Abstract
Protein kinase clients are recruited to the Hsp90 molecular chaperone system via Cdc37, which simultaneously binds Hsp90 and kinases and regulates the Hsp90 chaperone cycle. Pharmacological inhibition of Hsp90 in vivo results in degradation of kinase clients, with a therapeutic effect in dependent tumours. We show here that Cdc37 directly antagonises ATP binding to client kinases, suggesting a role for the Hsp90-Cdc37 complex in controlling kinase activity. Unexpectedly, we find that Cdc37 binding to protein kinases is itself antagonised by ATP-competitive kinase inhibitors including vemurafenib and lapatinib. In cancer cells these inhibitors deprive oncogenic kinases such as BRaf and ErbB2 of access to the Hsp90-Cdc37 complex, leading to their degradation. Our results suggest that at least part of the efficacy of ATP-competitive inhibitors of Hsp90-dependent kinases clients in tumour cells may be due to targeted chaperone deprivation.
Introduction
Protein kinases, which function as the major regulators and transducers of signalling in eukaryotic cells, constitute the largest coherent class of client proteins of the Hsp90 molecular chaperone system 1. Recruitment to the Hsp90 system is mediated by Cdc37 (also known as p50), which functions as a scaffold protein, binding Hsp90 and protein kinases simultaneously and facilitating their mutual interaction 2–4. While the pairwise interaction of Hsp90 and Cdc37 has been defined at the atomic level 5, the structural basis for specific interaction of Cdc37 and client protein kinases is unknown. Low resolution structural analysis suggests that all three proteins are involved in multiple contacts within an assembled Hsp90-Cdc37-kinase complex 6.
The biochemical effect of Cdc37 and Hsp90 on protein kinase client function is not well understood, but recruitment to the chaperone system appears to be critical for cellular stability. Pharmacological inhibition of the system in vivo by ATP-competitive inhibitors of the Hsp90 chaperone cycle, results in degradation of client kinases via the ubiquitin-dependant proteasome pathway 7,8. This provides the therapeutic rationale for the development of highly specific Hsp90 inhibitors that exert their strong anti-tumour activities by promoting depletion of oncogenic client protein kinases such as BRaf, ErbB2, Cdk4 and Bcr-Abl, for example, as well as other non-kinase Hsp90 clients such as the estrogen and androgen receptors 9,10. Whether this effect is due to inherent structural instability of the client proteins, or is a default targeted-destruction pathway unmasked by the absence of countervailing chaperone function, is not known.
While Hsp90 is essentially involved in the biological function of many different classes of proteins 11, Cdc37 is primarily associated with eukaryotic protein kinases 4,12. This suggests that Cdc37, rather than Hsp90, encapsulates the structural features that mediate recognition of the large, but highly specific subset of protein kinases whose biological function is tied to the Hsp90 chaperone system. The basis for this specificity has been the subject of considerable interest 13–18, but there is currently no definitive view as to which features of Cdc37 or of kinase clients are truly involved.
We have now established a system for producing complexes of Hsp90, Cdc37 and client protein kinases either co-expressed in cells, or assembled in vitro using purified proteins. We find that Cdc37 directly antagonises ATP binding to client protein kinases, and inhibits phosphorylation of kinase substrate proteins. Unexpectedly, we find that ATP-competitive inhibitors of protein kinases antagonise Cdc37 interaction with Hsp90-dependent kinases in vitro and in cells, and thereby deprive the client kinase of access to the Hsp90 molecular chaperone system, promoting its degradation via the ubiquitin-directed proteasome. These studies reveal an unanticipated role for the Hsp90-Cdc37 system in directly controlling the signalling activity of their client protein kinases. They further suggest that many of the protein kinase inhibitors in clinical use, while designed as ATP-competitors, may achieve part of their biological and therapeutic effects through chaperone deprivation.
Results
Assembly of Hsp90-Cdc37-BRaf complexes
We have previously described the expression and purification of a stable Hsp90-Cdc37-Cdk4 complex using a baculovirus system for overexpression of human Cdc37 and Cdk4, which recruit the insect cell Hsp90 6. We now developed a baculovirus system that permits expression and purification of a fully human in vivo assembled complex of Hsp90β, Cdc37 and the kinase domains of BRaf or its oncogenic variant BRafV600E, which have previously been shown to be Hsp90 clients 19,20 (Figure 1a). We also sought to reconstitute the Hsp90β-Cdc37-BRaf kinase domain complex from separately purified proteins in vitro. While Hsp90 and Cdc37 can be readily expressed in bacteria, 16 surface residues within the C-terminal lobe of the BRaf kinase domain had to be mutated to make it amenable for soluble bacterial expression21. When co-expressed with Hsp90 and Cdc37 in insect cells, the solubilised BRaf (sBRaf and sBRafV600E) formed stable Hsp90-Cdc37-kinase complexes that were indistinguishable from those formed by the wildtype BRaf kinase domain when similarly co-expressed (Figure 1a). Thus, the solubilising mutations, which are all distal to the ATP-binding site and activation segment of the kinase, do not affect its ability to associate with the Hsp90 chaperone machinery.
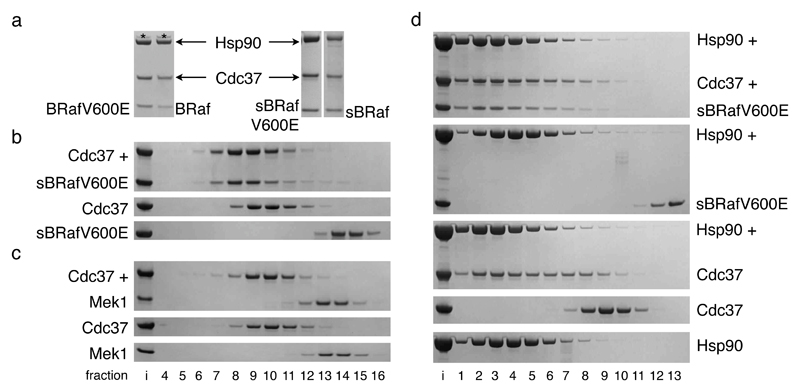
Figure 1. The BRaf catalytic domain associates with Cdc37-Hsp90.
a. Coomassie stained SDS-PAGE gels of purified complexes of BRaf kinase domain (wild-type, V600E, mutationally solubilised wild-type or V600E), Cdc37 and Hsp90β from Sf9 insect cells. BRaf recruitment to the Hsp90 system is mediated by the kinase domain. The solubilising mutations do not alter BRaf association with Cdc37-Hsp90. *: endogenous Sf9 Hsp90.
b. Coomassie stained SDS-PAGE gels of Superose 6 gel filtration fractions – ‘i’: input; top: sBRafV600E+Cdc37; middle: Cdc37 only; bottom: sBRafV600E only. sBRafV600E and Cdc37 form of a stable complex.
c. As b, but with the Mek1 kinase domain. Cdc37 and Mek1 do not interact stably.
d. As b – top to bottom: sBRafV600E+Cdc37+Hsp90β; sBRafV600E+Hsp90β; Cdc37+Hsp90β; Cdc37 only; Hsp90β only. Whereas sBRafV600E-Cdc37-Hsp90 β and Cdc37-Hsp90 β form ternary and binary complexes respectively, no stable interaction between sBRafV600E and Hsp90 is evident.
The purified BRaf kinase domain (native or solubilised) bound to Cdc37 in vitro, forming a stable complex in gel filtration (Figure 1b and Supplementary Figure 2a and b). Previous cell biology studies showed that the oncogenic BRafV600E mutant displays a greater dependency in vivo on the Hsp90 chaperone system than the wild-type protein 19,20. However, we observed no substantial difference in binding to Cdc37 between wild-type and oncogenic BRafV600E in vitro. Whatever the origin of the enhanced chaperone dependence in vivo conferred by the V600E mutation, it does not appear to result from any significant enhancement in affinity for Cdc37. We measured the Kd of the interaction of sBRafV600E with Cdc37 as 0.2 μM by isothermal titration calorimetry (Supplementary Figure 2c). Kinase binding to Cdc37 in vitro mirrored the dependency of different kinases on the Hsp90 system in vivo22: The known Hsp90 client BRafV600E19,20 did bind, whereas the kinase domain of its downstream substrate Mek1 did not (Figure 1c).
