Abstract
During the evolutionary progression of pancreatic ductal adenocarcinoma (PDAC), heterogeneous subclonal populations emerge that drive primary tumor growth, regional spread, distant metastasis, and patient death1–3. However, the genetics of metastases largely reflects that of the primary tumor in untreated patients, and PDAC driver mutations are shared by all subclones1. This raises the possibility than an epigenetic process might operate during metastasis. Here we detected striking epigenetic reprogramming of global chromatin modifications during the natural evolutionary history of distant metastasis. Genome-wide mapping revealed that global changes were targeted to thousands of large chromatin domains across the genome that collectively specified malignant traits, including euchromatin and large organized chromatin K9-modified (LOCK) heterochromatin. Remarkably, distant metastases co-evolved a dependence on the oxidative branch of the pentose phosphate pathway (oxPPP), and oxPPP inhibition selectively reversed malignant chromatin and expression states and blocked tumorigenicity. This suggests a model whereby linked metabolic-epigenetic programs are selected for enhanced tumorigenic fitness during the evolution of distant metastasis.
Despite significant progress in survival rates for most human cancers, PDAC remains nearly universally lethal and is projected to be the second-leading cause of cancer deaths in the western world by 20204. Primary PDACs have been shown to contain distinct subclonal populations. However, these subclones share identical driver mutations and the genetics of metastases largely reflects that of the primary tumor1. Furthermore, subclones are defined genetically by their unique progressor mutations, the vast majority if not all of which are thought to be passenger events1. This raises questions as to what mechanisms might drive progression and metastasis during the natural history of disease evolution.
One prometastatic candidate is epigenomic regulation. In particular, we wished to investigate the role of large-scale epigenomic changes during PDAC subclonal evolution and distant metastasis, especially within heterochromatin domains including large organized heterochromatin lysine (K)-9 modified domains (LOCKs)5 and large DNA hypomethylated blocks6. These regions could represent selectable targets for large-scale epigenetic reprogramming, since they occupy over half of the genome, partially overlap with one another, and are found in many human cancers including PDAC6–8. We therefore hypothesized that epigenomic dysregulation within these regions could be a major selective force for tumor progression, given the lack of any consistent metastasis-specific driver mutations.
RESULTS
Reprogramming of global epigenetic state during the evolution of distant metastasis
To test this hypothesis, we employed a large panel of uniquely matched primary and metastatic PDAC lesions from individual patients previously collected by rapid autopsy2,3. These samples represent a unique resource especially suited to study tumor evolution (detailed in the Supplementary Note), especially since the genetic progression of subclonal evolution has been defined by whole exome and sanger sequencing for coding mutations1,9, paired-end sequencing for rearrangements10, and whole genome sequencing for total mutations and copy number alterations11. Importantly, these samples were also chosen because no metastasis-specific driver mutations were identified1,11.
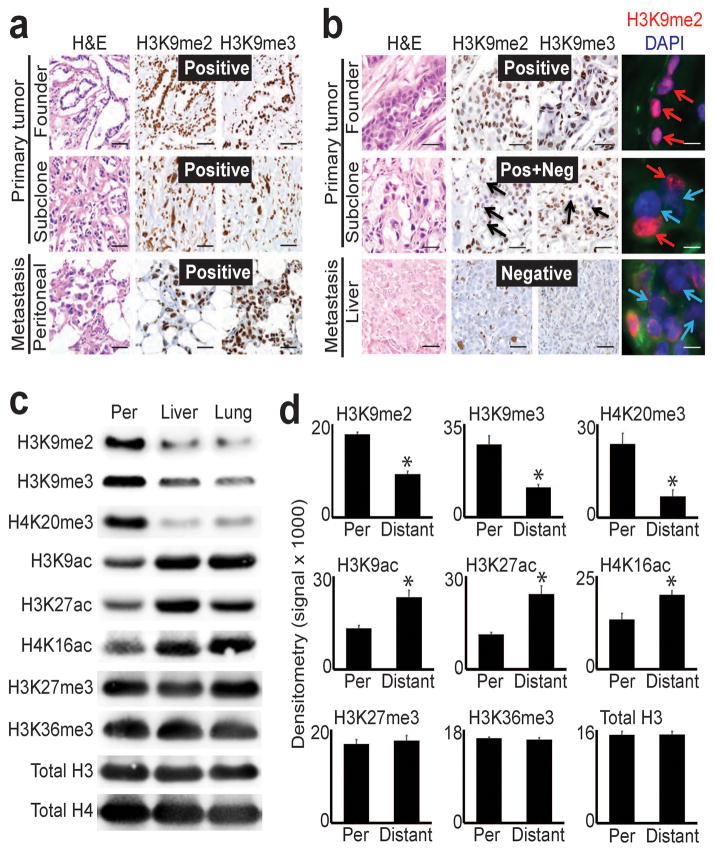
We began our analysis with immunostains performed on formalin-fixed patient tissue samples (totaling 16 uniquely matched, sequence-verified tumor sections from five patients). For each patient, sections were taken from different regions of the primary tumor that were occupied by distinct clones, including the common ancestor that founded the neoplasm (founder clone) and the subclone descendants that seeded metastasis. These were further matched to the corresponding metastases from the same patient(s), each of which shared identical driver mutations as the primary tumor (Supplementary Table 1). We performed immunostains against H3K9me2/3 across these sections to detect global changes from heterochromatin domains (including LOCKs5,12) that might be selectable targets during subclonal evolution. Immunostains revealed uniform, diffusely positive (>80%) H3K9me2/3 staining