in SHP-1, while SHIP-deficient B cells had only a mild decrease in Ets1.85 Loss of either CD22 or SiglecG resulted in a reduction of Ets1 levels in B cells, which was more profound in CD22 and SiglecG double knockouts.85 In contrast, individual loss of PIR-B, CD72, or FcγRIIB had no effect on Ets1 expression.85 Thus, an inhibitory pathway involving Lyn, SHP-1, CD22 and SiglecG normally maintains Ets1 levels in B cells (Figure 2).
FIGURE 2. Signaling pathways controlling Ets1 expression in B cells.
Signaling molecules outlined in solid lines are known to downregulate Ets1 expression, while those outlined in dotted lines have been shown to maintain Ets1 levels in B cells. Signaling molecules in italics have not been shown to control Ets1 levels, but are relevant to SLE and are likely to feed into pathways known to modulate Ets1 expression. * Polymorphisms in the gene encoding this molecule are associated with SLE. ** Expression is reduced in a subset of SLE B cells.
Upon antigen stimulation of B cells, positive signaling pathways prevail over Ets1-maintaining inhibitory signals and Ets1 mRNA and protein expression are downregulated.85 A combination of gain- and loss-of-function approaches and the use of small molecule inhibitors of BCR signaling components in both primary murine B cells and mouse B cell lines has revealed that BCR-induced Ets1 downregulation is mediated by PI3K, Btk, IKK2, and JNK, but not Akt, p38 or MEK (Figure 2).85 Treatment of B cells with the TLR ligands LPS (TLR4) or CpG DNA (TLR9) also downregulates Ets1 expression, and low level TLR9 signaling cooperates with low level BCR signaling to downregulate Ets1.85 Intriguingly, nucleic acid containing antigens, a common target of autoantibodies in lupus, can activate B cells via both the BCR and endosomal nucleic acid sensing TLRs,107–109 with dual signaling by the BCR and TLR7 being particularly efficient at driving plasma cell differentiation.110 Nucleic acid-specific B cells may thus be particularly efficient at downregulating Ets1 and differentiating into autoantibody-secreting plasma cells even in response to low levels of antigen. In contrast, stimuli that mimic T cell help (anti-CD40, IL-4, IL-21), promote B cell survival (BAFF), or would be encountered in an inflammatory environment (IL-6) have no effect on Ets1 levels.85 The inability of T cell-derived signals to downregulate Ets1 may prevent the inappropriate differentiation of bystander B cells activated in a non-cognate manner.
As described above, transduction of TLR-stimulated primary murine B cells with a retrovirus expressing Ets1 prevents differentiation into plasma cells and secretion of antibody.77,86 Similarly, enforced Ets1 expression ameliorates the enhanced terminal differentiation of Lyn-deficient or SHP-1-deficient B cells that occurs in response to TLR engagement.85 Thus, Ets1 downregulation is likely required for plasma cell differentiation during normal humoral immune responses, and inappropriate loss of Ets1 expression likely contributes to the unrestrained accumulation of autoreactive antibody-secreting cells that occurs in mice with impaired inhibitory signaling via the Lyn/SHP-1 pathway. The development of systems to prevent Ets1 downregulation in vivo at various times during immunization protocols or during the development of autoimmune disease is ongoing and will formally test this hypothesis.
Other types of genetic approaches have confirmed that a strict balance of the Btk-dependent activating signals and Lyn-mediated inhibitory signals that converge on Ets1 is required to maintain normal steady-state plasma cell numbers. First, mice heterozygous for both Lyn and Ets1 have increased IgM autoantibodies compared to Lyn+/− or Ets1+/− mice alone, although they do not develop full blown autoimmune disease.106 This indicates that Lyn and Ets1 do indeed work together in a common signaling pathway to limit B cell differentiation and that partial disruption of this pathway is sufficient for an initial break in B cell tolerance. Second, the excessive downregulation of Ets1 in the absence of inhibitory signals depends on Btk. Lyn−/−Btklo mice, which express a reduced level of Btk, demonstrate normal levels of Ets1 in their B cells85 and do not accumulate plasma cells or autoantibodies.111,112 Finally, Btk signaling to Ets1 also controls steady-state plasma cell numbers when Lyn-dependent inhibitory signaling pathways are intact. Btk−/− mice demonstrate reduced splenic IgM-secreting cells and low serum IgM levels; this defect is normalized in the absence of Ets1.106 These observations suggest that manipulations that shift the balance between Ets1-downregulating activating signals and Ets1-maintaining inhibitory signals may be useful therapeutic approaches to promote or dampen antibody responses as desired.
VII. Ets1 functions in CD4+ T cells
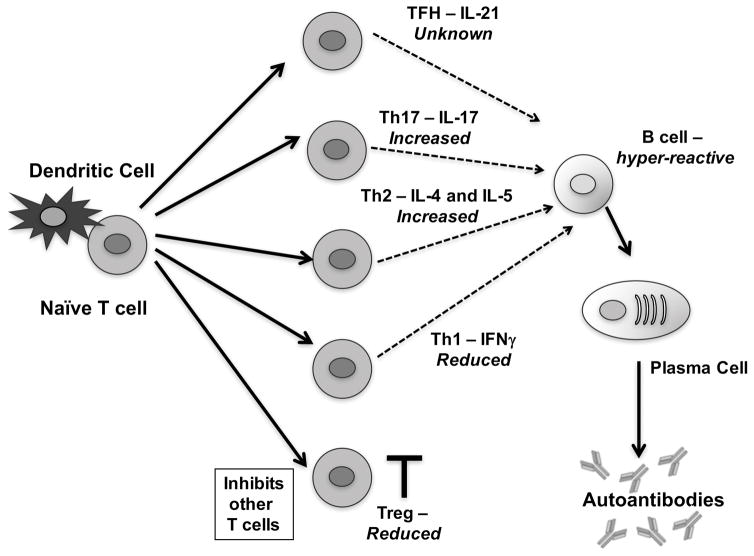
In addition to its roles in B cells, Ets1 also regulates the function of T cells. Of particular interest is the function of Ets1 in the CD4+ T cell subset, as these cells are important in lupus pathogenesis and function in part by regulating the responses of autoreactive B cells. Ets1 is highly expressed in human and mouse T cells including CD4+ T cells.9,113–116 Loss of Ets1 in mice results in a variety of aberrations in CD4 T cell differentiation, as we have described in previous reviews.117,118 Here we briefly describe the major known alterations in the CD4 lineage and discuss how they may be relevant in lupus pathogenesis (summarized in Figure 3).
FIGURE 3. T cell aberrations in Ets1-deficient mice.
Schematic of T cell differentiation with major subsets that have been implicated in lupus shown. Alterations in T cell subsets in Ets1-deficient mice are indicated in italics. Naïve autoreactive T cells interact with antigen-presenting dendritic cells and subsequently differentiate into various T cell subsets, depending on the cytokine environment in which they become activated. T cells can differentiate into T follicular helper (TFH) cells, T helper 17 (Th17) cells, T helper 2 (Th2), T helper 1 (Th1) or regulatory T cells (Treg). Tregs function to inhibit the other T cell subsets and limit autoimmune disease. Tregs are reduced in Ets1-deficient mice. The other T cell subsets produce various cytokines (as shown in the figure) and can interact with B cells to promote proliferation, germinal center formation, isotype-switching, affinity maturation and plasma cell formation. Ets1-deficiency results in skewing towards Th17 and Th2 cytokines and away from Th1 cytokines, while the effect on TFH cells is unknown. Depending on the type of T cells that interact in vivo with B cells, differences in B cell outcomes are possible. B cells from Ets1 knockout mice are also hyper-responsive and have an intrinsic propensity to differentiate into plasma cells.
A. Th1 Cells
Ets1 was first implicated in the regulating the differentiation of and cytokine production by Th1 and Th2 subsets of CD4+ T cells 114. CD4+ T cells isolated from Ets1−/− mice and cultured in vitro under conditions that promote Th1 differentiation showed greatly reduced production of IFNγ.114 IFNγ promotes lupus development and pathogenesis in multiple mouse models of the disease.119–130 IFNγ levels are increased in human patients with lupus and are correlated with more severe disease.131–136 Furthermore, a SNP in the IFNγ gene has been shown to be associated with lupus susceptibility.137 Clearly Ets1-deficiency leads to a loss of self-tolerance in mice regardless of reduced IFNγ production by Th1 cells. Perhaps, as described below, the overall balance of T cell cytokines remains sufficiently inflammatory despite a reduction in Th1 responses in the absence of Ets1. Alternatively, cells other than Th1 cells may be an important source of IFNγ in the context of reduced or absent Ets1 expression. However, the fact that Ets1−/− CD4+ T cells produce reduced amounts of IFNγ may result in less tissue damage in Ets1−/− mice. Ets1−/− mice develop high titers of autoantibodies with immune complexes deposit in the kidney,18,19 but proteinuria in this strain is weak.19 This might be due to reduced IFNγ-mediated kidney damage, since IFNγ is required for nephritis and impaired kidney function in NZB/W and MRL/lpr lupus-prone mice.119,129,138
B. Th2 Cells
Ets1−/− CD4+ T cells also produce less IL-4 when cultured under Th2 conditions.114 Further analysis indicated that secretion of the Th2 cytokines IL-5 and IL-13 was also reduced in Ets1−/− CD4+ T cells.139 The IL-4, IL-5 and IL-13 genes are clustered in the genome and are coordinately regulated.140,141 Ets1 binds to several sites within this Th2 locus to stimulate expression of the Th2 cytokines.139 Although IL-4, IL-5 and IL-13 were all reduced in Ets1−/− CD4+ cells cultured under Th2 conditions, contradictory results were obtained when assessing IL-4, IL-5 and IL-13 levels in freshly-isolated CD4+ T cells from the spleens of Ets1−/− mice, where each of these cytokines was over-produced rather than reduced in levels.18
IL-4 and IL-5 are implicated in promoting B cell responses such as isotype switching and plasma cell formation. IL-4 contributes to disease pathogenesis in NZB/W, MRL/lpr and NZM2410 lupus-prone mice,127,142–144 but is not required for lupus in the BXSB mouse strain.145 Recently, IL-4 was shown to also promote autoimmunity in Lyn-deficient mice.146 IL-4 is elevated in some human SLE patients147,148 and SNPs in the IL-4 locus are associated with increased susceptibility to lupus.149,150 In keeping with increased levels of Th2 cytokines in vivo in Ets1−/− mice, B cells from these animals show increased isotype-switching to IgG1 and IgE and increased numbers of plasma cells (18,77 and unpublished data). Although it is not yet clear whether the IgG1 or IgE produced by Ets1−/− mice is pathogenic, it is interesting to note that pathogenic IgE autoantibodies have recently been described in Lyn knockout mice that share many similar phenotypic features with Ets1 knockout mice (see above).146 IgE autoantibodies are also found in another mouse model of lupus, Fcgr2b−/−Yaa mice,151 and recently IgE autoantibodies have been detected in human SLE patients and are associated with more severe disease.152
C. IL-10
Ets1−/− CD4+ T cells cultured under either Th1 or Th2 conditions over-produce IL-10.114,153 IL-10 suppresses autoimmune symptoms in MRL/lpr and Sle1.2.3 lupus-prone mice.154,155 However, in NZB/W mice, anti-IL-10 antibody therapy results in reduced rather than increased disease symptoms.156 IL-10 stimulates human B cell proliferation and antibody secretion157,158 and is elevated in human patients with SLE.159–163 SNPs in the human IL-10 gene promoter are associated with lupus susceptibility.164,165 The over-production of IL-10 in Ets1-deficient mice might contribute to their autoimmune phenotype.