Addition of Hsp90 resulted in formation of a stable ternary complex, with sBRafV600E, Cdc37 and Hsp90 co-migrating in gel filtration (Figure 1d). In contrast to Hsp90 and Cdc37, which form a stable binary complex5, Hsp90 and sBRafV600E did not co-elute in the absence of Cdc37, showing that the co-chaperone is essential for the establishment of an Hsp90-kinase interaction in vitro.
Cdc37 blocks ATP binding and utilisation by protein kinase clients
The apparent ability of Cdc37 to interact with a very wide range of protein kinase clients suggests that it recognizes a common if not universal feature of protein kinase structure. A defining feature of all protein kinases is a bilobal structure in which the largely β-strand N-lobe is separated from the predominantly α-helical C-lobe by a deep cleft that provides the binding site for ATP 23. While there is sufficient evolutionary variability in this region to permit the development of selective ATP-competitive inhibitors 24, it provides a number of invariant features that make it a good candidate for specific interaction with Cdc37. This hypothesis is supported by mutational studies14,15,17,25 which have consistently implicated features in and around the ATP-binding cleft as being involved in kinase recruitment to the Hsp90-Cdc37 system.
To determine if Cdc37 affects the ATP-binding cleft, we utilized a fluorescently labelled ADP analogue, MANT-ADP, fluorescence of which is specifically enhanced by binding to ATP-binding sites in proteins 26. When added to sBRafV600E, we observed a significant increase at 440nm consistent with MANT-ADP binding to the ATP-binding cleft of the kinase 27 (Figure 2a). This fluorescence enhancement was reduced when Cdc37 was also present, suggesting that Cdc37 antagonised binding of MANT-ADP to sBRafV600E (Figure 2a and b and Supplementary Figure 3a-c). In a parallel experiment we immobilised sBRafV600E on an ATP-agarose resin, and observed that it could be eluted by addition of Cdc37, ATP or the ATP-competitive BRaf inhibitors vemurafenib or sorafenib (Figure 2c). The effect in both experiments was substantially reduced with a Cdc37 mutant that failed to co-migrate with sBRafV600E in gel filtration (Supplementary Figure 3d and e). Taken together, these data show that binding of Cdc37 is directly antagonistic to binding of adenine nucleotides to BRaf, either by allosteric remodelling of the structure of the ATP-binding cleft or by its direct competitive blockade.
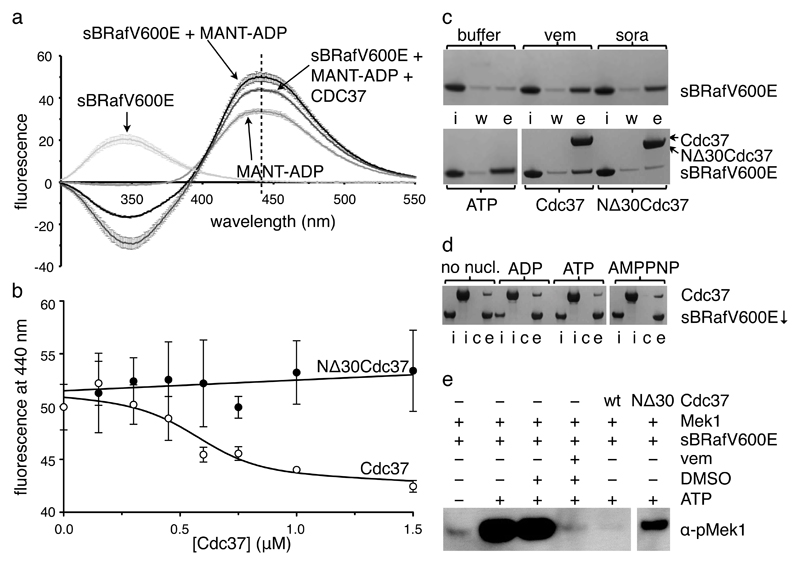
Figure 2. Cdc37 antagonises nucleotide binding to BRaf.
a. Fluorescence emission spectra of MANT-ADP (120 μM) and sBRafV600E (1 μM) alone and incubated together as well as of sBRafV600E•MANT-ADP incubated with 1 μM of Cdc37. MANT-ADP shows enhanced fluorescence at the emission maximum of 440 nm in the presence of sBRafV600E. Presence of Cdc37 decreases the fluorescence enhancement at 440nm.Standard deviations of at least three independent experiments are indicated.
b. sBRafV600E•MANT-ADP fluorescence at 440 nm as function of the concentration of Cdc37. Fluorescence decreases with increasing concentration of full length Cdc37 (○), but not of a Cdc37 N-terminal deletion construct that is impaired in kinase binding (●). Standard deviations of three independent experiments are shown.
c. Elution of sBRafV600E from ATP-agarose beads, visualised by Coomassie stained SDS-PAGE. In each case ‘i’: input; ‘w’: final buffer wash; ‘e’: elution with competitor. Vemurafenib, sorafenib, ATP and full length Cdc37 are effective at competing sBRafV600E off the beads, whereas buffer or NΔ30Cdc37 are not.
d. Pull-down of Cdc37 with IMAC-beads via N-terminally His6-tagged sBRafV600E visualised by Coomassie stained SDS-PAGE. In each case ‘i’: input sBRafV600E or Cdc37; ‘c’: control pull-down in absence of His6-sBRafV600E; ‘e’: elution in presence of His6-sBRafV600E. Presence of 1 mM ADP, ATP or AMPPNP has no effect.
e. Kinase activity of sBRafV600E visualised by western blotting of phosphorylated full-length Mek1. Phosphorylation of Mek1 is inhibited by vemurafenib and equimolar amounts of full-length Cdc37. NΔ30Cdc37 is substantially less effective in inhibiting Mek1 phosphorylation.
In the inverse experiment, we found that none of ATP, ADP or the non-hydrolysable ATP analogue AMP-PNP were able to prevent Cdc37 binding to sBRafV600E even when present in a ~175 fold excess over Cdc37 (Figure 2d). This is consistent with the affinity of sBRafV600E for Cdc37 (KD~0.2μM) being significantly higher than that of BRafV600E for ATP (KM~65μM)28 and suggests that antagonism between Cdc37 and ATP is likely to be one-way in the cell, with Cdc37 inhibiting ATP binding to BRaf.
The observation that Cdc37 antagonises nucleotide binding suggests that Cdc37 might inhibit the ATP-dependent catalytic activity of a client kinase such as BRaf. To test this we utilized a standard kinase assay in which specific phosphorylation of the downstream biological target of BRaf, Mek1, is detected by a phosphospecific antibody (see Methods). We were able to detect phosphorylation of Mek1 by sBRafV600E in the presence of ATP (Figure 2e). As expected, Mek1 phosphorylation was inhibited by the specific BRaf ATP-competitive inhibitor vemurafenib 21,29, but was also substantially inhibited by the addition of Cdc37. BRaf inhibition by Cdc37 was unaffected by phosphorylation of Cdc37 on Ser13 30 and by the presence or absence of the activity-stimulating oncogenic BRafV600E or of the fully activating BRaf T599E V600E S602D31 triple mutation (Supplementary Figure 4). Taken together these data suggest that Cdc37 binding does not distinguish between active and inactive conformations of the kinase domain.
