Abstract
We report here the main characteristics of “Streptomyces massilialgeriensis” strain S35T (CSUR = P3927), a new bacterial species within the Streptomyces genus, isolated from an extremely saline soil sample collected from the site of Garaet Ank Djemel in the Wilaya of Oum El Bouaghi, Algeria.
Keywords: Culturomics, dry lake, saline soil, “Streptomyces massilialgeriensis”, taxono-genomics
A culturomics approach has enabled the isolation of >295 new bacterial species from the human gut [1]. Here, using culture conditions containing a high salt concentration [2], we isolated strain S35T from an extremely saline soil sample collected from the dry lake of Ank el Djamel in Algeria. This strain could not be identified by our systematic matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF-MS) screening (http://mediterranee-infection.com/article.php?laref=256&titre=urms-database) on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3].
Strain S35T was isolated after 14 days of incubation at 30°C on International Streptomyces Project 4 medium (ISP4) (Becton Dickinson, Le Pont-de-Claix, France) modified by the addition of 50 g/L of NaCl and with pH adjusted to 8. Then, the growing colonies were sub-cultured on 5% sheep's blood-enriched Columbia agar (bioMérieux, Marcy l’Etoile, France) for 7 days in an aerobic atmosphere at 37°C.
Strain S35T is a Gram-positive, filamentous-shape, ranging from 0.4 to 0.6 μm in diameter. The colonies obtained after 7 days of incubation appear opaque, brown with a wrinkled surface with a mean diameter of 2 mm. Strain S35T exhibited catalase but not oxidase activity. This strain is mesophilic and strictly aerobic, growing optimally at 37°C and pH 7.5.
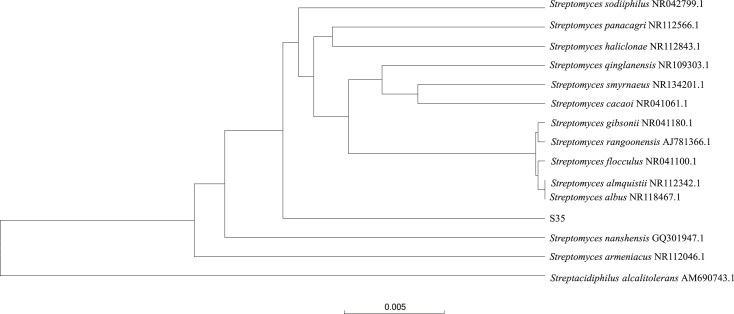
Identification based on 16S rDNA demonstrates that strain S35T exhibits 97.26% sequence identity with Streptomyces albus ATCC 25426 (GenBank Accession number DQ026669.1), its most closely related species with standing in nomenclature [4] (Fig. 1). This value is under the threshold of 98.7% separating new species established by Stackebrandt and Ebers [5]. Consequently, we propose the creation of “Streptomyces massilialgeriensis” sp. nov., a new bacterial species belonging to the Streptomyces genus. Strain S35T is the type strain of “Streptomyces massilialgeriensis” sp. nov. (ma.sil.io.al.ge.ri.en'sis. L. gen. masc. n. massilioalgeriensis, a combination of massili for Massilia, the Roman name of Marseille where the type strain S35T was identified and sequenced, and algeriensis, for Algeria, the country in North Africa, where the strain was isolated).
Fig. 1.
Phylogenetic tree showing the position of “Streptomyces massilialgeriensis” strain S35T relative to other phylogenetically close neighbours. The 16S rRNA gene sequences were aligned using CLUSTAL W, and phylogenetic inferences were obtained using the maximum-likelihood method within the MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Only bootstrap scores of at least 95 were retained. The scale bar indicates a 0.005% nucleotide sequence divergence.
Actinobacteria are considered as providers of molecules of medical interest with antitumour, antiviral and antiparasitic activities. In particular, species belonging to the genus Streptomyces are producing the majority of the antibiotics used in human therapy. It has been estimated that out of 16 500 known antibiotics, 8700 are produced by Actinobacteria, including 6550 by Streptomyces species [6]. These bacteria actually have a large number of “hidden” or “cryptic” metabolites. If we could induce the production of these metabolites, it would be possible to obtain several new molecules of therapeutic interest.
MALDI-TOF spectrum The MALDI-TOF-MS spectrum of “Streptomyces massilialgeriensis” is available in: http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database.
Nucleotide sequences accession numbers The 16S rRNA gene sequence of “Streptomyces massilialgeriensis” was deposited in GenBank under Accession number: LT838406.
Deposit in a culture collection The strain S35T was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) under number P3927.
Conflict of interest
There is no conflict of interest.
Acknowledgements
This work was funded by the Mediterranean-Infection Foundation.
References
- 1.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 2.Khelaifia S., Croce O., Lagier J.C., Robert C., Couderc C., Di Pinto F. Noncontiguous finished genome sequence and description of Virgibacillus massiliensis sp. nov., a moderately halophilic bacterium isolated from human gut. New Microbes New Infect. 2015;8:78–88. doi: 10.1016/j.nmni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.-C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons A.J., Jr., Pridham T.G. Proposal to designate strain ATCC 3004 (IMRU 3004) as the neotype strain of Streptomyces albus (rossi-doria) Waksman and Henrici. J Bacteriol. 1962;83:370–380. doi: 10.1128/jb.83.2.370-380.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 6.Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]