D. Th17 Cells
IL-17 has been extensively implicated as a pathogenic factor in multiple autoimmune diseases.166,167 Ets1-deficient CD4+ T cells cultured under Th17-skewing conditions or without polarizing cytokines produce increased levels of IL-17.115,168 IL-17 mRNA levels are elevated in freshly-isolated lung tissue from Ets1−/− mice, consistent with an elevation of Th17 cells in vivo as well.115 Viral-driven over-expression of Ets1 can also block development of IL-17 secreting cells from naïve wild-type precursors.115 The presence of two lupus-associated SNPs in the human ETS1 locus (rs10893872 or rs1128334) has been shown to correlate with the serum level of IL-17.61 This is consistent with the fact that SNP rs1128334 has been shown to decrease Ets1 mRNA levels.7 In Th17 cells, the microRNA miR155 targets Ets1 and the absence of miR155 leads to increased Ets1 protein in Th17 cells.169 High levels of Ets1 in Th17 cells lacking miR155 inhibits their function, because knocking down Ets1 in these cells leads to improved expression of typical Th17 transcripts such as IL-17A, IL-22 and IL23R.169
Serum levels of IL-17 are elevated in several autoimmune prone mouse strains170–172 and IL-17 plays important roles in the pathogenesis of lupus in BXD2 and Fcgr2b−/− mice172,173 and in pristane-induced lupus.174 Furthermore, IL-17 deficient mice are resistant to the induction of lupus nephritis caused by the injection of DNA from concanavalin A-activated lymphocytes.170 However, IL-17 is not required for lupus nephritis in MRL/lpr or NZB/NZW mice.175 IL-17 is also elevated in the serum of human lupus patients.176–180 Some studies have shown a correlation between serum IL-17 levels and disease activity as measured by the Systemic Lupus Erythematosus disease activity index (SLEDAI),176,178 but other studies have failed to find this association.179,180 It is likely that increased IL-17 in Ets1−/− mice plays a role in the autoimmune disease pathogenesis.
E. Treg Cells
In addition to the defects described above, CD4+ T cells from Ets1−/− mice have also been shown to develop less efficiently into regulatory T cells, with a reduced percentage of CD4+CD25+FoxP3+ cells in the spleens, reduced levels of FoxP3 within those cells and reduced functional capacity in suppressing inflammation.18 The defect in Treg production from Ets1-deficient progenitors is cell-intrinsic and in vitro culture of Ets1-deficient naïve CD4+ T cells under Treg skewing conditions results in reduced Treg production.18 Finally, transferring wild-type Tregs into Ets1−/− mice reduces splenomegaly and reverses certain aberrations in B cells.18 Ets1 binds to FoxP3 gene regulatory sequences.18,181,182
In MRL/lpr and NZB/W lupus-prone mouse models there are reduced numbers and/or functionality of Tregs.183–187 In human lupus, the contribution of Tregs is confusing with some studies reporting reduced numbers or impaired function,188–191 but others finding no abnormalities192,193 or even increased numbers.194 Effector T cells in lupus patients may also be resistant to Treg-mediated suppression.193,195 The levels of Ets1 and FoxP3 are correlated in T cells isolated from lupus patients and patients with Hashimoto’s thyroiditis, with low Ets1 and low FoxP3 in patient samples as compared to normal controls.63,196
F. IL-2
IL-2 is produced mainly by T cells and functions to support T cell activation and also the survival of regulatory T cells. IL-2 also suppresses the development of inflammatory Th17 cells.197 Ets1−/− CD4+ T cells make less IL-2 when cultured under Th1 conditions.114 There is also reduced IL-2 production when the cells are cultured under Th2 conditions, but this is less obvious since CD4+ T cells make relatively lower amounts of IL-2 when cultured in Th2 promoting conditions.114 Reduced production of IL-2 by Ets1−/− T cells was shown to be due to a role for Ets1 in recruiting the transcription factor NFAT to the IL-2 promoter.97 On the other hand, elevated levels of Blimp1 in Ets1−/− CD4+ T cells do not appear to contribute to reduced IL-2 production, since reducing T-cell expressed Blimp1 by crossing Ets1−/− mice to mice with a CD4-specific deletion of the Prdm1 gene does not reverse the IL-2 defect.97 As described above, CD4+ T cells from Ets1 knockout mice develop more robustly into Th17 cells when cultured under appropriate conditions.115 This is in part due to reduced IL-2 production in the Ets1 knockout background and in part due to resistance of Ets1−/− CD4+ T cells to the effects of IL-2.115
Mice carrying the lpr mutation of Fas on a B6 background develop lupus and deletion of IL-2 from this strain results in reduced lupus development,198 despite the fact that IL-2 is required to support Treg survival. This is likely due to the role for IL-2 in promoting T cell activation and proliferation, which are reduced in these mice. While this study indicates that some IL-2 is required for lupus, the amount of IL-2 that T cells produce can determine whether or not those T cells are pathogenic. Similar to T cells from Ets1−/− mice, lupus patients’ T cells make less IL-2 than healthy controls.199,200 A recent early phase and short term trial of low dose IL-2 therapy in SLE patients showed a normalization of Th17 and T follicular helper (Tfh) cell to Treg ratios (fewer Th17 and Tfh and more Tregs post treatment) and the Tregs were more functional.201 This suggests that low levels of T-cell derived IL-2 in SLE contribute to a potentially pathogenic skewing of T cell subsets in a similar way that Ets1 deficiency does in mice.
Thus, multiple T cell defects in Ets1−/− mice have the potential to contribute to autoantibody production and autoimmune pathology. A comparison of B-cell and T-cell specific Ets1 knockout mice would be informative in delineating the relative importance of B-cell vs. T-cell intrinsic functions of Ets1 in the development of autoimmune disease.
VIII. Concluding remarks and future directions
As reviewed above, several lines of evidence suggest that Ets1 plays an important role in limiting autoantibody production in SLE patients. Polymorphisms in the Ets1 gene have been repeatedly identified as being associated with SLE and other autoimmune diseases, and Ets1 levels are reduced in PBMCs and Tregs from SLE patients. Mice deficient in Ets1 develop lupus-like autoimmunity, characterized by excessive plasma cell accumulation, autoantibody production against lupus associated auto-antigens, immune complex deposition in the kidneys, and infiltration of lymphocytes into several tissues. This is likely due, at least in part, to a critical role for Ets1 in maintaining B cell tolerance and preventing plasma cell differentiation by regulating the expression of a cohort of genes involved in B cell immune responses and inhibiting Blimp1 function in a B cell-intrinsic manner. Consistent with these roles of Ets1 in limiting autoimmunity, its expression is under tight control by the balance of activating and inhibitory signaling in B cells.
Despite the substantial evidence linking Ets1 to autoantibody production in mice and the association of Ets1 polymorphisms with SLE, little is known about Ets1 expression or function in B cells from SLE patients. Intriguingly, polymorphisms and signaling defects that affect inhibitory signaling in B cells are associated with SLE in humans, suggesting that control of Ets1 expression by these pathways might be altered in lupus patients (Figure 2). For example, Lyn expression is reduced and its subcellular localization is altered in B cells from a subset of SLE patients,202,203 and polymorphisms in LYN are associated with SLE.204 An SLE-associated polymorphism in CSK, a negative regulator of Lyn, results in reduced Lyn activity and increased B cell activation.205 Expression of mRNA from the PTPN6 gene, which encodes SHP-1, is decreased in some SLE B cells.206 Additional changes in lupus B cells that do not directly affect known Ets1-maintaining inhibitory pathways85 may also contribute to control of Ets1 expression (Figure 2). For example, reduced expression of PTEN, an inhibitor of PI3K signaling, has also been observed in SLE B cells,207 and PI3K mediates BCR-induced downregulation of Ets1.85 While events downstream of IKK2 that downregulate Ets1 have not been defined, it is likely that these include NFκB.85 Intriguingly, SLE associated polymorphisms in UBE2L3 result in elevated NFκB activity in B cells and an increase in plasmablasts and plasma cells.208 Thus, even in the absence of ETS1 risk alleles, Ets1 expression may be reduced in SLE B cells by other polymorphisms or defects, increasing the propensity of autoreactive B cells to differentiate and produce potentially pathogenic autoantibodies. The elevated autoantibodies in compound heterozygotes of Lyn and Ets1106 suggest that polymorphisms in more than one component of Ets1-regulating pathways may result in enhanced B cell defects in SLE. A subset of SLE patients have a particularly striking plasma cell phenotype;34,37 these individuals may be more likely to have disruptions in Ets1 or its regulators. Therapeutic strategies that promote signaling through Ets1-maintaining inhibitory pathways, or block signaling through Ets1-downregulating activating pathways, may reduce pathogenic autoantibodies in SLE patients.
Ets1 may also act in T cells to promote autoantibody production or contribute to the pathogenesis of SLE, perhaps by limiting IL-2 expression, promoting Th2 or Th17 differentiation, and/or inhibiting Treg differentiation. Numerous ITIM-containing inhibitory receptors exist on T cells, including CTLA-4 and PD-1, and can recruit SHP-1 to restrain T cell activation.209, 210 The T cell receptor (TCR) signaling pathway also involves molecules similar to that found in the BCR pathway, including Lck and Fyn (homologs of the Lyn tyrosine kinase) and Itk and Tek (homologs of Btk).211,212 In fact, TCR signaling is already known to downregulate Ets1 in T cells.9 These similarities suggest that corresponding pathways in T cells may regulate Ets1 levels in a fashion similar to that found in B cells. This would imply that positive signaling via the TCR and Itk/Tek would function to downregulate Ets1 in T cells, while inhibitory signaling via ITIM-containing receptors would maintain Ets1. In fact, aberrations in TCR signaling are found in SLE patients.213 Normal levels of Ets1 may be needed to maintain normal T cell functional differentiation. Further study will be required to better define the pathways in T cells that maintain Ets1 under unstimulated conditions and lead to its downregulation upon stimulation.
Acknowledgments
Anne Satterthwaite and Lee-Ann Garrett Sinha are supported by NIH Grant AI122720. Lee-Ann Garrett-Sinha is also supported by a Novel Research Grant from the Lupus Research Alliance. Anne Satterthwaite is a Southwestern Medical Foundation Scholar in Biomedical Research and holds the Peggy Chavellier Professorship for Arthritis Research and Treatment.