ATP-competitive kinase inhibitors antagonise Cdc37 interaction with kinases in vitro
Based on our observation that Cdc37 antagonises nucleotide binding to BRaf kinase domain and inhibits its kinase activity, we were keen to discover if small molecule inhibitors that function by competitive inhibition of ATP binding to kinases are in turn antagonistic to binding of their respective target kinases Cdc37. We found that clinically-approved selective BRaf inhibitor vemurafenib substantially inhibited the interaction of Cdc37 with sBRafV600E, observed both with sBRafV600E immobilized on an Octet Biosensor and in gel filtration (Figure 3a and b). Incubation of a preassembled BRafV600E-Cdc37 complex with vemurafenib resulted in the release of some BRafV600E; this effect was more pronounced with BRaf without the oncogenic mutation (Supplementary Figure 5a and b). Sorafenib and SB-590885, BRaf inhibitors that preferentially bind to the inactive and active conformation of the kinase domain respectively32,33, similarly inhibited sBRafV600E binding to Cdc37 (Supplementary Figure 5c-e). This reinforces the conclusion that Cdc37 binding does not distinguish between active and inactive conformations of the kinase domain.
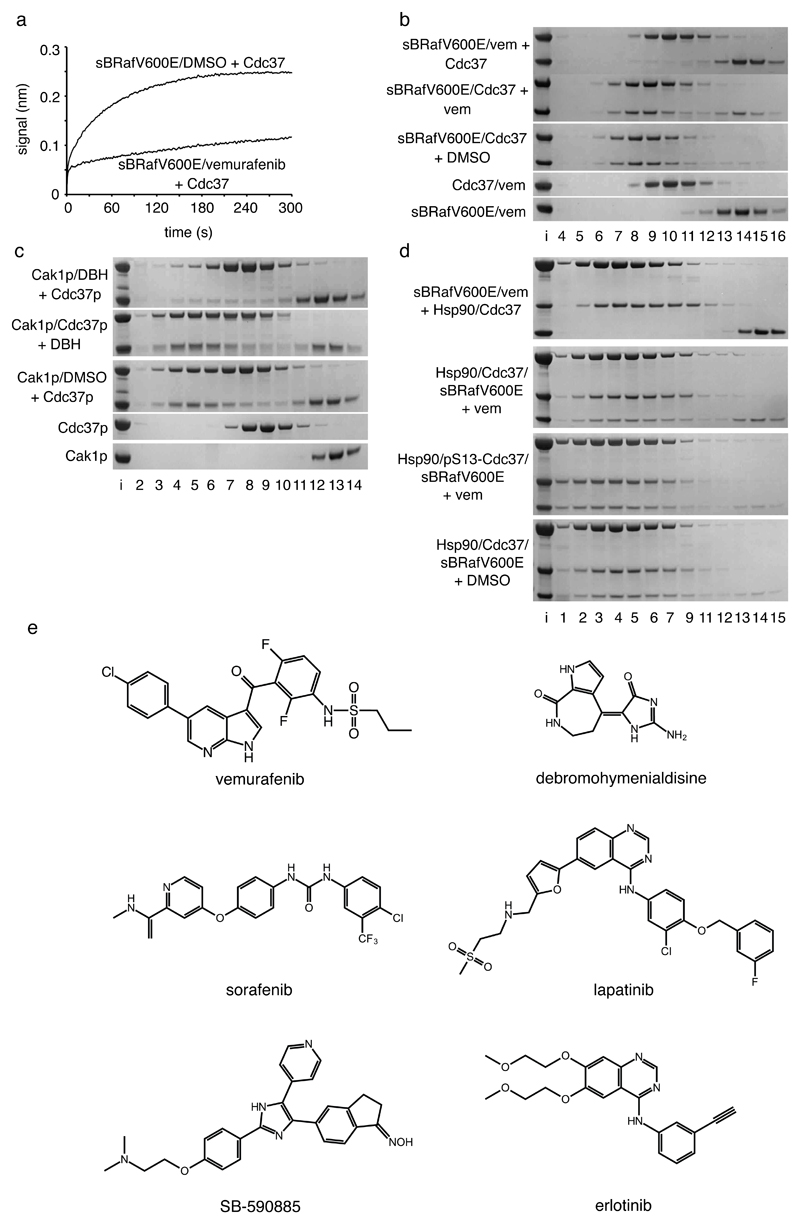
Figure 3. ATP-competitive inhibitors antagonise Cdc37 binding to kinases.
a. Octet Biosensor association curve for Cdc37 binding to immobilised His6-sBRafV600E in the absence or presence of vemurafenib. Binding is substantially inhibited by vemurafenib.
b. Coomassie stained SDS-PAGE gels showing Superose 6 elution profiles (top to bottom): Cdc37 incubated with a pre-formed sBRafV600E-vemurafenib complex; sBRafV600E-Cdc37 complex incubated with vemurafenib; sBRafV600E-Cdc37 complex incubated with DMSO; Cdc37 incubated with vemurafenib; sBRafV600E incubated with vemurafenib. Vemurafenib prevents sBRafV600E recruitment to Cdc37 and is able to partially displace sBRafV600E from a pre-formed Cdc37-kinase complex.
c. As b, but with the budding yeast kinase Cak1p, budding yeast Cdc37p and the broad specificity kinase inhibitor debromohymenialdisine. DBH prevents Cak1p binding to Cdc37p, but is less able to disrupt a pre-formed Cak1p-Cdc37p complex.
d. As b – top to bottom: Hsp90β and Cdc37 incubated with a pre-formed sBRafV600E-vemurafenib complex; Hsp90β-Cdc37-sBRafV600E complex incubated with vemurafenib; Hsp90β-sBRafV600E complex with pSer13-Cdc37 incubated with vemurafenib; Hsp90β-Cdc37-sBRafV600E complex incubated with DMSO. Vemurafenib prevents the recruitment of sBRafV600E to the Hsp90β-Cdc37 complex and is able to partially displace sBRafV600E from the Hsp90β-Cdc37 complex. Ser13 phosphorylation stabilises the ternary complex towards vemurafenib relative to the unphosphorylated complex.
e. ATP-competitive kinase inhibitors that all antagonise Cdc37 binding to their target protein kinases, in vitro and/or in cells.
We also looked at a protein kinase evolutionarily very distant from human BRaf, the cyclin-dependent kinase activating kinase Cak1p, from budding yeast 34. As with the human BRaf and Cdc37 proteins, we found that Cak1p formed a complex with yeast Cdc37p in analytical gel filtration in presence of DMSO (Figure 3c). We identified the low-specificity ATP-competitive protein kinase inhibitor debromohymenialdisine as able to bind to Cak1p (KD~1.7 μM, Supplementary Figure 6) and found that it inhibited formation of a Cak1p complex with Cdc37p (Figure 3c). Thus, Cdc37 antagonism by ATP-competitive inhibitors appears to be a general effect amongst Hsp90 protein kinase clients and not restricted to BRaf or to human Cdc37. A preassembled Cak1p-Cdc37p complex was more resistant to disruption by debromohymenialdisine than the preassembled BRafV600E-Cdc37 complex was to disruption by the tighter binding vemurafenib.
Preincubation of sBRafV600E with vemurafenib not only prevented recruitment to Cdc37 but also to an Hsp90-Cdc37 complex, and addition of vemurafenib to a preassembled Hsp90-Cdc37-sBRafV600E complex promoted dissociation of the kinase (Figure 3d). However, the Hsp90-Cdc37-BRafV600E complex was stabilised to disruption by vemurafenib when Cdc37 had previously been phosphorylated on Ser13 (Figure 3d and Supplementary Figure 9), consistent with the role proposed for this modification in regulating the stability of client kinase complexes35.
ATP-competitive kinase inhibitors deprive kinases of access to the Hsp90-Cdc37 system in cells
Next, based on the in vitro observations on kinase-chaperone complexes, we asked whether kinase inhibitors would also antagonise kinase interaction with the Hsp90-Cdc37 system in intact cells. We used a human colon cancer cell line (HT29) that expresses the oncogenic BRafV600E and examined the co-immunoprecipitation of Cdc37 and Hsp90. Incubation of cells with vemurafenib (Figure 4a) or sorafenib (Supplementary Figure 7) led to a marked, time- and concentration-dependent decrease in the amounts of Cdc37 co-precipitating with BRafV600E. Vemurafenib exhibited a clear effect after 8 hours exposure to a concentration that causes 50 % inhibition of cell growth (GI50) with complete antagonism of the Cdc37-BRafV600E interaction at 24 hours. BRafV600E underwent complete chaperone deprivation, with loss of association with both Cdc37 and Hsp90, after 24 hours exposure to 10 x GI50 (Figure 4a).
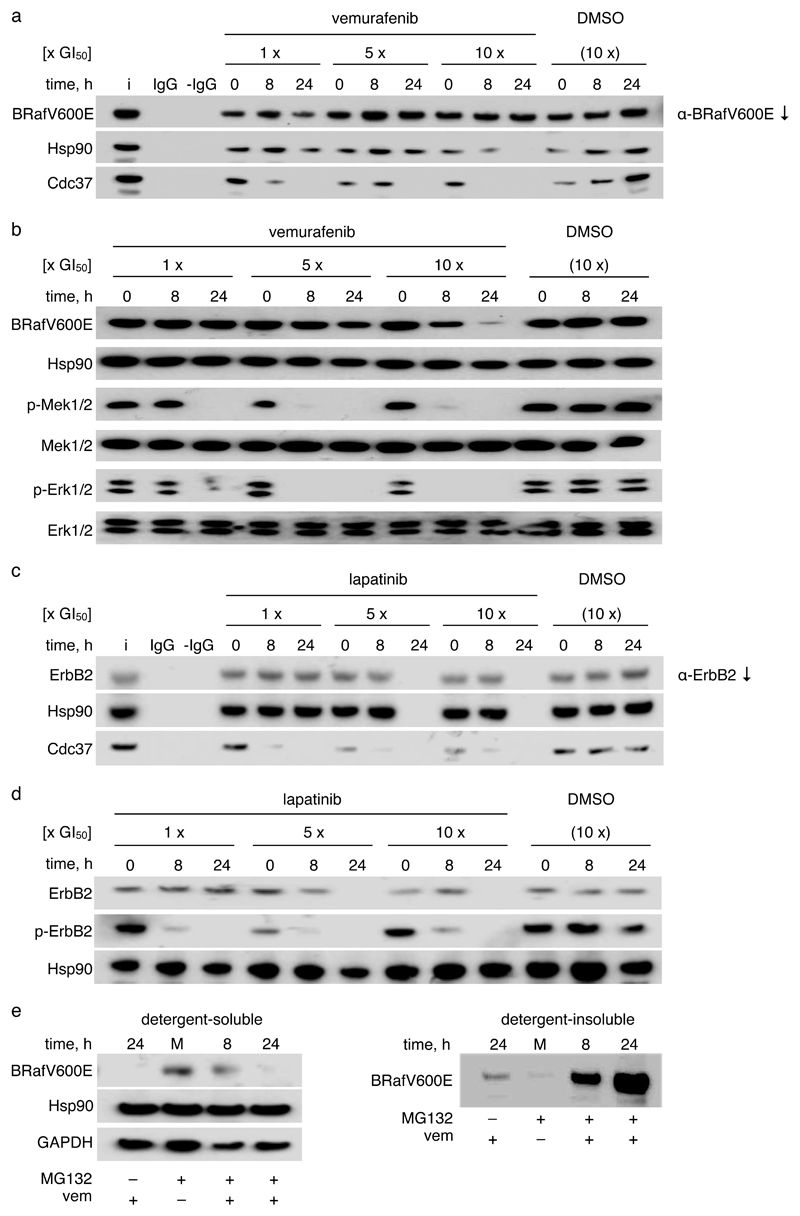
Figure 4. Kinase inhibitors prevent association with Cdc37-Hsp90 in human tumour cells.
a. Western blots of BRafV600E immunoprecipitated from HT29 human colon cancer cells incubated with different concentrations of vemurafenib for different times. The GI50 for 96 h exposure was determined as 0.46 μM. Controls are ‘i’: cell lysate input; IgG: α-BRafV600E IgG but no protein; -IgG: protein but no α-BRafV600E IgG. DMSO vehicle controls are at DMSO concentrations used in the 10x GI50 incubation with vemurafenib. All experiments were performed in triplicate, representative results are shown. Chaperone deprivation, defined by loss of co-immunoprecipitating Cdc37 and Hsp90, is apparent.
b. Western blots of cell lysates from a. Phosphorylation of BRafV600E targets Mek and Erk is decreased by vemurafenib. High levels of drug exposure decrease cellular BRafV600E levels.
c. As a, but for ErbB2 immunoprecipitation from BT474 human breast cancer cells exposed to different concentrations of lapatinib. GI50 for 96 h exposure to lapatinib: 30 nM. After 24 h of incubation with 5x or 10x GI50 lapatinib all ErbB2 is degraded so that no detectable protein is immunoprecipitated. Chaperone deprivation is again apparent.
d. Western blots of cell lysates from c. ErbB2 kinase activity measured by auto-phosphorylation is decreased by lapatinib concentrations that significantly reduce Cdc37 levels in ErbB2 co-immunoprecipitates. High levels of drug exposure deplete cellular ErbB2.
e. Western blots of cell lysates from HT29 colon cancer cells incubated with 5x GI50 MG132 (200 nM) for 1 h prior to exposure to 10x GI50 vemurafenib for the indicated time spans – ‘M’: 24 h DMSO vehicle exposure. Treatment with vemurafenib following proteasome inhibition leads to BRafV600E accumulation in the detergent-insoluble fraction.
As expected, exposure to vemurafenib caused a significant decrease in phosphorylation of Mek and Erk in HT29 cells, consistent with competitive blockade of the ATP-binding site of BRafV600E (Figure 4b). Importantly, this occurred at drug exposures where depletion of Cdc37 was already evident in BRafV600E co-immunoprecipitates (Figure 4a). Furthermore, at higher exposure levels where little or no Cdc37 or Hsp90 were co-immunoprecipitated with BRafV600E, we observed a marked decrease in the level of cellular BRafV600E protein, whereas total Mek and Erk levels were unaffected.
Next, to determine whether our observations for the BRafV600E/vemurafenib system were applicable to other Hsp90-dependent kinases and their specific inhibitors, we first treated BT474 human breast cancer cells with the clinically approved ATP-competitive tyrosine kinase inhibitor lapatinib, and found that this resulted in the displacement of Cdc37 and Hsp90 from the target kinase ErbB2 (Figure 4c). Cdc37 displacement correlated strongly with a decrease in ErbB2 auto-phosphorylation attributable to competitive blockade of the ErbB2 ATP-binding site by lapatinib (Figure 4d). At higher drug exposures, Cdc37 and Hsp90 were undetectable in ErbB2 co-immunoprecipitates and we observed a substantial decrease in cellular ErbB2 levels. Consistent with this cellular effect, an Hsp90-Cdc37-ErbB2 complex purified from Sf9 insect cells was destabilised in vitro by incubation with lapatinib (Supplementary Figure 5f).
Finally, as a further example, we examined mutant Egfr G719S in SW48 human colon adenocarcinoma cells treated with the clinically approved drug erlotinib. We observed a similar pattern of Cdc37/Hsp90 displacement and kinase degradation (Supplementary Figure 8).
Chaperone deprivation by ATP-competitive inhibitors promotes degradation of client kinases
When we treated HT29 cells with vemurafenib at 10x GI50 for 24 hours in the presence of the proteasome inhibitor MG132, BRafV600E protein was still substantially depleted from the detergent-soluble cell lysate fraction, but accumulated at a significant level in the insoluble fraction (Figure 4e). This indicates that BRafV600E depletion accompanying chaperone deprivation by vemurafenib proceeds via proteasomal degradation, rather than inhibition of biosynthesis. While a substantial level of insoluble BRafV600E had accumulated at 8 hours, the soluble pool of BRafV600E was relatively undiminished. This suggests that – rather than disrupting existing Hsp90-Cdc37-kinase complexes – the kinase inhibitors are acting primarily to deprive nascent kinase of access to Cdc37, thereby exposing it to an Hsp90-independent degradation pathway36. The protein kinase pool stabilized by association with functional and cycling Hsp90 is progressively depleted via association with downstream partners and/or the Hsp90-dependent degradation pathway. This model also explains the time lag observed between loss of Cdc37 and Hsp90 from BRafV600E in the presence of vemurafenib (Figure 4a), since Cdc37 is only part of early Hsp90-kinase complexes 37.