Abbreviations
- BCR
B cell receptor
- ChIP-seq
chromatin immunoprecipitation sequencing
- DAS28
rheumatoid arthritis disease activity score 28
- GWAS
genome-wide association studies
- HEL
hen egg lysozyme
- IFN
interferon
- Ig
immunoglobulin
- IL
interleukin
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- PBMCs
peripheral blood mononuclear cells
- SLE
Systemic Lupus Erythematosus
- SLEDAI
Systemic Lupus Erythematosus disease activity index
- SNP
single nucleotide polymorphism
- TCR
T cell receptor
- Tfh
T follicular helper
- Th
T helper
- TLR
Toll-like receptor
- Tregs
regulatory T cells
- UTR
untranslated region
References
- 1.Carter EE, Barr SG, Clarke AE. The global burden of SLE: prevalence, health disparities and socioeconomic impact. Nat Rev Rheumatol. 2016 Oct;12(10):605–20. doi: 10.1038/nrrheum.2016.137. [DOI] [PubMed] [Google Scholar]
- 2.Durcan L, Petri M. Why targeted therapies are necessary for systemic lupus erythematosus. Lupus. 2016 Sep;25(10):1070–9. doi: 10.1177/0961203316652489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DJ. The evolution of drug discovery in systemic lupus erythematosus. Nat Rev Rheumatol. 2015 Oct;11(10):616–20. doi: 10.1038/nrrheum.2015.86. [DOI] [PubMed] [Google Scholar]
- 4.Mohan C, Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat Rev Nephrol. 2015 Jun;11(6):329–41. doi: 10.1038/nrneph.2015.33. [DOI] [PubMed] [Google Scholar]
- 5.Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann Rheum Dis. 2013 Apr;72(Suppl 2):ii56–61. doi: 10.1136/annrheumdis-2012-202351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009 Oct 18;41(11):1234–7. doi: 10.1038/ng.472. Epub 2009/10/20. Eng. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, Leung AM, Li XP, Avihingsanon Y, Wong CM, Lee TL, Ho MH, Lee PP, Chang YK, Li PH, Li RJ, Zhang L, Wong WH, Ng IO, Lau CS, Sham PC, Lau YL. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genetics. 2010 Feb;6(2):e1000841. doi: 10.1371/journal.pgen.1000841. Epub 2010/02/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessard CJ, Sajuthi S, Zhao J, Kim K, Ice JA, Li H, Ainsworth H, Rasmussen A, Kelly JA, Marion M, Bang SY, Bin Joo Y, Choi J, Lee HS, Mo Kang Y, Suh CH, Tae Chung W, Lee SK, Choe JY, Cheol Shim S, Hee Oh J, Jin Kim Y, Han BG, Shen N, Siew Howe H, Wakeland EK, Li QZ, Wook Song Y, Gaffney PM, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Vyse TJ, Harley JB, Sivils KL, Bae SC, Langefeld CD, Tsao BP. Identification of a Systemic Lupus Erythematosus Risk Locus Spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol. 2016 May;68(5):1197–209. doi: 10.1002/art.39548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat NK, Thompson CB, Lindsten T, June CH, Fujiwara S, Koizumi S, Fisher RJ, Papas TS. Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: regulatory role for the protooncogene ETS1. Proceedings of the National Academy of Sciences of the United States of America. 1990 May;87(10):3723–7. doi: 10.1073/pnas.87.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grund EM, Spyropoulos DD, Watson DK, Muise-Helmericks RC. Interleukins 2 and 15 regulate Ets1 expression via ERK1/2 and MNK1 in human natural killer cells. J Biol Chem. 2005 Feb 11;280(6):4772–8. doi: 10.1074/jbc.M408356200. [DOI] [PubMed] [Google Scholar]
- 11.Overbeck BM, Martin-Subero JI, Ammerpohl O, Klapper W, Siebert R, Giefing M. ETS1 encoding a transcription factor involved in B-cell differentiation is recurrently deleted and down-regulated in classical Hodgkin’s lymphoma. Haematologica. 2012 Oct;97(10):1612–4. doi: 10.3324/haematol.2012.061770. Epub 2012/05/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacchi N, de Klein A, Showalter SD, Bigi G, Papas TS. High expression of ets-1 gene in human thymocytes and immature T leukemic cells. Leukemia. 1988 Jan;2(1):12–8. [PubMed] [Google Scholar]
- 13.Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, Oostendorp RA, Leung W. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood. 2014 Dec 18;124(26):3905–13. doi: 10.1182/blood-2014-06-582965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Sun LD, Lu WS, Hu WL, Gao JP, Cheng YL, Yu ZY, Yao S, He CF, Liu JL, Cui Y, Yang S. Expression analysis of ETS1 gene in peripheral blood mononuclear cells with systemic lupus erythematosus by real-time reverse transcription PCR. Chin Med J (Engl) 2010 Aug;123(16):2287–8. Epub 2010/09/08. eng. [PubMed] [Google Scholar]
- 15.Shan S, Dang J, Li J, Yang Z, Zhao H, Xin Q, Ma X, Liu Y, Bian X, Gong Y, Liu Q. ETS1 variants confer susceptibility to ankylosing spondylitis in Han Chinese. Arthritis Research & Therapy. 2014 Apr 4;16(2):R87. doi: 10.1186/ar4530. Epub 2014/04/09. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, Xiang Q, Zhou Y, Kijlstra A, Yang P. MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS ONE. 2014;9(3):e91199. doi: 10.1371/journal.pone.0091199. Epub 2014/03/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen Z, Xu L, Chen X, Xu W, Yin Z, Gao X, Xiong S. Autoantibody induction by DNA-containing immune complexes requires HMGB1 with the TLR2/microRNA-155 pathway. J Immunol. 2013 Jun 1;190(11):5411–22. doi: 10.4049/jimmunol.1203301. Epub 2013/04/26. eng. [DOI] [PubMed] [Google Scholar]
- 18.Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-Defranoux O, Bandeira A, Bories JC. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010 Sep 27;207(10):2113–25. doi: 10.1084/jem.20092153. Epub 2010/09/22. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int Immunol. 2005 Sep;17(9):1179–91. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012 Jun 06;18(6):871–82. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsokos GC, Lo MS, Costa Reis P, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016 Nov 22;12(12):716–30. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 22.Rother N, van der Vlag J. Disturbed T Cell Signaling and Altered Th17 and Regulatory T Cell Subsets in the Pathogenesis of Systemic Lupus Erythematosus. Front Immunol. 2015;6:610. doi: 10.3389/fimmu.2015.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comte D, Karampetsou MP, Tsokos GC. T cells as a therapeutic target in SLE. Lupus. 2015 Apr;24(4–5):351–63. doi: 10.1177/0961203314556139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gensous N, Schmitt N, Richez C, Ueno H, Blanco P. T follicular helper cells, interleukin-21 and systemic lupus erythematosus. Rheumatology (Oxford) 2016 Aug 07; doi: 10.1093/rheumatology/kew297. [DOI] [PubMed] [Google Scholar]
- 25.Alunno A, Bartoloni E, Bistoni O, Nocentini G, Ronchetti S, Caterbi S, Valentini V, Riccardi C, Gerli R. Balance between regulatory T and Th17 cells in systemic lupus erythematosus: the old and the new. Clin Dev Immunol. 2012;2012:823085. doi: 10.1155/2012/823085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard KM, Cauvi DM, Toomey CB, Morris KV, Kono DH. Interferon-gamma and systemic autoimmunity. Discov Med. 2013 Sep;16(87):123–31. [PMC free article] [PubMed] [Google Scholar]
- 27.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003 Oct 16;349(16):1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 28.Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005 Mar 07;201(5):703–11. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cappione A, 3rd, Anolik JH, Pugh-Bernard A, Barnard J, Dutcher P, Silverman G, Sanz I. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005 Nov;115(11):3205–16. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, Gonzalez JB, Pascual V, Stichweh D, Wardemann H, Nussenzweig MC. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proceedings of the National Academy of Sciences of the United States of America. 2008 Jul 15;105(28):9727–32. doi: 10.1073/pnas.0803644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Jacobi AM, Wang T, Diamond B. Pathogenic autoantibodies in systemic lupus erythematosus are derived from both self-reactive and non-self-reactive B cells. Mol Med. 2008 Nov-Dec;14(11–12):675–81. doi: 10.2119/2008-00066.Zhang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, Wakeland EK. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proceedings of the National Academy of Sciences of the United States of America. 2000 Jun 06;97(12):6670–5. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arce E, Jackson DG, Gill MA, Bennett LB, Banchereau J, Pascual V. Increased frequency of pre-germinal center B cells and plasma cell precursors in the blood of children with systemic lupus erythematosus. J Immunol. 2001 Aug 15;167(4):2361–9. doi: 10.4049/jimmunol.167.4.2361. [DOI] [PubMed] [Google Scholar]
- 34.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika AM, Acs P, Turner J, Anguiano E, Vinod P, Kahn S, Obermoser G, Blankenship D, Wakeland E, Nassi L, Gotte A, Punaro M, Liu YJ, Banchereau J, Rossello-Urgell J, Wright T, Pascual V. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016 Apr 21;165(3):551–65. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odendahl M, Jacobi A, Hansen A, Feist E, Hiepe F, Burmester GR, Lipsky PE, Radbruch A, Dorner T. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. J Immunol. 2000 Nov 15;165(10):5970–9. doi: 10.4049/jimmunol.165.10.5970. [DOI] [PubMed] [Google Scholar]
- 36.Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, Schieferl S, Hom J, Jenks S, Feldman RJ, Mehr R, Wei C, Lee FE, Cheung WC, Rosenberg AF, Sanz I. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015 Jul;16(7):755–65. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garaud JC, Schickel JN, Blaison G, Knapp AM, Dembele D, Ruer-Laventie J, Korganow AS, Martin T, Soulas-Sprauel P, Pasquali JL. B cell signature during inactive systemic lupus is heterogeneous: toward a biological dissection of lupus. PLoS One. 2011;6(8):e23900. doi: 10.1371/journal.pone.0023900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan KE, Piliero LM, Dharia T, Goldman D, Petri MA. 3′ polymorphisms of ETS1 are associated with different clinical phenotypes in SLE. Hum Mutat. 2000;16(1):49–53. doi: 10.1002/1098-1004(200007)16:1<49::AID-HUMU9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 39.Zhong H, Li XL, Li M, Hao LX, Chen RW, Xiang K, Qi XB, Ma RZ, Su B. Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Research & Therapy. 2011;13(6):R186. doi: 10.1186/ar3514. Epub 2011/11/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molineros JE, Chua KH, Sun C, Lian LH, Motghare P, Kim-Howard X, Nath SK. Evaluation of SLE Susceptibility Genes in Malaysians. Autoimmune Diseases. 2014;2014:305436. doi: 10.1155/2014/305436. Epub 2014/04/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis NA, Lareau CA, White BC, Pandey A, Wiley G, Montgomery CG, Gaffney PM, McKinney BA. Encore: Genetic Association Interaction Network centrality pipeline and application to SLE exome data. Genet Epidemiol. 2013 Sep;37(6):614–21. doi: 10.1002/gepi.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freudenberg J, Lee HS, Han BG, Shin HD, Kang YM, Sung YK, Shim SC, Choi CB, Lee AT, Gregersen PK, Bae SC. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011 Apr;63(4):884–93. doi: 10.1002/art.30235. Epub 2011/04/01. eng. [DOI] [PubMed] [Google Scholar]
- 43.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Kawaguchi T, Stahl EA, Kurreeman FA, Nishida N, Ohmiya H, Myouzen K, Takahashi M, Sawada T, Nishioka Y, Yukioka M, Matsubara T, Wakitani S, Teshima R, Tohma S, Takasugi K, Shimada K, Murasawa A, Honjo S, Matsuo K, Tanaka H, Tajima K, Suzuki T, Iwamoto T, Kawamura Y, Tanii H, Okazaki Y, Sasaki T, Gregersen PK, Padyukov L, Worthington J, Siminovitch KA, Lathrop M, Taniguchi A, Takahashi A, Tokunaga K, Kubo M, Nakamura Y, Kamatani N, Mimori T, Plenge RM, Yamanaka H, Momohara S, Yamada R, Matsuda F, Yamamoto K. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012 May;44(5):511–6. doi: 10.1038/ng.2231. Epub 2012/03/27. eng. [DOI] [PubMed] [Google Scholar]
- 44.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. Altered sequence of the ETS1 transcription factor may predispose to rheumatoid arthritis susceptibility. Scandinavian Journal of Rheumatology. 2013;42(1):11–4. doi: 10.3109/03009742.2012.711367. Epub 2012/10/30. eng. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Huang Z, Yang B, Cai B, Su Z, Wang L. Association of E26 Transformation Specific Sequence 1 Variants with Rheumatoid Arthritis in Chinese Han Population. PLoS ONE. 2015;10(8):e0134875. doi: 10.1371/journal.pone.0134875. Epub 2015/08/05. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, Kim TH, Jun JB, Yoo DH, Kang YM, Kim SK, Suh CH, Shim SC, Lee SS, Lee J, Chung WT, Choe JY, Shin HD, Lee JY, Han BG, Nath SK, Eyre S, Bowes J, Pappas DA, Kremer JM, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Arlestig L, Okada Y, Diogo D, Liao KP, Karlson EW, Raychaudhuri S, Rantapaa-Dahlqvist S, Martin J, Klareskog L, Padyukov L, Gregersen PK, Worthington J, Greenberg JD, Plenge RM, Bae SC. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann Rheum Dis. 2015 Mar;74(3):e13. doi: 10.1136/annrheumdis-2013-204749. Epub 2014/02/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, Pappas DA, Jiang L, Yin J, Ye L, Su DF, Yang J, Xie G, Keystone E, Westra HJ, Esko T, Metspalu A, Zhou X, Gupta N, Mirel D, Stahl EA, Diogo D, Cui J, Liao K, Guo MH, Myouzen K, Kawaguchi T, Coenen MJ, van Riel PL, van de Laar MA, Guchelaar HJ, Huizinga TW, Dieude P, Mariette X, Bridges SL, Jr, Zhernakova A, Toes RE, Tak PP, Miceli-Richard C, Bang SY, Lee HS, Martin J, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Rantapaa-Dahlqvist S, Arlestig L, Choi HK, Kamatani Y, Galan P, Lathrop M, Eyre S, Bowes J, Barton A, de Vries N, Moreland LW, Criswell LA, Karlson EW, Taniguchi A, Yamada R, Kubo M, Liu JS, Bae SC, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Raychaudhuri S, Stranger BE, De Jager PL, Franke L, Visscher PM, Brown MA, Yamanaka H, Mimori T, Takahashi A, Xu H, Behrens TW, Siminovitch KA, Momohara S, Matsuda F, Yamamoto K, Plenge RM. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014 Feb 20;506(7488):376–81. doi: 10.1038/nature12873. Epub 2014/01/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, Kang HM, Allen MH, McManus R, Novelli G, Samuelsson L, Schalkwijk J, Stahle M, Burden AD, Smith CH, Cork MJ, Estivill X, Bowcock AM, Krueger GG, Weger W, Worthington J, Tazi-Ahnini R, Nestle FO, Hayday A, Hoffmann P, Winkelmann J, Wijmenga C, Langford C, Edkins S, Andrews R, Blackburn H, Strange A, Band G, Pearson RD, Vukcevic D, Spencer CC, Deloukas P, Mrowietz U, Schreiber S, Weidinger S, Koks S, Kingo K, Esko T, Metspalu A, Lim HW, Voorhees JJ, Weichenthal M, Wichmann HE, Chandran V, Rosen CF, Rahman P, Gladman DD, Griffiths CE, Reis A, Kere J, Nair RP, Franke A, Barker JN, Abecasis GR, Elder JT, Trembath RC. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012 Dec;44(12):1341–8. doi: 10.1038/ng.2467. Epub 2012/11/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart PE, Nair RP, Tsoi LC, Tejasvi T, Das S, Kang HM, Ellinghaus E, Chandran V, Callis-Duffin K, Ike R, Li Y, Wen X, Enerback C, Gudjonsson JE, Koks S, Kingo K, Esko T, Mrowietz U, Reis A, Wichmann HE, Gieger C, Hoffmann P, Nothen MM, Winkelmann J, Kunz M, Moreta EG, Mease PJ, Ritchlin CT, Bowcock AM, Krueger GG, Lim HW, Weidinger S, Weichenthal M, Voorhees JJ, Rahman P, Gregersen PK, Franke A, Gladman DD, Abecasis GR, Elder JT. Genome-wide Association Analysis of Psoriatic Arthritis and Cutaneous Psoriasis Reveals Differences in Their Genetic Architecture. Am J Hum Genet. 2015 Dec 03;97(6):816–36. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin X, Low HQ, Wang L, Li Y, Ellinghaus E, Han J, Estivill X, Sun L, Zuo X, Shen C, Zhu C, Zhang A, Sanchez F, Padyukov L, Catanese JJ, Krueger GG, Duffin KC, Mucha S, Weichenthal M, Weidinger S, Lieb W, Foo JN, Li Y, Sim K, Liany H, Irwan I, Teo Y, Theng CT, Gupta R, Bowcock A, De Jager PL, Qureshi AA, de Bakker PI, Seielstad M, Liao W, Stahle M, Franke A, Zhang X, Liu J. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat Commun. 2015 Apr 23;6:6916. doi: 10.1038/ncomms7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates T(H)-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009 Oct 18;10(12):1252–9. doi: 10.1038/ni.1798. Epub 2009/10/20. Eng. [DOI] [PubMed] [Google Scholar]
- 52.Lill CM, Luessi F, Alcina A, Sokolova EA, Ugidos N, de la Hera B, Guillot-Noel L, Malhotra S, Reinthaler E, Schjeide BM, Mescheriakova JY, Mashychev A, Wohlers I, Akkad DA, Aktas O, Alloza I, Antiguedad A, Arroyo R, Astobiza I, Blaschke P, Boyko AN, Buttmann M, Chan A, Dorner T, Epplen JT, Favorova OO, Fedetz M, Fernandez O, Garcia-Martinez A, Gerdes LA, Graetz C, Hartung HP, Hoffjan S, Izquierdo G, Korobko DS, Kroner A, Kubisch C, Kumpfel T, Leyva L, Lohse P, Malkova NA, Montalban X, Popova EV, Rieckmann P, Rozhdestvenskii AS, Schmied C, Smagina IV, Tsareva EY, Winkelmann A, Zettl UK, Binder H, Cournu-Rebeix I, Hintzen R, Zimprich A, Comabella M, Fontaine B, Urcelay E, Vandenbroeck K, Filipenko M, Matesanz F, Zipp F, Bertram L. Genome-wide significant association with seven novel multiple sclerosis risk loci. Journal of Medical Genetics. 2015 Dec;52(12):848–55. doi: 10.1136/jmedgenet-2015-103442. Epub 2015/10/18. eng. [DOI] [PubMed] [Google Scholar]
- 53.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U, Davey-Smith G, Timpson NJ, Tung JY. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013 Aug;45(8):907–11. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium EGaLEEE. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015 Dec;47(12):1449–56. doi: 10.1038/ng.3424. Epub 2015/10/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, de la Concha EG, de Almeida RC, Dias KR, van Diemen CC, Dubois PC, Duerr RH, Edkins S, Franke L, Fransen K, Gutierrez J, Heap GA, Hrdlickova B, Hunt S, Izurieta LP, Izzo V, Joosten LA, Langford C, Mazzilli MC, Mein CA, Midah V, Mitrovic M, Mora B, Morelli M, Nutland S, Nunez C, Onengut-Gumuscu S, Pearce K, Platteel M, Polanco I, Potter S, Ribes-Koninckx C, Ricano-Ponce I, Rich SS, Rybak A, Santiago JL, Senapati S, Sood A, Szajewska H, Troncone R, Varade J, Wallace C, Wolters VM, Zhernakova A, Thelma BK, Cukrowska B, Urcelay E, Bilbao JR, Mearin ML, Barisani D, Barrett JC, Plagnol V, Deloukas P, Wijmenga C, van Heel DA. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011 Dec;43(12):1193–201. doi: 10.1038/ng.998. Epub 2011/11/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Adany R, Aromaa A, Bardella MT, van den Berg LH, Bockett NA, de la Concha EG, Dema B, Fehrmann RS, Fernandez-Arquero M, Fiatal S, Grandone E, Green PM, Groen HJ, Gwilliam R, Houwen RH, Hunt SE, Kaukinen K, Kelleher D, Korponay-Szabo I, Kurppa K, MacMathuna P, Maki M, Mazzilli MC, McCann OT, Mearin ML, Mein CA, Mirza MM, Mistry V, Mora B, Morley KI, Mulder CJ, Murray JA, Nunez C, Oosterom E, Ophoff RA, Polanco I, Peltonen L, Platteel M, Rybak A, Salomaa V, Schweizer JJ, Sperandeo MP, Tack GJ, Turner G, Veldink JH, Verbeek WH, Weersma RK, Wolters VM, Urcelay E, Cukrowska B, Greco L, Neuhausen SL, McManus R, Barisani D, Deloukas P, Barrett JC, Saavalainen P, Wijmenga C, van Heel DA. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010 Apr;42(4):295–302. doi: 10.1038/ng.543. Epub 2010/03/02. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C, Ahlford A, Jarvinen TM, Nordmark G, Eloranta ML, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Truedsson L, Eriksson C, Rantapaa-Dahlqvist S, Sjowall C, Julkunen H, Criswell LA, Graham RR, Behrens TW, Kere J, Ronnblom L, Syvanen AC, Sandling JK. Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. European Journal of Human Genetics : EJHG. 2013 Sep;21(9):994–9. doi: 10.1038/ejhg.2012.277. Epub 2012/12/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW, Martin J, Fairfax BP, Knight JC, Chen L, Replogle J, Syvanen AC, Ronnblom L, Graham RR, Wither JE, Rioux JD, Alarcon-Riquelme ME, Vyse TJ. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015 Dec;47(12):1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, Chen L, Cunninghame Graham DS, Bentham J, Roberts AL, Chen R, Zuo X, Wang T, Wen L, Yang C, Liu L, Yang L, Li F, Huang Y, Yin X, Yang S, Ronnblom L, Furnrohr BG, Voll RE, Schett G, Costedoat-Chalumeau N, Gaffney PM, Lau YL, Zhang X, Yang W, Cui Y, Vyse TJ. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet. 2016 Aug;48(8):940–6. doi: 10.1038/ng.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, Quan C, Sun LD, Zheng HF, Zuo XB, Xu SX, Sheng YJ, Yao S, Hu WL, Li Y, Yu ZY, Yin XY, Zhang XJ, Cui Y, Yang S. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010 Sep;19(10):1181–6. doi: 10.1177/0961203310367918. Epub 2010/06/03. eng. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Zhang Y, Zhang L, Yang J, Ying D, Zeng S, Lee TL, Lau CS, Chan TM, Leung AM, Mok CC, Wong SN, Lee KW, Ho MH, Lee PP, Chung BH, Chong CY, Wong RW, Mok MY, Wong WH, Lau YL, Yang W. Epistatic Interaction between Genetic Variants in Susceptibility Gene ETS1 Correlates with IL-17 Levels in SLE Patients. Annals of Human Genetics. 2013 Apr 24; doi: 10.1111/ahg.12018. Epub 2013/04/26. Eng. [DOI] [PubMed] [Google Scholar]