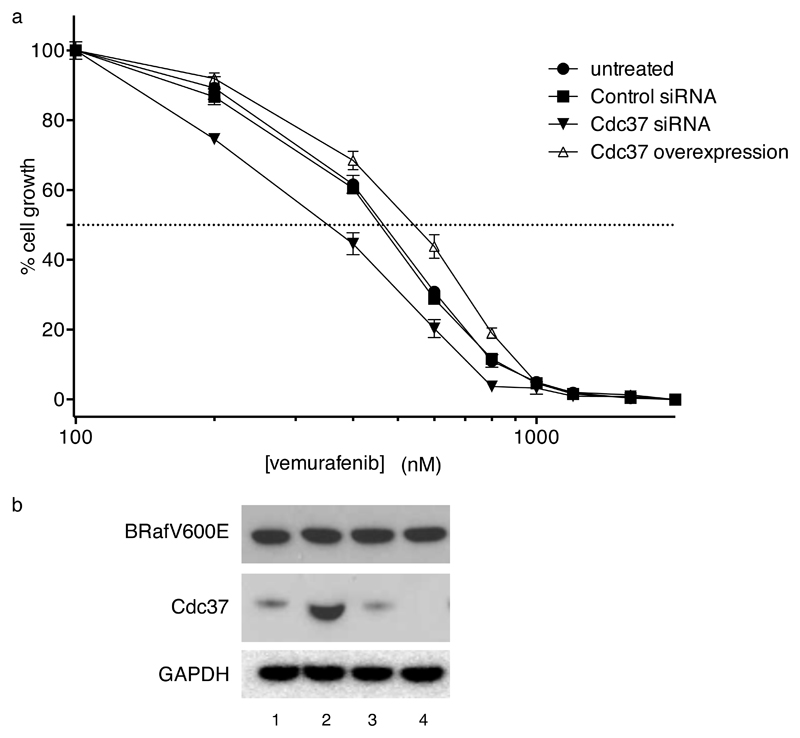
The in vitro and cellular antagonism we observe between kinase inhibitors and Cdc37 for kinase binding, suggests that the pharmacological behaviour of the inhibitor should be sensitive to the levels of Cdc37 in the cell. To test this, we compared the GI50 for vemurafenib in HT29 cells with normal Cdc37 expression with that in cells in which Cdc37 was either knocked down by siRNA or overexpressed (Figure 5a,b). We observed statistically significant differences between the untreated cells and the Cdc37 knock-down or overexpressing lines, with a ~1.5-fold higher vemurafenib concentration required for inhibition of cell growth in Cdc37 overexpressing cells compared to Cdc37 knock-down cells. These data confirm the cellular antagonism between Cdc37 and kinase inhibitors, and suggest that at least part of the pharmacological efficacy of vemurafenib may be determined by its ability to deprive BRafV600E of access to the Hsp90-Cdc37 chaperone system.
Figure 5. Manipulation of Cdc37 levels alter sensitivity of HT29 human colon cancer cells to growth inhibition by vemurafenib.
a. GI50 for vemurafenib after 96-hours exposure in parent HT29 (circles), cells treated with a non-targeting control siRNA (squares), cells treated with a specific siRNA to Cdc37 (full triangle), and cells over-expressing Cdc37 (open triangle). Data are means of four measurements; standard errors are shown. Knock-down and over-expressing HT29 cell lines are as described in 36,44 Over-expression of Cdc37 was 6-10-fold compared to parent HT29 (data not shown). GI50 values determined from three independently replicated experiments by non-linear curve fitting to a sigmoid dose-response model are: untreated - 431.4 (SD 8.0) nM; control siRNA - 422.8 (SD 14.1) nM; Cdc37 siRNA (oligo O4 from Smith et al, Oncogene. 2009, 28, 157-169) - 338.0 (SD 4.4) nM; Cdc37 over-expression - 494.6 (SD 22.8) nM. R2 for all curve fittings are >0.98. Differences between knock-down, over-expression and control are all highly significant at P< 0.0001.
b. Western blots showing levels of BRafV600E and Cdc37 protein in HT29 cells as follows: 1 - untreated; 2 - Cdc37 over-expressing; 3 - control siRNA; 4 - Cdc37 siRNA as above. GAPDH is provided as a loading control. Steady-state BRafV600E protein levels are not significantly altered by Cdc37 knock-down or over-expression, so that the observed differences in GI50 for vemurafenib can be attributed to the altered cellular Cdc37 levels. Overexpression of Cdc37 was 6-10 fold compared to parent HT29.
Discussion
We have shown here that Cdc37 antagonises binding of ATP to protein kinase clients such as BRaf, ErbB2 and Egfr G719S. The measured affinity of Cdc37 for BRafV600E (~200nM) is significantly higher than the Km of BRafV600E for ATP (65μM), which is typical of most protein kinases, so that an important biological effect of recruitment to Cdc37 would be the effective silencing of the catalytic activity of a potentially active protein kinase. Thus, Cdc37 appears to play a direct role in controlling the activity of its clientele (Supplementary Figure 1).). Interestingly, the Km of BRaf for ATP is reported to be 5 μM, 13 fold lower than for BRafV600E 28. This significantly lower affinity of BRafV600E for ATP might be the reason for the enhanced in vivo Hsp90/Cdc37 association of BRafV600E compared to wild-type BRaf19,20.
In vitro and in cells we find that Cdc37 binding to different protein kinase clients is antagonised by a range of different protein kinase inhibitors of widely divergent structure, including clinically important drugs such as vemurafenib, sorafenib, lapatinib and erlotinib. These compounds, developed on the basis of their ability to compete with ATP binding to the kinase active site, have the unexpected additional ability to deprive the kinase of access to the Hsp90 chaperone system, resulting in client kinase degradation. While formally we have only demonstrated this unanticipated mechanism for a few agents, chaperone deprivation by ATP-competitive kinase inhibitors is likely to be a general phenomenon amongst the large Hsp90-dependent subset of the kinome that is recruited via association with Cdc37 38. Disruption of Hsp90 association and/or degradation of c-Kit, ErbB2 and Bcr-Abl kinases have been reported following treatment with kinase inhibitors38–40. However the molecular basis for the phenomenon, and in particular the critical mechanism of Cdc37 antagonism we describe here, was not recognised.
The in vitro mode of action of protein kinase inhibitors such as vemurafenib, sorafenib, lapatinib and erlotinib, and the major driver in their medicinal chemistry design and development, is their ability to compete effectively with ATP for the binding site in the cleft between the N and C-lobes of the kinase domain of an oncogenic protein kinase. Given sufficiently high affinity, typically in the mid-low nanomolar range, their presumed mode of action as molecularly targeted antitumour drugs in vivo, is to prevent the target protein kinase accessing its ATP co-substrate, and thereby preventing phosphorylation of downstream targets that promote tumour cell division, growth, survival and other cancer hallmarks.