- 62.Leng RX, Wang W, Cen H, Zhou M, Feng CC, Zhu Y, Yang XK, Yang M, Zhai Y, Li BZ, Wang XS, Li R, Chen GM, Chen H, Pan HF, Ye DQ. Gene-gene and gene-sex epistatic interactions of MiR146a, IRF5, IKZF1, ETS1 and IL21 in systemic lupus erythematosus. PLoS ONE. 2012;7(12):e51090. doi: 10.1371/journal.pone.0051090. Epub 2012/12/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang N, Li XP, Li XM, Wang GS, Tao JH, Pan HF, Fang X, Ma Q, Yu N. Expression of Ets-1 and FOXP3 mRNA in CD4CD25 T regulatory cells from patients with systemic lupus erythematosus. Clinical and Experimental Medicine. 2013 Nov 13; doi: 10.1007/s10238-013-0263-4. In press Epub 2013/11/14. Eng. [DOI] [PubMed] [Google Scholar]
- 64.Lu X, Zoller EE, Weirauch MT, Wu Z, Namjou B, Williams AH, Ziegler JT, Comeau ME, Marion MC, Glenn SB, Adler A, Shen N, Nath SK, Stevens AM, Freedman BI, Tsao BP, Jacob CO, Kamen DL, Brown EE, Gilkeson GS, Alarcon GS, Reveille JD, Anaya JM, James JA, Sivils KL, Criswell LA, Vila LM, Alarcon-Riquelme ME, Petri M, Scofield RH, Kimberly RP, Ramsey-Goldman R, Joo YB, Choi J, Bae SC, Boackle SA, Graham DC, Vyse TJ, Guthridge JM, Gaffney PM, Langefeld CD, Kelly JA, Greis KD, Kaufman KM, Harley JB, Kottyan LC. Lupus Risk Variant Increases pSTAT1 Binding and Decreases ETS1 Expression. Am J Hum Genet. 2015 May 7;96(5):731–9. doi: 10.1016/j.ajhg.2015.03.002. Epub 2015/04/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Molecular and Cellular Biology. 1995 Dec;15(12):6961–70. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, Gilkeson GS, Zhang XK. Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clin Exp Immunol. 2010 Nov;162(2):362–71. doi: 10.1111/j.1365-2249.2010.04245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, Watson DK, Gilkeson G. Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. J Immunol. 2004 Nov 15;173(10):6481–9. doi: 10.4049/jimmunol.173.10.6481. [DOI] [PubMed] [Google Scholar]
- 68.Chan VW, Meng F, Soriano P, DeFranco AL, Lowell CA. Characterization of the B lymphocyte populations in Lyn-deficient mice and the role of Lyn in signal initiation and down-regulation. Immunity. 1997 Jul;7(1):69–81. doi: 10.1016/s1074-7613(00)80511-7. [DOI] [PubMed] [Google Scholar]
- 69.Hibbs ML, Tarlinton DM, Armes J, Grail D, Hodgson G, Maglitto R, Stacker SA, Dunn AR. Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell. 1995 Oct 20;83(2):301–11. doi: 10.1016/0092-8674(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 70.Lamagna C, Hu Y, DeFranco AL, Lowell CA. B cell-specific loss of Lyn kinase leads to autoimmunity. J Immunol. 2014 Feb 01;192(3):919–28. doi: 10.4049/jimmunol.1301979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishizumi H, Taniuchi I, Yamanashi Y, Kitamura D, Ilic D, Mori S, Watanabe T, Yamamoto T. Impaired proliferation of peripheral B cells and indication of autoimmune disease in lyn-deficient mice. Immunity. 1995 Nov;3(5):549–60. doi: 10.1016/1074-7613(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 72.Pao LI, Lam KP, Henderson JM, Kutok JL, Alimzhanov M, Nitschke L, Thomas ML, Neel BG, Rajewsky K. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 2007 Jul;27(1):35–48. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 73.Tsui HW, Siminovitch KA, de Souza L, Tsui FW. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993 Jun;4(2):124–9. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- 74.Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995 Oct 19;377(6550):635–8. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 75.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995 Oct 19;377(6550):639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 76.Eyquem S, Chemin K, Fasseu M, Chopin M, Sigaux F, Cumano A, Bories JC. The development of early and mature B cells is impaired in mice deficient for the Ets-1 transcription factor. Eur J Immunol. 2004 Nov;34(11):3187–96. doi: 10.1002/eji.200425352. [DOI] [PubMed] [Google Scholar]
- 77.John S, Russell L, Chin SS, Luo W, Oshima R, Garrett-Sinha LA. Transcription factor ets1, but not the closely related factor ets2, inhibits antibody-secreting cell differentiation. Molecular and Cellular Biology. 2014 Feb;34(3):522–32. doi: 10.1128/MCB.00612-13. Epub 2013/11/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998 Oct;9(4):555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen HV, Mouly E, Chemin K, Luinaud R, Despres R, Fermand JP, Arnulf B, Bories JC. The Ets-1 transcription factor is required for Stat1-mediated T-bet expression and IgG2a class switching in mouse B cells. Blood. 2012 May 3;119(18):4174–81. doi: 10.1182/blood-2011-09-378182. Epub 2012/03/23. eng. [DOI] [PubMed] [Google Scholar]
- 80.Devey ME, Lee SR, Le Page S, Feldman R, Isenberg DA. Serial studies of the IgG subclass and functional affinity of DNA antibodies in systemic lupus erythematosus. J Autoimmun. 1988 Oct;1(5):483–94. doi: 10.1016/0896-8411(88)90069-8. [DOI] [PubMed] [Google Scholar]
- 81.Jacobi AM, Odendahl M, Reiter K, Bruns A, Burmester GR, Radbruch A, Valet G, Lipsky PE, Dorner T. Correlation between circulating CD27high plasma cells and disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2003 May;48(5):1332–42. doi: 10.1002/art.10949. [DOI] [PubMed] [Google Scholar]
- 82.Feldman D, Feldman D, Ginzler E, Kaplan D. Rheumatoid factor in patients with systemic lupus erythematosus. J Rheumatol. 1989 May;16(5):618–22. [PubMed] [Google Scholar]
- 83.Egg R, Reindl M, Deisenhammer F, Linington C, Berger T. Anti-MOG and anti-MBP antibody subclasses in multiple sclerosis. Mult Scler. 2001 Oct;7(5):285–9. doi: 10.1177/135245850100700503. [DOI] [PubMed] [Google Scholar]
- 84.Link H, Baig S, Olsson O, Jiang YP, Hojeberg B, Olsson T. Persistent anti-myelin basic protein IgG antibody response in multiple sclerosis cerebrospinal fluid. J Neuroimmunol. 1990 Aug;28(3):237–48. doi: 10.1016/0165-5728(90)90017-h. [DOI] [PubMed] [Google Scholar]
- 85.Luo W, Mayeux J, Gutierrez T, Russell L, Getahun A, Muller J, Tedder T, Parnes J, Rickert R, Nitschke L, Cambier J, Satterthwaite AB, Garrett-Sinha LA. A balance between B cell receptor and inhibitory receptor signaling controls plasma cell differentiation by maintaining optimal Ets1 levels. J Immunol. 2014 Jul 15;193(2):909–20. doi: 10.4049/jimmunol.1400666. Epub 2014/06/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.John SA, Clements JL, Russell LM, Garrett-Sinha LA. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J Biol Chem. 2008 Jan 11;283(2):951–62. doi: 10.1074/jbc.M705262200. Epub 2007/11/06. eng. [DOI] [PubMed] [Google Scholar]
- 87.Hartley SB, Cooke MP, Fulcher DA, Harris AW, Cory S, Basten A, Goodnow CC. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993 Feb 12;72(3):325–35. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 88.Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, Goodnow CC. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991 Oct 24;353(6346):765–9. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 89.Russell L, John S, Cullen J, Luo W, Shlomchik MJ, Garrett-Sinha LA. Requirement for Transcription Factor Ets1 in B Cell Tolerance to Self-Antigens. J Immunol. 2015 Oct 15;195(8):3574–83. doi: 10.4049/jimmunol.1500776. Epub 2015/09/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cooke MP, Heath AW, Shokat KM, Zeng Y, Finkelman FD, Linsley PS, Howard M, Goodnow CC. Immunoglobulin signal transduction guides the specificity of B cell-T cell interactions and is blocked in tolerant self-reactive B cells. J Exp Med. 1994 Feb 1;179(2):425–38. doi: 10.1084/jem.179.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988 Aug 25;334(6184):676–82. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 92.Schmidt KN, Cyster JG. Follicular exclusion and rapid elimination of hen egg lysozyme autoantigen-binding B cells are dependent on competitor B cells, but not on T cells. J Immunol. 1999 Jan 1;162(1):284–91. [PubMed] [Google Scholar]
- 93.Hannum LG, Ni D, Haberman AM, Weigert MG, Shlomchik MJ. A disease-related rheumatoid factor autoantibody is not tolerized in a normal mouse: implications for the origins of autoantibodies in autoimmune disease. J Exp Med. 1996 Oct 1;184(4):1269–78. doi: 10.1084/jem.184.4.1269. Epub 1996/10/01. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koenig-Marrony S, Soulas P, Julien S, Knapp AM, Garaud JC, Martin T, Pasquali JL. Natural autoreactive B cells in transgenic mice reproduce an apparent paradox to the clonal tolerance theory. J Immunol. 2001 Feb 1;166(3):1463–70. doi: 10.4049/jimmunol.166.3.1463. Epub 2001/02/13. eng. [DOI] [PubMed] [Google Scholar]
- 95.Saelee P, Kearly A, Nutt SL, Garrett-Sinha LA. Genome-wide Identification of Target Genes for the Key B Cell Transcription Factor Ets1. Front Immunol. 2017 doi: 10.3389/fimmu.2017.00383. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Molecular and Cellular Biology. 2003 Mar;23(6):1946–60. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsao HW, Tai TS, Tseng W, Chang HH, Grenningloh R, Miaw SC, Ho IC. Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proceedings of the National Academy of Sciences of the United States of America. 2013 Sep 24;110(39):15776–81. doi: 10.1073/pnas.1304343110. Epub 2013/09/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nitschke L. The role of CD22 and other inhibitory co-receptors in B-cell activation. Curr Opin Immunol. 2005 Jun;17(3):290–7. doi: 10.1016/j.coi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 99.Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity. 2005 Jan;22(1):9–18. doi: 10.1016/j.immuni.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 100.Bolland S, Ravetch JV. Spontaneous autoimmune disease in Fc(gamma)RIIB-deficient mice results from strain-specific epistasis. Immunity. 2000 Aug;13(2):277–85. doi: 10.1016/s1074-7613(00)00027-3. [DOI] [PubMed] [Google Scholar]
- 101.Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. CD22 × Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol. 2010 Apr 01;184(7):3618–27. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- 102.Kubo T, Uchida Y, Watanabe Y, Abe M, Nakamura A, Ono M, Akira S, Takai T. Augmented TLR9-induced Btk activation in PIR-B-deficient B-1 cells provokes excessive autoantibody production and autoimmunity. J Exp Med. 2009 Aug 31;206(9):1971–82. doi: 10.1084/jem.20082392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Q, Oliveira-Dos-Santos AJ, Mariathasan S, Bouchard D, Jones J, Sarao R, Kozieradzki I, Ohashi PS, Penninger JM, Dumont DJ. The inositol polyphosphate 5-phosphatase ship is a crucial negative regulator of B cell antigen receptor signaling. J Exp Med. 1998 Oct 05;188(7):1333–42. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Neill SK, Getahun A, Gauld SB, Merrell KT, Tamir I, Smith MJ, Dal Porto JM, Li QZ, Cambier JC. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity. 2011 Nov 23;35(5):746–56. doi: 10.1016/j.immuni.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu M, Hou R, Sato-Hayashizaki A, Man R, Zhu C, Wakabayashi C, Hirose S, Adachi T, Tsubata T. Cd72(c) is a modifier gene that regulates Fas(lpr)-induced autoimmune disease. J Immunol. 2013 Jun 01;190(11):5436–45. doi: 10.4049/jimmunol.1203576. [DOI] [PubMed] [Google Scholar]
- 106.Mayeux J, Skaug B, Luo W, Russell LM, John S, Saelee P, Abbasi H, Li QZ, Garrett-Sinha LA, Satterthwaite AB. Genetic Interaction between Lyn, Ets1, and Btk in the Control of Antibody Levels. J Immunol. 2015 Sep 01;195(5):1955–63. doi: 10.4049/jimmunol.1500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, Marshak-Rothstein A. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005 Nov 07;202(9):1171–7. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002 Apr 11;416(6881):603–7. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 109.Viglianti GA, Lau CM, Hanley TM, Miko BA, Shlomchik MJ, Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003 Dec;19(6):837–47. doi: 10.1016/s1074-7613(03)00323-6. [DOI] [PubMed] [Google Scholar]
- 110.Nundel K, Green NM, Shaffer AL, Moody KL, Busto P, Eilat D, Miyake K, Oropallo MA, Cancro MP, Marshak-Rothstein A. Cell-intrinsic expression of TLR9 in autoreactive B cells constrains BCR/TLR7-dependent responses. J Immunol. 2015 Mar 15;194(6):2504–12. doi: 10.4049/jimmunol.1402425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gutierrez T, Halcomb KE, Coughran AJ, Li QZ, Satterthwaite AB. Separate checkpoints regulate splenic plasma cell accumulation and IgG autoantibody production in Lyn-deficient mice. Eur J Immunol. 2010 Jul;40(7):1897–905. doi: 10.1002/eji.200940043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Whyburn LR, Halcomb KE, Contreras CM, Lowell CA, Witte ON, Satterthwaite AB. Reduced dosage of Bruton’s tyrosine kinase uncouples B cell hyperresponsiveness from autoimmunity in lyn−/− mice. J Immunol. 2003 Aug 15;171(4):1850–8. doi: 10.4049/jimmunol.171.4.1850. [DOI] [PubMed] [Google Scholar]
- 113.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126(14):3131–48. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 114.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005 Feb 21;201(4):615–26. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007 Nov 26;204(12):2825–35. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Q, Eppolito C, Odunsi K, Shrikant PA. Antigen-induced Erk1/2 activation regulates Ets-1-mediated sensitization of CD8+ T cells for IL-12 responses. J Leukoc Biol. 2010 Feb;87(2):257–63. doi: 10.1189/jlb.0409221. Epub 2009/10/22. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Russell L, Garrett-Sinha LA. Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine. 2010 Sep;51(3):217–26. doi: 10.1016/j.cyto.2010.03.006. Epub 2010/04/10. eng. [DOI] [PubMed] [Google Scholar]
- 118.Garrett-Sinha LA. Review of Ets1 structure, function, and roles in immunity. Cellular and molecular life sciences : CMLS. 2013 Jan 5;70:3375–90. doi: 10.1007/s00018-012-1243-7. Epub 2013/01/05. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balomenos D, Rumold R, Theofilopoulos AN. Interferon-gamma is required for lupus-like disease and lymphoaccumulation in MRL-lpr mice. J Clin Invest. 1998 Jan 15;101(2):364–71. doi: 10.1172/JCI750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haas C, Ryffel B, Le Hir M. IFN-gamma receptor deletion prevents autoantibody production and glomerulonephritis in lupus-prone (NZB × NZW)F1 mice. J Immunol. 1998 Apr 15;160(8):3713–8. [PubMed] [Google Scholar]
- 121.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012 Nov 16;37(5):880–92. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 122.Seery JP, Carroll JM, Cattell V, Watt FM. Antinuclear autoantibodies and lupus nephritis in transgenic mice expressing interferon gamma in the epidermis. J Exp Med. 1997 Nov 03;186(9):1451–9. doi: 10.1084/jem.186.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hasegawa K, Hayashi T, Maeda K. Promotion of lupus in NZB × NZWF1 mice by plasmids encoding interferon (IFN)-gamma but not by those encoding interleukin (IL)-4. J Comp Pathol. 2002 Jul;127(1):1–6. doi: 10.1053/jcpa.2002.0556. [DOI] [PubMed] [Google Scholar]
- 124.Richards HB, Satoh M, Jennette JC, Croker BP, Yoshida H, Reeves WH. Interferon-gamma is required for lupus nephritis in mice treated with the hydrocarbon oil pristane. Kidney Int. 2001 Dec;60(6):2173–80. doi: 10.1046/j.1523-1755.2001.00045.x. [DOI] [PubMed] [Google Scholar]
- 125.Lawson BR, Prud’homme GJ, Chang Y, Gardner HA, Kuan J, Kono DH, Theofilopoulos AN. Treatment of murine lupus with cDNA encoding IFN-gammaR/Fc. J Clin Invest. 2000 Jul;106(2):207–15. doi: 10.1172/JCI10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacob CO, van der Meide PH, McDevitt HO. In vivo treatment of (NZB X NZW)F1 lupus-like nephritis with monoclonal antibody to gamma interferon. J Exp Med. 1987 Sep 01;166(3):798–803. doi: 10.1084/jem.166.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Peng SL, Moslehi J, Craft J. Roles of interferon-gamma and interleukin-4 in murine lupus. J Clin Invest. 1997 Apr 15;99(8):1936–46. doi: 10.1172/JCI119361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schwarting A, Wada T, Kinoshita K, Tesch G, Kelley VR. IFN-gamma receptor signaling is essential for the initiation, acceleration, and destruction of autoimmune kidney disease in MRL-Fas(lpr) mice. J Immunol. 1998 Jul 01;161(1):494–503. [PubMed] [Google Scholar]
- 129.Haas C, Ryffel B, Le Hir M. IFN-gamma is essential for the development of autoimmune glomerulonephritis in MRL/Ipr mice. J Immunol. 1997 Jun 01;158(11):5484–91. [PubMed] [Google Scholar]
- 130.Takahashi S, Fossati L, Iwamoto M, Merino R, Motta R, Kobayakawa T, Izui S. Imbalance towards Th1 predominance is associated with acceleration of lupus-like autoimmune syndrome in MRL mice. J Clin Invest. 1996 Apr 01;97(7):1597–604. doi: 10.1172/JCI118584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979 Jul 05;301(1):5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 132.Csiszar A, Nagy G, Gergely P, Pozsonyi T, Pocsik E. Increased interferon-gamma (IFN-gamma), IL-10 and decreased IL-4 mRNA expression in peripheral blood mononuclear cells (PBMC) from patients with systemic lupus erythematosus (SLE) Clin Exp Immunol. 2000 Dec;122(3):464–70. doi: 10.1046/j.1365-2249.2000.01369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harigai M, Kawamoto M, Hara M, Kubota T, Kamatani N, Miyasaka N. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. J Immunol. 2008 Aug 01;181(3):2211–9. doi: 10.4049/jimmunol.181.3.2211. [DOI] [PubMed] [Google Scholar]
- 134.Funauchi M, Sugishima H, Minoda M, Horiuchi A. Serum level of interferon-gamma in autoimmune diseases. Tohoku J Exp Med. 1991 Aug;164(4):259–67. doi: 10.1620/tjem.164.259. [DOI] [PubMed] [Google Scholar]
- 135.Akahoshi M, Nakashima H, Tanaka Y, Kohsaka T, Nagano S, Ohgami E, Arinobu Y, Yamaoka K, Niiro H, Shinozaki M, Hirakata H, Horiuchi T, Otsuka T, Niho Y. Th1/Th2 balance of peripheral T helper cells in systemic lupus erythematosus. Arthritis Rheum. 1999 Aug;42(8):1644–8. doi: 10.1002/1529-0131(199908)42:8<1644::AID-ANR12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 136.Masutani K, Akahoshi M, Tsuruya K, Tokumoto M, Ninomiya T, Kohsaka T, Fukuda K, Kanai H, Nakashima H, Otsuka T, Hirakata H. Predominance of Th1 immune response in diffuse proliferative lupus nephritis. Arthritis Rheum. 2001 Sep;44(9):2097–106. doi: 10.1002/1529-0131(200109)44:9<2097::AID-ART360>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 137.Leng RX, Pan HF, Liu J, Yang XK, Zhang C, Tao SS, Wang DG, Li XM, Li XP, Yang W, Ye DQ. Evidence for genetic association of TBX21 and IFNG with systemic lupus erythematosus in a Chinese Han population. Sci Rep. 2016 Feb 26;6:22081. doi: 10.1038/srep22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ozmen L, Roman D, Fountoulakis M, Schmid G, Ryffel B, Garotta G. Experimental therapy of systemic lupus erythematosus: the treatment of NZB/W mice with mouse soluble interferon-gamma receptor inhibits the onset of glomerulonephritis. Eur J Immunol. 1995 Jan;25(1):6–12. doi: 10.1002/eji.1830250103. [DOI] [PubMed] [Google Scholar]
- 139.Strempel JM, Grenningloh R, Ho IC, Vercelli D. Phylogenetic and functional analysis identifies Ets-1 as a novel regulator of the Th2 cytokine gene locus. J Immunol. 2010 Feb 01;184(3):1309–16. doi: 10.4049/jimmunol.0804162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol. 2000 Sep 15;165(6):2982–6. doi: 10.4049/jimmunol.165.6.2982. [DOI] [PubMed] [Google Scholar]
- 141.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002 Sep 01;169(5):2498–506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 142.Nakajima A, Hirose S, Yagita H, Okumura K. Roles of IL-4 and IL-12 in the development of lupus in NZB/W F1 mice. J Immunol. 1997 Feb 01;158(3):1466–72. [PubMed] [Google Scholar]