In our cellular assays, at concentrations and exposures that inhibited Mek/Erk, ErbB2 or Egfr phosphorylation, we find that ATP-competitive kinase inhibitors also promote substantial displacement of Cdc37 and Hsp90 from BRafV600E, ErbB2 or Egfr, with higher exposures not only inducing chaperone deprivation but also causing depletion of the client kinases in a very similar way to a bone fide Hsp90 inhibitor 7–10. Thus, the kinase inhibitors appear to have a dual mode of action, firstly as conventional ATP-competitive inhibitors, but also additionally as protein-protein interaction antagonists, blocking kinase recruitment to the Hsp90-Cdc37 molecular chaperone system – an interaction that may be essential for the kinase’s cellular stability. Consistent with this we find that manipulation of cellular Cdc37 is reflected in changes in the pharmacological efficacy of vemurafenib in BRafV600E-containing cancer cells. As ATP-blockade and kinase degradation consequent to chaperone deprivation both have the desired effect of decreasing downstream phosphorylation and signalling by the target kinase, unpicking the relative importance of these two activities of the present generation of ATP-competitive kinase inhibitors is difficult. However, as degradation of client protein kinases elicited by bone fide Hsp90 inhibitors is very effective in reducing downstream signalling in tumours and is responsible for their therapeutic effects on cancer cells 9,10, it would not be surprising if promotion of degradation following chaperone deprivation by ATP-competitive protein kinase inhibitors did not play some role in their antitumour activity.
Tumour responsiveness to BRaf inhibitors, in particular, is highly complex. Recent studies have shown that in human tumour xenograft models which acquired resistance following treatment with vemurafenib, the cancers actually became dependent on the drug for their continued proliferation, and regressed only when vemurafenib was withdrawn 41. Doses of vemurafenib that abolish detectable MAP kinase signalling in melanoma biopsies failed to elicit tumour regression and paradoxically promoted growth of keratocanthomas and squamous cell carcinomas in nearly a third of patients treated42. Very high levels of exposure, considerably in excess of those required to achieve pathway inhibition and >18-fold higher than the concentration required for chaperone deprivation in cell culture, were required to cause significant tumour regression in patients43. While BRaf protein levels were not reported in these clinical studies, the results are fully consistent with our observations that higher levels of vemurafenib exposure promote chaperone deprivation with consequent cellular destabilisation of BRaf protein. Thus, we hypothesize that chaperone deprivation and consequent destabilisation and degradation of the client oncoprotein may be an important component in the clinical response to vemurafenib and other ATP-competitive drugs that target Hsp90-dependent protein kinases. Clinical studies are required to test this intriguing possibility.
Online Methods
Expression and purification of proteins
Bacterial expression and purification of the solubilised His6-BRaf kinase domains (sBRaf and sBRafV600E)21, Mek1-His6 kinase domain45, His6-Hsp90β46, His6-Cdc37p and His6-Prescission-Cdc3747 have been previously described. The N-terminal Cdc37 deletion mutant NΔ30Cdc37 (amino acids 31-378) was expressed with an N-terminal, Prescission protease-cleavable His6-tag from pRSF in BL21(DE3) (Merck Millipore). The constitutively active BRaf mutant sBRaf T599E V600E S602D was expressed with an N-terminal, Prescission protease-cleavable His6-tag from pET17b in BL21(DE3). Cak1p was expressed with an N-terminal, TEV protease-cleavable His6-tag in S. cerevisiae. Briefly, all proteins were purified using a combination of metal affinity chromatography on Talon resin (Clontech), ion-exchange chromatography on HiTrapQ (GE Healthcare) or HiTrapS (GE Healthcare, for sBRaf, sBRafV600E and sBRaf T599E V600E S602D) and Superdex 75 or 200 gel filtration chromatography (GE Healthcare) in 20mM Tris pH 7.520°C, 200mM NaCl, 0.5mM EDTA, 0.5mM TCEP, 10% glycerol. If applicable, the His6-tag was cleaved off after the Talon column. All proteins were concentrated to ~10mg/ml and snap frozen in liquid nitrogen.
Phosphorylated Cdc37 was prepared by incubating 45μM Cdc37 with 9μM GST-CKII and 500μM ATP in TNET buffer (20mM Tris pH 7.520°C, 150mM NaCl, 0.5mM EDTA, 0.5mM TCEP) supplemented with 10mM MgCl2 at 30°C for 2h. GST-CKII was expressed from p3E (in house modified version of pGEX-6P-1) in Rosetta 2(DE3)pLysS (Merck Millipore) and purified via GSTrap FF (GE Healthcare) and Superdex 200 chromatography. The kinase was removed from the phosphorylation mixture by GSTrap FF chromatography and successful phosphorylation of Cdc37 was confirmed with western blotting using a phospho-Ser13-Cdc37 antibody35.
The BRaf-Cdc37-Hsp90β, BRafV600E-Cdc37-Hsp90β and ErbB2-Cdc37-Hsp90β complexes were expressed from pFBDM derivatives in Sf9 insect cells. Hsp90β was expressed with an N-terminal, Prescission protease-cleavable Strep-tag and its linker region (amino acids 217-279) was deleted. The kinase domains (BRaf constructs: amino acids 433-726, ErbB2 construct: amino acids 704-1029) were modified by an N-terminal, Prescission protease-cleavable His6-tag. The purification was performed similarly as described before6 with sequential Talon, Strep-Tactin Superflow (Quiagen) and Superdex 200 HR16/60 chromatography steps. The sBRaf-Cdc37-Hsp90β and sBRafV600E-Cdc37-Hsp90β complexes were expressed and purified accordingly, with the modifications that the Hsp90βΔ217-279 constructs were tagged with a C-terminal, Prescission protease-cleavable double Strep-tag and the Cdc37 construct with a C-terminal, Prescission protease-cleavable CBP (calmodulin binding peptide)-tag. The Sf9 expressed sBRaf was purified as a by-product of the sBRaf-Cdc37-Hsp90β complex purification.
BRaf-Cdc37-Sf9 Hsp90 and BRafV600E-Cdc37-Sf9 Hsp90 complexes were expressed and purified as described before6 in absence of sodium molybdate. After a Talon chromatography step, the ternary and binary complexes as well as small amounts of free BRaf or BRafV600E could be isolated by Superdex 200 HR 26/60 chromatography.
Superose 6 gel filtration experiments
20μM kinase, 20μM Cdc37 variant and 40μM Hsp90β were incubated at 4°C for 30min in a total volume of 150μl and applied to an analytical Superose 6 column (24ml, GE Healthcare) operated in TNET buffer at 4°C. Fractions of 500μl were analyzed by SDS-PAGE and Coomassie staining, 2μl of the input were used as loading control.
To test the influence of ATP-competitive kinase inhibitors on complex formation, vemurafenib (Selleck), sorafenib (Selleck), SB-590885 (Tocris) and debromohymenialdisine (Alexis Biochemicals) were used at 40μM and a final DMSO concentration of 2%. Kinase-inhibitor (a), kinase-Cdc37 (b) or kinase-Cdc37-Hsp90β (c) complexes were pre-incubated at 4°C for 30min, Cdc37 (a) or inhibitor (b and c) were added and incubated at 4°C for 30min (Fig. 3c and Supplementary Fig. 5d) or overnight (Fig. 3b and d and Supplementary Fig. 5a, b and e). The results for 30min or overnight incubation were very similar. 2% DMSO were used in control experiments. To partially disassemble an ErbB2-Cdc37-Hsp90β complex purified from Sf9 insect cells, 20μM of the complex were incubated with 1mM lapatinib (Selleck) at a final DMSO concentration of 2% at 4°C over night. The gel filtration fractions were analyzed by SDS-PAGE and silver staining.
Pull down assays
Pull down assays were performed in 20mM Tris pH 7.5, 150mM NaCl, 0.5mM TCEP, 5mM imidazole, 1mM MgCl2 (PD buffer) at 20°C. 100μl of equilibrated Talon resin were incubated in a total volume of 400μl with 50μg of His6-sBRafV600E or, for negative control, buffer (4°C, 60min). After washing with 200μl of PD buffer, the resin was incubated with Cdc37 at an 1.5 molar excess with regard to the loaded sBRafV600E (4°C, 90min). The resin was washed five times with 200μl of PD buffer supplemented with 0.1% NP-40. Elution was performed by boiling in 1x SDS gel loading buffer, and 5% of the protein inputs as well as 10% of the eluates were analyzed by SDS-PAGE.
To test the influence of nucleotides on the Cdc37-sBRafV600E complex, pull down assays were performed in PD buffer supplemented with 1mM ADP, 1mM ATP or 1mM AMPPNP.
Octet assay
The binding of Cdc37 to sBRafV600E in presence of ATP-competitive kinase inhibitors was analysed on an Octet Red96 instrument (FortéBio) at 26°C using 20mM Tris pH 7.520°C, 150mM NaCl, 0.5mM EDTA as buffer. The buffer-equilibrated anti-penta-His biosensors were loaded with 100μg/ml (3μM) His6-sBrafV600E for 400s, washed in buffer for 180s, incubated with 3.6μM sorafenib or vemurafenib (in 1% DMSO) or 1% DMSO for 60s and washed in buffer for 180s. Association and dissociation of 10μM Cdc37 were followed for 300s each. In a control experiment, the sensors were incubated with buffer instead of Cdc37. The Cdc37 association curves were corrected by the respective buffer curves.
Isothermal titration calorimetry and KD determinations
The heat of interaction between sBRafV600E and Cdc37 was measured on an ITC200 microcalorimeter (Microcal). Both proteins were in TNET buffer supplemented with 5% glycerol. Ten 3.8μl aliquots of 250μM sBRaf V600E were injected into 200μl of 20μM Cdc37 at 20°C. The heat of dilution was determined in a separate experiment by diluting sBRafV600E into buffer. The corrected data were fitted using a nonlinear least square curve-fitting algorithm (Microcal Origin) with three floating variables: stoichiometry, binding constant and change in enthalpy of interaction.
The heat of interaction between debromohymenialdisine and Cak1p was determined in an equivalent experiment. The buffer was 20mM Tris pH 820°C, 100mM NaCl, 0.5mM EDTA, 0.5mM TCEP, 2% DMSO. 15 2.5μl aliquots of 300μM Cak1p were injected into 200μl of 20μM debromohymenialdisine at 30°C. The respective heat of dilution was determined by diluting Cak1p into buffer.
MANT-ADP release assay
The release of MANT-ADP (Invitrogen) from sBRafV600E was measured with a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) at 20°C. All spectra were recorded in TNET buffer supplemented with 10mM MgCl2. 1μM sBRafV600E was pre-incubated with 120μM MANT-ADP at 4°C for 30min, 0, 0.15, 0.3, 0.45, 0.6, 0.75, 1 or 2μM of the Cdc37 construct were added and the spectra were recorded in triplicate after 1h (excitation wavelength 290nm, slit widths 5nm). Alternatively, sBRafV600E and Cdc37 were pre-incubated at 4°C for 30min, MANT-ADP was added, and the spectra were measured after 1h. Each experiment was repeated independently for at least three times.
The spectra were corrected as follows: The sBRafV600E and MANT-ADP spectra were corrected by a buffer spectrum. The sBRafV600E/MANT-ADP spectrum was corrected by the sBRafV600E spectrum. The spectra of the sBRafV600E/MANT-ADP/Cdc37 construct mixtures were corrected by the corresponding sBRafV600E/Cdc37 spectra.
To ensure that the Cdc37 constructs do not quench MANT-ADP fluorescence on their own, spectra of Cdc37 construct/MANT-ADP mixtures were measured and corrected by the corresponding Cdc37 construct spectra.
ATP-agarose elution assay
The ATP-agarose elution assay was performed in TNET buffer supplemented with 10mM MgCl2. 50μg of sBRafV600E were incubated with 100μl of equilibrated 2’/3’-EDA-ATP-agarose (immobilized 2’/3’-O-(2-Aminoethyl-carbamoyl)-ATP, Jena Bioscience) at 4°C for 3h in a total volume of 400μl. The resin was washed three times with 200μl of buffer and sBRafV600E was eluted for 40min at 4°C with 200μl 2% DMSO, 200μM vemurafenib or sorafenib (both in 2% DMSO), 100mM ATP/MgCl2, 3.75μM Cdc37 or NΔ30Cdc37 (molar ratio of 1:2 with regard to loaded sBRafV600E). 5% of the loaded sBRafV600E as well as 10% of the last wash fraction and the eluate were analyzed by SDS-PAGE.
BRaf kinase assay
sBRafV600E kinase activity was tested by incubating 10μM sBRafV600E or sBRaf T599E V600E S602D with 0.39μM Mek1, inactive (1100ng, Merck Millipore) and 20μM ATP/MgCl2 (Merck Millipore) at 30°C and 400RPM for 30min. The reaction was performed in a total volume of 40μl in 20mM MOPS pH 7.2, 25mM β-glycerol phosphate, 5mM EGTA, 1mM sodium orthovanadate, 1mM DTT. After SDS-PAGE and transfer to PVDF, the phosphorylation state of Mek1 was probed with a phospho-Mek1Ser218/222/Mek2Ser222/226 antibody (1h, RT, 05-747, Merck Millipore). Mek1 was visualized using a donkey anti-rabbit HRP-conjugated secondary antibody and an ECL plus western blotting detection system (GE Healthcare). To test the effect of inhibitors, sBRafV600E was pre-incubated with 250μM vemurafenib (final DMSO concentration 0.5%), 0.5% DMSO, 10μM Cdc37 or 10μM NΔ30Cdc37 at 4°C for 20min before addition of Mek1 and ATP. For negative control, the activity test was performed in absence of ATP.
Due to differences in their activities, the BRaf constructs were probed at different concentrations. While E. coli expressed sBRaf was poorly active at 20μM (data not shown), Sf9 expressed sBRaf was tested at 0.38μM and at inhibitor concentrations of 50μM vemurafenib (final DMSO concentration 0.1%), 0.1% DMSO, 5μM Cdc37, 5μM NΔ30Cdc37 and 5μM Cdc37/10μM Hsp90β.
The activities of Sf9-assembled BRaf, BRaf-Cdc37, BRaf-Cdc37-Sf9 Hsp90, BRafV600E, BRafV600E-Cdc37 and BRafV600E-Cdc37-Sf9 Hsp90 were measured at 75nM and at inhibitor concentrations of 50μM vemurafenib (final DMSO concentration 0.1%), 0.1% DMSO, 5μM Cdc37, 5μM NΔ30Cdc37, 5μM pS13-Cdc37, 5μM Cdc37/10μM Hsp90β and 5μM pS13-Cdc37/10μM Hsp90β. All proteins utilised for inhibition were expressed in E. coli. Mek1 was used at 0.18μM (500ng).
Cell experiments
Human HT29 colon cancer cells, BT474 breast cancer cells and SW48 colon adenocarcinoma cells were cultured as a monolayer in DMEM (Invitrogen) supplemented with 10% (v/v) foetal calf serum (PAA Laboratories), 2mM L-glutamine and non-essential amino acids. Cells were maintained at 37°C in a non-humidified incubator with 5% CO2. Sulforhodamine blue staining was used to determine the 50% growth inhibition value (GI50) for continuous exposure of the respective cells to vemurafenib, sorafenib, lapatinib, erlotinib (all from Selleck) or MG132 (Calbiochem) for 96h 48. To examine kinase:chaperone interactions, soluble proteins from control and drug-treated cells were extracted and co-immunoprecipitated from 350μg protein lysate in 150μl modified RIPA buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1% sodium deoxycholate, 1% NP-40 alternative, 0.02% SDS, complete protease inhibitor (Roche), phosphatase inhibitor cocktails 1 and 2 (Sigma)) with 1:1000 BRaf (F-7, Santa Cruz Biotechnology), Egfr (2232, Cell Signalling Technology) or ErbB2 (2242, Cell Signalling Technology) antibody overnight at 4°C. These conditions were established by exploring co-immunoprecipitation of a number of Hsp90 client proteins with Hsp90 using RIPA buffers with different detergent and salt concentrations and with or without the addition of molybdate. The chosen RIPA buffer conditions maintained the interaction between multiple client proteins and Hsp90 while preventing non-specific binding of Hsp90 to the protein G beads (data not shown). Antigen-antibody complexes were purified using Protein G sepharose, pre-blocked with 1% BSA/RIPA buffer, for 4h at 4°C. After washing of the beads with RIPA buffer, bound proteins were eluted by boiling in 1x SDS gel loading buffer and analysed by immunoblotting. Equal amounts of protein were separated by electrophoresis through SDS-PAGE and proteins were transferred onto nitrocellulose membranes using the Xcell 2 system and protocol (Invitrogen). Mek1/2 (9122), phospho-MekSer217/221(41G9), Erk1/2 (9102), phospho-Erk1/2Thr202/Tyr204 (9101), phospho-EgfrTyr1045 (2237) and phospho-ErbB2Tyr1221/1222 (2249) antibodies were from Cell Signalling Technology; Hsp90α/β (H-114), Cdc37 (H-271) and GAPDH antibodies were from Santa Cruz Biotechnology. All antibodies were used as recommended by the manufacturer and specific antigen-antibody interaction was detected with an HRP-conjugated secondary antibody using ECL western blotting detection reagents (GE Healthcare).