- 143.Schorlemmer HU, Dickneite G, Kanzy EJ, Enssle KH. Modulation of the immunoglobulin dysregulation in GvH- and SLE-like diseases by the murine IL-4 receptor (IL-4-R) Inflamm Res. 1995 Aug;44(Suppl 2):S194–6. doi: 10.1007/BF01778328. [DOI] [PubMed] [Google Scholar]
- 144.Singh RR, Saxena V, Zang S, Li L, Finkelman FD, Witte DP, Jacob CO. Differential contribution of IL-4 and STAT6 vs STAT4 to the development of lupus nephritis. J Immunol. 2003 May 01;170(9):4818–25. doi: 10.4049/jimmunol.170.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kono DH, Balomenos D, Park MS, Theofilopoulos AN. Development of lupus in BXSB mice is independent of IL-4. J Immunol. 2000 Jan 01;164(1):38–42. doi: 10.4049/jimmunol.164.1.38. [DOI] [PubMed] [Google Scholar]
- 146.Charles N, Hardwick D, Daugas E, Illei GG, Rivera J. Basophils and the T helper 2 environment can promote the development of lupus nephritis. Nat Med. 2010 Jun;16(6):701–7. doi: 10.1038/nm.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Funauchi M, Ikoma S, Enomoto H, Horiuchi A. Decreased Th1-like and increased Th2-like cells in systemic lupus erythematosus. Scandinavian journal of rheumatology. 1998;27(3):219–24. doi: 10.1080/030097498440859. [DOI] [PubMed] [Google Scholar]
- 148.Wong CK, Ho CY, Li EK, Lam CW. Elevation of proinflammatory cytokine (IL-18, IL-17, IL-12) and Th2 cytokine (IL-4) concentrations in patients with systemic lupus erythematosus. Lupus. 2000;9(8):589–93. doi: 10.1191/096120300678828703. [DOI] [PubMed] [Google Scholar]
- 149.Yu HH, Liu PH, Lin YC, Chen WJ, Lee JH, Wang LC, Yang YH, Chiang BL. Interleukin 4 and STAT6 gene polymorphisms are associated with systemic lupus erythematosus in Chinese patients. Lupus. 2010 Sep;19(10):1219–28. doi: 10.1177/0961203310371152. [DOI] [PubMed] [Google Scholar]
- 150.Mahmoudi M, Tahghighi F, Ziaee V, Harsini S, Rezaei A, Soltani S, Sadr M, Moradinejad MH, Aghighi Y, Rezaei N. Interleukin-4 single nucleotide polymorphisms in juvenile systemic lupus erythematosus. Int J Immunogenet. 2014 Dec;41(6):512–7. doi: 10.1111/iji.12152. [DOI] [PubMed] [Google Scholar]
- 151.Dema B, Charles N, Pellefigues C, Ricks TK, Suzuki R, Jiang C, Scheffel J, Hasni S, Hoffman V, Jablonski M, Sacre K, Gobert D, Papo T, Daugas E, Crampton S, Bolland S, Rivera J. Immunoglobulin E plays an immunoregulatory role in lupus. J Exp Med. 2014 Oct 20;211(11):2159–68. doi: 10.1084/jem.20140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dema B, Pellefigues C, Hasni S, Gault N, Jiang C, Ricks TK, Bonelli MM, Scheffel J, Sacre K, Jablonski M, Gobert D, Papo T, Daugas E, Illei G, Charles N, Rivera J. Autoreactive IgE is prevalent in systemic lupus erythematosus and is associated with increased disease activity and nephritis. PLoS One. 2014;9(2):e90424. doi: 10.1371/journal.pone.0090424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee CG, Kwon HK, Sahoo A, Hwang W, So JS, Hwang JS, Chae CS, Kim GC, Kim JE, So HS, Hwang ES, Grenningloh R, Ho IC, Im SH. Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J Immunol. 2012 Mar 01;188(5):2244–53. doi: 10.4049/jimmunol.1101614. [DOI] [PubMed] [Google Scholar]
- 154.Yin Z, Bahtiyar G, Zhang N, Liu L, Zhu P, Robert ME, McNiff J, Madaio MP, Craft J. IL-10 regulates murine lupus. J Immunol. 2002 Aug 15;169(4):2148–55. doi: 10.4049/jimmunol.169.4.2148. [DOI] [PubMed] [Google Scholar]
- 155.Blenman KR, Duan B, Xu Z, Wan S, Atkinson MA, Flotte TR, Croker BP, Morel L. IL-10 regulation of lupus in the NZM2410 murine model. Lab Invest. 2006 Nov;86(11):1136–48. doi: 10.1038/labinvest.3700468. [DOI] [PubMed] [Google Scholar]
- 156.Ishida H, Muchamuel T, Sakaguchi S, Andrade S, Menon S, Howard M. Continuous administration of anti-interleukin 10 antibodies delays onset of autoimmunity in NZB/W F1 mice. J Exp Med. 1994 Jan 01;179(1):305–10. doi: 10.1084/jem.179.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rousset F, Garcia E, Defrance T, Peronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1992 Mar 01;89(5):1890–3. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rousset F, Peyrol S, Garcia E, Vezzio N, Andujar M, Grimaud JA, Banchereau J. Long-term cultured CD40-activated B lymphocytes differentiate into plasma cells in response to IL-10 but not IL-4. Int Immunol. 1995 Aug;7(8):1243–53. doi: 10.1093/intimm/7.8.1243. [DOI] [PubMed] [Google Scholar]
- 159.Grondal G, Gunnarsson I, Ronnelid J, Rogberg S, Klareskog L, Lundberg I. Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin Exp Rheumatol. 2000 Sep-Oct;18(5):565–70. [PubMed] [Google Scholar]
- 160.Llorente L, Richaud-Patin Y, Wijdenes J, Alcocer-Varela J, Maillot MC, Durand-Gasselin I, Fourrier BM, Galanaud P, Emilie D. Spontaneous production of interleukin-10 by B lymphocytes and monocytes in systemic lupus erythematosus. Eur Cytokine Netw. 1993 Nov-Dec;4(6):421–7. [PubMed] [Google Scholar]
- 161.Llorente L, Richaud-Patin Y, Fior R, Alcocer-Varela J, Wijdenes J, Fourrier BM, Galanaud P, Emilie D. In vivo production of interleukin-10 by non-T cells in rheumatoid arthritis, Sjogren’s syndrome, and systemic lupus erythematosus. A potential mechanism of B lymphocyte hyperactivity and autoimmunity. Arthritis Rheum. 1994 Nov;37(11):1647–55. doi: 10.1002/art.1780371114. [DOI] [PubMed] [Google Scholar]
- 162.Chun HY, Chung JW, Kim HA, Yun JM, Jeon JY, Ye YM, Kim SH, Park HS, Suh CH. Cytokine IL-6 and IL-10 as biomarkers in systemic lupus erythematosus. J Clin Immunol. 2007 Sep;27(5):461–6. doi: 10.1007/s10875-007-9104-0. [DOI] [PubMed] [Google Scholar]
- 163.Houssiau FA, Lefebvre C, Vanden Berghe M, Lambert M, Devogelaer JP, Renauld JC. Serum interleukin 10 titers in systemic lupus erythematosus reflect disease activity. Lupus. 1995 Oct;4(5):393–5. doi: 10.1177/096120339500400510. [DOI] [PubMed] [Google Scholar]
- 164.Liu P, Song J, Su H, Li L, Lu N, Yang R, Peng Z. IL-10 gene polymorphisms and susceptibility to systemic lupus erythematosus: a meta-analysis. PLoS One. 2013;8(7):e69547. doi: 10.1371/journal.pone.0069547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001 Mar 15;166(6):3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 166.Singh RP, Hasan S, Sharma S, Nagra S, Yamaguchi DT, Wong DT, Hahn BH, Hossain A. Th17 cells in inflammation and autoimmunity. Autoimmun Rev. 2014 Dec;13(12):1174–81. doi: 10.1016/j.autrev.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 167.Konya C, Paz Z, Apostolidis SA, Tsokos GC. Update on the role of Interleukin 17 in rheumatologic autoimmune diseases. Cytokine. 2015 Oct;75(2):207–15. doi: 10.1016/j.cyto.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 168.Nagaleekar VK, Diehl SA, Juncadella I, Charland C, Muthusamy N, Eaton S, Haynes L, Garrett-Sinha LA, Anguita J, Rincon M. IP3 receptor-mediated Ca2+ release in naive CD4 T cells dictates their cytokine program. J Immunol. 2008 Dec 15;181(12):8315–22. doi: 10.4049/jimmunol.181.12.8315. Epub 2008/12/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O’Connell RM. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013 Jun 15;190(12):5972–80. doi: 10.4049/jimmunol.1300351. Epub 2013/05/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wen Z, Xu L, Xu W, Yin Z, Gao X, Xiong S. Interleukin-17 expression positively correlates with disease severity of lupus nephritis by increasing anti-double-stranded DNA antibody production in a lupus model induced by activated lymphocyte derived DNA. PLoS ONE. 2013;8(3):e58161. doi: 10.1371/journal.pone.0058161. Epub 2013/03/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lech M, Weidenbusch M, Kulkarni OP, Ryu M, Darisipudi MN, Susanti HE, Mittruecker HW, Mak TW, Anders HJ. IRF4 deficiency abrogates lupus nephritis despite enhancing systemic cytokine production. J Am Soc Nephrol. 2011 Aug;22(8):1443–52. doi: 10.1681/ASN.2010121260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hsu HC, Yang P, Wang J, Wu Q, Myers R, Chen J, Yi J, Guentert T, Tousson A, Stanus AL, Le TV, Lorenz RG, Xu H, Kolls JK, Carter RH, Chaplin DD, Williams RW, Mountz JD. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008 Feb;9(2):166–75. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 173.Pisitkun P, Ha HL, Wang H, Claudio E, Tivy CC, Zhou H, Mayadas TN, Illei GG, Siebenlist U. Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis. Immunity. 2012 Dec 14;37(6):1104–15. doi: 10.1016/j.immuni.2012.08.014. Epub 2012/11/06. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Amarilyo G, Lourenco EV, Shi FD, La Cava A. IL-17 promotes murine lupus. J Immunol. 2014 Jul 15;193(2):540–3. doi: 10.4049/jimmunol.1400931. [DOI] [PubMed] [Google Scholar]
- 175.Schmidt T, Paust HJ, Krebs CF, Turner JE, Kaffke A, Bennstein SB, Koyro T, Peters A, Velden J, Hunemorder S, Haag F, Steinmetz OM, Mittrucker HW, Stahl RA, Panzer U. Function of the Th17/interleukin-17A immune response in murine lupus nephritis. Arthritis Rheumatol. 2015 Feb;67(2):475–87. doi: 10.1002/art.38955. [DOI] [PubMed] [Google Scholar]
- 176.Wong CK, Lit LC, Tam LS, Li EK, Wong PT, Lam CW. Hyperproduction of IL-23 and IL-17 in patients with systemic lupus erythematosus: implications for Th17-mediated inflammation in auto-immunity. Clin Immunol. 2008 Jun;127(3):385–93. doi: 10.1016/j.clim.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 177.Tanasescu C, Balanescu E, Balanescu P, Olteanu R, Badea C, Grancea C, Vagu C, Bleotu C, Ardeleanu C, Georgescu A. IL-17 in cutaneous lupus erythematosus. Eur J Intern Med. 2010 Jun;21(3):202–7. doi: 10.1016/j.ejim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 178.Chen XQ, Yu YC, Deng HH, Sun JZ, Dai Z, Wu YW, Yang M. Plasma IL-17A is increased in new-onset SLE patients and associated with disease activity. J Clin Immunol. 2010 Mar;30(2):221–5. doi: 10.1007/s10875-009-9365-x. Epub 2010/01/29. eng. [DOI] [PubMed] [Google Scholar]
- 179.Zhao XF, Pan HF, Yuan H, Zhang WH, Li XP, Wang GH, Wu GC, Su H, Pan FM, Li WX, Li LH, Chen GP, Ye DQ. Increased serum interleukin 17 in patients with systemic lupus erythematosus. Mol Biol Rep. 2010 Jan;37(1):81–5. doi: 10.1007/s11033-009-9533-3. [DOI] [PubMed] [Google Scholar]
- 180.Vincent FB, Northcott M, Hoi A, Mackay F, Morand EF. Clinical associations of serum interleukin-17 in systemic lupus erythematosus. Arthritis Research & Therapy. 2013 Aug 23;15(4):R97. doi: 10.1186/ar4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fayyad-Kazan H, Rouas R, Merimi M, El Zein N, Lewalle P, Jebbawi F, Mourtada M, Badran H, Ezzeddine M, Salaun B, Romero P, Burny A, Martiat P, Badran B. Valproate treatment of human cord blood CD4-positive effector T cells confers on them the molecular profile (microRNA signature and FOXP3 expression) of natural regulatory CD4-positive cells through inhibition of histone deacetylase. J Biol Chem. 2010 Jul 2;285(27):20481–91. doi: 10.1074/jbc.M110.119628. Epub 2010/04/30. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med. 2010 Oct;88(10):1029–40. doi: 10.1007/s00109-010-0642-1. Epub 2010/06/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scalapino KJ, Tang Q, Bluestone JA, Bonyhadi ML, Daikh DI. Suppression of disease in New Zealand Black/New Zealand White lupus-prone mice by adoptive transfer of ex vivo expanded regulatory T cells. J Immunol. 2006 Aug 01;177(3):1451–9. doi: 10.4049/jimmunol.177.3.1451. [DOI] [PubMed] [Google Scholar]
- 184.Humrich JY, Morbach H, Undeutsch R, Enghard P, Rosenberger S, Weigert O, Kloke L, Heimann J, Gaber T, Brandenburg S, Scheffold A, Huehn J, Radbruch A, Burmester GR, Riemekasten G. Homeostatic imbalance of regulatory and effector T cells due to IL-2 deprivation amplifies murine lupus. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jan 05;107(1):204–9. doi: 10.1073/pnas.0903158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu HY, Staines NA. A deficiency of CD4+CD25+ T cells permits the development of spontaneous lupus-like disease in mice, and can be reversed by induction of mucosal tolerance to histone peptide autoantigen. Lupus. 2004;13(3):192–200. doi: 10.1191/0961203303lu1002oa. [DOI] [PubMed] [Google Scholar]
- 186.Yang CH, Tian L, Ling GS, Trendell-Smith NJ, Ma L, Lo CK, Stott DI, Liew FY, Huang FP. Immunological mechanisms and clinical implications of regulatory T cell deficiency in a systemic autoimmune disorder: roles of IL-2 versus IL-15. Eur J Immunol. 2008 Jun;38(6):1664–76. doi: 10.1002/eji.200838190. [DOI] [PubMed] [Google Scholar]
- 187.Parietti V, Monneaux F, Decossas M, Muller S. Function of CD4+, CD25+ Treg cells in MRL/lpr mice is compromised by intrinsic defects in antigen-presenting cells and effector T cells. Arthritis Rheum. 2008 Jun;58(6):1751–61. doi: 10.1002/art.23464. [DOI] [PubMed] [Google Scholar]
- 188.Lyssuk EY, Torgashina AV, Soloviev SK, Nassonov EL, Bykovskaia SN. Reduced number and function of CD4+CD25highFoxP3+ regulatory T cells in patients with systemic lupus erythematosus. Adv Exp Med Biol. 2007;601:113–9. [PubMed] [Google Scholar]
- 189.Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007 Feb 15;178(4):2579–88. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- 190.Bonelli M, Savitskaya A, von Dalwigk K, Steiner CW, Aletaha D, Smolen JS, Scheinecker C. Quantitative and qualitative deficiencies of regulatory T cells in patients with systemic lupus erythematosus (SLE) Int Immunol. 2008 Jul;20(7):861–8. doi: 10.1093/intimm/dxn044. [DOI] [PubMed] [Google Scholar]
- 191.Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, Nochy D, Debre P, Piette JC, Gorochov G. Global natural regulatory T cell depletion in active systemic lupus erythematosus. J Immunol. 2005 Dec 15;175(12):8392–400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- 192.Alvarado-Sanchez B, Hernandez-Castro B, Portales-Perez D, Baranda L, Layseca-Espinosa E, Abud-Mendoza C, Cubillas-Tejeda AC, Gonzalez-Amaro R. Regulatory T cells in patients with systemic lupus erythematosus. J Autoimmun. 2006 Sep;27(2):110–8. doi: 10.1016/j.jaut.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 193.Vargas-Rojas MI, Crispin JC, Richaud-Patin Y, Alcocer-Varela J. Quantitative and qualitative normal regulatory T cells are not capable of inducing suppression in SLE patients due to T-cell resistance. Lupus. 2008 Apr;17(4):289–94. doi: 10.1177/0961203307088307. [DOI] [PubMed] [Google Scholar]
- 194.Lin SC, Chen KH, Lin CH, Kuo CC, Ling QD, Chan CH. The quantitative analysis of peripheral blood FOXP3-expressing T cells in systemic lupus erythematosus and rheumatoid arthritis patients. Eur J Clin Invest. 2007 Dec;37(12):987–96. doi: 10.1111/j.1365-2362.2007.01882.x. [DOI] [PubMed] [Google Scholar]
- 195.Venigalla RK, Tretter T, Krienke S, Max R, Eckstein V, Blank N, Fiehn C, Ho AD, Lorenz HM. Reduced CD4+, CD25− T cell sensitivity to the suppressive function of CD4+, CD25high, CD127−/low regulatory T cells in patients with active systemic lupus erythematosus. Arthritis Rheum. 2008 Jul;58(7):2120–30. doi: 10.1002/art.23556. [DOI] [PubMed] [Google Scholar]
- 196.Tokic S, Stefanic M, Glavas-Obrovac L, Jaman S, Novosadova E, Petrkova J, Navratilova Z, Suver Stevic M, Petrek M. The Expression of T Cell FOXP3 and T-Bet Is Upregulated in Severe but Not Euthyroid Hashimoto’s Thyroiditis. Mediators Inflamm. 2016;2016:3687420. doi: 10.1155/2016/3687420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007 Mar;26(3):371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 198.Xiao S, Sung SS, Fu SM, Ju ST. Combining Fas mutation with interleukin-2 deficiency prevents Colitis and Lupus: implicating interleukin-2 for auto-reactive T cell expansion and Fas ligand for colon epithelial cell death. J Biol Chem. 2003 Dec 26;278(52):52730–8. doi: 10.1074/jbc.M308707200. [DOI] [PubMed] [Google Scholar]
- 199.Alcocer-Varela J, Alarcon-Segovia D. Decreased production of and response to interleukin-2 by cultured lymphocytes from patients with systemic lupus erythematosus. J Clin Invest. 1982 Jun;69(6):1388–92. doi: 10.1172/JCI110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.de Faucal P, Godard A, Peyrat MA, Moreau JF, Soulillou JP. Impaired IL2 production by lymphocytes of patients with systemic lupus erythematosus. Ann Immunol (Paris) 1984 Sep-Oct;135D(2):161–72. doi: 10.1016/s0769-2625(84)81108-3. [DOI] [PubMed] [Google Scholar]
- 201.von Spee-Mayer C, Siegert E, Abdirama D, Rose A, Klaus A, Alexander T, Enghard P, Sawitzki B, Hiepe F, Radbruch A, Burmester GR, Riemekasten G, Humrich JY. Low-dose interleukin-2 selectively corrects regulatory T cell defects in patients with systemic lupus erythematosus. Ann Rheum Dis. 2016 Jul;75(7):1407–15. doi: 10.1136/annrheumdis-2015-207776. [DOI] [PubMed] [Google Scholar]
- 202.Flores-Borja F, Kabouridis PS, Jury EC, Isenberg DA, Mageed RA. Decreased Lyn expression and translocation to lipid raft signaling domains in B lymphocytes from patients with systemic lupus erythematosus. Arthritis Rheum. 2005 Dec;52(12):3955–65. doi: 10.1002/art.21416. Epub 2005/12/02. eng. [DOI] [PubMed] [Google Scholar]
- 203.Liossis SN, Solomou EE, Dimopoulos MA, Panayiotidis P, Mavrikakis MM, Sfikakis PP. B-cell kinase lyn deficiency in patients with systemic lupus erythematosus. J Investig Med. 2001 Mar;49(2):157–65. doi: 10.2310/6650.2001.34042. Epub 2001/04/06. eng. [DOI] [PubMed] [Google Scholar]
- 204.Lu R, Vidal GS, Kelly JA, Delgado-Vega AM, Howard XK, Macwana SR, Dominguez N, Klein W, Burrell C, Harley IT, Kaufman KM, Bruner GR, Moser KL, Gaffney PM, Gilkeson GS, Wakeland EK, Li QZ, Langefeld CD, Marion MC, Divers J, Alarcon GS, Brown EE, Kimberly RP, Edberg JC, Ramsey-Goldman R, Reveille JD, McGwin G, Jr, Vila LM, Petri MA, Bae SC, Cho SK, Bang SY, Kim I, Choi CB, Martin J, Vyse TJ, Merrill JT, Harley JB, Alarcon-Riquelme ME, Biolupus, Nath SK, James JA, Guthridge JM Collaborations GM. Genetic associations of LYN with systemic lupus erythematosus. Genes Immun. 2009 Jul;10(5):397–403. doi: 10.1038/gene.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Manjarrez-Orduno N, Marasco E, Chung SA, Katz MS, Kiridly JF, Simpfendorfer KR, Freudenberg J, Ballard DH, Nashi E, Hopkins TJ, Cunninghame Graham DS, Lee AT, Coenen MJ, Franke B, Swinkels DW, Graham RR, Kimberly RP, Gaffney PM, Vyse TJ, Behrens TW, Criswell LA, Diamond B, Gregersen PK. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat Genet. 2012 Nov;44(11):1227–30. doi: 10.1038/ng.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Becker AM, Dao KH, Han BK, Kornu R, Lakhanpal S, Mobley AB, Li QZ, Lian Y, Wu T, Reimold AM, Olsen NJ, Karp DR, Chowdhury FZ, Farrar JD, Satterthwaite AB, Mohan C, Lipsky PE, Wakeland EK, Davis LS. SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature. PLoS One. 2013;8(6):e67003. doi: 10.1371/journal.pone.0067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu XN, Ye YX, Niu JW, Li Y, Li X, You X, Chen H, Zhao LD, Zeng XF, Zhang FC, Tang FL, He W, Cao XT, Zhang X, Lipsky PE. Defective PTEN regulation contributes to B cell hyperresponsiveness in systemic lupus erythematosus. Sci Transl Med. 2014 Jul 23;6(246):246ra99. doi: 10.1126/scitranslmed.3009131. [DOI] [PubMed] [Google Scholar]
- 208.Lewis MJ, Vyse S, Shields AM, Boeltz S, Gordon PA, Spector TD, Lehner PJ, Walczak H, Vyse TJ. UBE2L3 polymorphism amplifies NF-kappaB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am J Hum Genet. 2015 Feb 05;96(2):221–34. doi: 10.1016/j.ajhg.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee KM, Chuang E, Griffin M, Khattri R, Hong DK, Zhang W, Straus D, Samelson LE, Thompson CB, Bluestone JA. Molecular basis of T cell inactivation by CTLA-4. Science. 1998 Dec 18;282(5397):2263–6. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 210.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999 Aug;11(2):141–51. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 211.Mustelin T, Tasken K. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J. 2003 Apr 01;371(Pt 1):15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Qi Q, August A. Keeping the (kinase) party going: SLP-76 and ITK dance to the beat. Sci STKE. 2007 Jul 24;2007(396):pe39. doi: 10.1126/stke.3962007pe39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Barrera-Vargas A, Gomez-Martin D, Alcocer-Varela J. T cell receptor-associated protein tyrosine kinases: the dynamics of tolerance regulation by phosphorylation and its role in systemic lupus erythematosus. Hum Immunol. 2014 Sep;75(9):945–52. doi: 10.1016/j.humimm.2014.08.207. [DOI] [PubMed] [Google Scholar]