To test whether cellular BRafV600E is degraded by the proteasome upon vemurafenib exposure, HT29 cells were treated with 5x GI50 MG132 (+) or DMSO (-) for 1h prior to incubation with 10x GI50 vemurafenib (+) or DMSO (-). Cells were lysed in modified RIPA buffer at the indicated time-points. Following centrifugation at 17,000x g for 30min, the supernatant was collected as the soluble protein fraction. For collection of the insoluble protein, the remaining pellet was resuspended in modified RIPA buffer and sonicated for 10s at 4°C using an MSE Soniprep150.
Supplementary Material
Acknowledgments
We are grateful to Lihong Zhou for assistance with insect cell expression, to Rhodri M.L. Morgan, Sarah Parry-Morris, Stefan H. Millson, Charles G. Grummitt and Jennifer R. Smith for providing materials, and to Antony W. Oliver and Mark Roe for useful discussions. PW, PAC and RSS acknowledge funding from the National Health Service to the NIHR Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden Hospital. LHP acknowledges funding for the Genome Damage and Stability Centre from the Medical Research Council. This work was supported by Cancer Research UK Programme Grant C309/A8274 (PW), an EMBO Long Term Fellowship (SP), and a Wellcome Trust Senior Investigator Award (LHP). PW is a Cancer Research UK Life Fellow.
Footnotes
Author Contributions
SP designed the study, performed the in vitro experiments, analysed the data, prepared the figures and co-authored the manuscript. RSS and PAC performed the in vivo experiments and analysed the data. PW designed the study, analysed the data and co-authored the manuscript. CP designed the study, performed in vitro experiments and analysed the data. LHP designed the study, analysed the data, prepared the figures and co-authored the manuscript. All authors commented on the manuscript.
Competing Financial Interests
The authors have no competing financial interests.
References
- 1.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–94. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 2.Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Karnitz LM, Felts SJ. Cdc37 regulation of the kinome: when to hold 'em and when to fold 'em. Sci STKE. 2007;2007:22. doi: 10.1126/stke.3852007pe22. [DOI] [PubMed] [Google Scholar]
- 5.Roe SM, et al. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 6.Vaughan CK, et al. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 8.Schneider C, et al. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci U S A. 1996;93:14536–41. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581:3758–69. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 10.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410:439–53. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 11.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochimica et biophysica acta. 2012;1823:674–82. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Hunter T, Poon RYC. Cdc37: A protein kinase chaperone? Trends in Cell Biology. 1997;7:157–161. doi: 10.1016/S0962-8924(97)01027-1. [DOI] [PubMed] [Google Scholar]
- 13.Terasawa K, Minami Y. A client-binding site of Cdc37. Febs J. 2005;272:4684–90. doi: 10.1111/j.1742-4658.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q, Boschelli F, Caplan AJ, Arndt KT. Identification of a conserved sequence motif that promotes Cdc37 and cyclin D1 binding to Cdk4. J Biol Chem. 2004;279:12560–4. doi: 10.1074/jbc.M308242200. [DOI] [PubMed] [Google Scholar]
- 15.Prince T, Matts RL. Definition of protein kinase sequence motifs that trigger high affinity binding of Hsp90 and Cdc37. J Biol Chem. 2004;279:39975–81. doi: 10.1074/jbc.M406882200. [DOI] [PubMed] [Google Scholar]
- 16.Shao J, Irwin A, Hartson SD, Matts RL. Functional dissection of cdc37: characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry. 2003;42:12577–88. doi: 10.1021/bi035138j. [DOI] [PubMed] [Google Scholar]
- 17.Prince T, Matts RL. Exposure of protein kinase motifs that trigger binding of Hsp90 and Cdc37. Biochem Biophys Res Commun. 2005;338:1447–54. doi: 10.1016/j.bbrc.2005.10.100. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, et al. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat Struct Mol Biol. 2005;12:120–6. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 19.da Rocha Dias S, et al. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–91. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 20.Grbovic OM, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp SY, et al. Inhibition of the heat shock protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues. Mol Cancer Ther. 2007;6:1198–211. doi: 10.1158/1535-7163.MCT-07-0149. [DOI] [PubMed] [Google Scholar]
- 23.Johnson LN, Noble MEM, Owen DJ. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 24.Johnson LN. Protein kinase inhibitors: contributions from structure to clinical compounds. Quarterly reviews of biophysics. 2009;42:1–40. doi: 10.1017/S0033583508004745. [DOI] [PubMed] [Google Scholar]
- 25.Terasawa K, et al. Cdc37 interacts with the glycine-rich loop of Hsp90 client kinases. Molecular and Cellular Biology. 2006;26:3378–89. doi: 10.1128/MCB.26.9.3378-3389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–79. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Ni Q, Shaffer J, Adams JA. Insights into nucleotide binding in protein kinase A using fluorescent adenosine derivatives. Protein science : a publication of the Protein Society. 2000;9:1818–27. doi: 10.1110/ps.9.9.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 29.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyata Y, Nishida E. CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol Cell Biol. 2004;24:4065–74. doi: 10.1128/MCB.24.9.4065-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. Embo J. 2000;19:5429–39. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 33.King AJ, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66:11100–5. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 34.Farrell A, Morgan DO. Cdc37 promotes the stability of protein kinases Cdc28 and Cak1. Molecular and Cellular Biology. 2000;20:749–754. doi: 10.1128/mcb.20.3.749-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan CK, et al. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31:886–95. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JR, Clarke PA, de Billy E, Workman P. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene. 2009;28:157–69. doi: 10.1038/onc.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu W, et al. Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Molecular Cell. 2012;47:434–43. doi: 10.1016/j.molcel.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taipale M, et al. Quantitative analysis of hsp90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakatani H, et al. STI571 (Glivec) inhibits the interaction between c-KIT and heat shock protein 90 of the gastrointestinal stromal tumor cell line, GIST-T1. Cancer Sci. 2005;96:116–9. doi: 10.1111/j.1349-7006.2005.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Citri A, et al. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. Embo J. 2002;21:2407–17. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013 doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith JR. PhD Thesis; University of London: 2008. [Google Scholar]
- 45.Meier C, et al. Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting. Journal of Structural Biology. 2012;177:329–34. doi: 10.1016/j.jsb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Vaughan CK, Piper PW, Pearl LH, Prodromou C. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. Febs J. 2009;276:199–209. doi: 10.1111/j.1742-4658.2008.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siligardi G, et al. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. Journal of Biological Chemistry. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 48.Holford J, Sharp SY, Murrer BA, Abrams M, Kelland LR. In vitro circumvention of cisplatin resistance by the novel sterically hindered platinum complex AMD473. British journal of cancer. 1998;77:366–73. doi: 10.1038/bjc.1998.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.