of PDAC nuclei across all tumor sections from patients with regional (peritoneal) metastatic disease (Fig. 1a and Supplementary Fig. 1a). In contrast, samples from patients who presented with widespread distant metastases (liver, lung) displayed progressive loss of H3K9me2/3 during subclonal evolution. This manifested as heterogeneous (positive + negative) staining in primary tumor subclones followed by either diffusely negative staining (<20%) or retention of heterogeneous staining in the metastases (Fig. 1b, Supplementary Fig. 1b). As in the individual patients above, we observed similar results for matched primary tumor subclones that seeded either peritoneal or distant metastases within the same patient (Supplementary Fig. 1c). Thus, global heterochromatin modifications remained stable during peritoneal spread. In contrast, global reprogramming initiated in primary tumor subclones that seeded distant metastases, and these changes were inherited or even accentuated in the metastases themselves.
Figure 1. Global epigenetic reprogramming during the evolution of distant metastasis.
(a) IHC stains against H3K9me2/3 performed on tumor sections as indicated for patient A124 (regional/peritoneal spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A124PrF; subclone: A124PrS; peritoneal metastasis: A124Per). Staining was diffusely positive (brown nuclei) across all tumor sections. (b) IHC and IF stains on tumor sections from patient A125 (distant metastatic spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A125PrF; subclone: A125PrS; liver metastasis: A125Lv1). There was progressive loss of nuclear staining that initiated in primary tumor subclones that seeded metastases (middle panels) with diffuse loss in the liver metastases (scale bars=100μm for IHC, 20μm for IF; IF colors: red=H3K9me2; blue=DAPI; green=vimentin). (c) Western blots on cells lines collected from a peritoneal subclone (Per) and distant (liver, lung) metastatic subclones from same patient (pA38, see Supplementary Fig. 1c for corresponding IHC) also showed reduced levels of heterochromatin modifications with increased acetylation in distant metastatic subclones compared to peritoneal. (d) Densitometry summary of western blot findings for the indicated histone modifications across cell lines from distant metastatic subclones compared to peritoneal carcinomatosis (derived from blots shown in Supplementary Fig. 1e–f, n=8 biological replicates, error bars: s.e.m., *p<0.01 by two-tailed t-tests).
To expand our analysis and test the generality of findings, we employed twelve low-passaged cell lines collected from eight rapid autopsy patients (Supplementary Table 1), including a subset that corresponded to the patient tissues (from Supplementary Fig. 1c). Cell lines were isolated from nine distant metastases, a peritoneal metastasis with matched liver and lung metastases from the same patient, and two primary tumor subclones matched to a lung metastasis from another patient. Importantly, six of the distant metastatic cell lines were previously whole exome sequenced9, and mutations were also present in the corresponding patient tissues1. PDAC cell lines from other sources of regional disease were also included: malignant ascites fluid from two patients (AsPC113, HPAFII14), and a primary tumor from a long-term survivor without distant metastases (Capan215).
We examined whether the global changes in chromatin detected in patient tissues were maintained in cell lines, which could reflect a reprogrammed epigenomic state. We began by performing western blots for eight histone modifications with well-understood functions16. Comparison of regional PDAC samples showed minimal or non-recurrent global changes across all histone modifications tested (Supplementary Fig. 1d), similar to the evolution of peritoneal disease observed in patient tissues. In contrast, distant metastases displayed striking reprogramming of methylation and acetylation that was targeted to specific histone residues (Fig. 1c–d, Supplementary Fig. 1e), including between matched peritoneal and distant metastatic subclones (Fig. 1c) that corresponded to patient tissues presented in Supplementary Fig. 1c. This manifested as recurrent reductions in H3K9me2/3 and H4K20me3 that was coupled to increased H3K9ac and H3K27ac in distant metastases (Fig. 1d). H3K9 methylation encodes large heterochromatin domains including LOCKs into the genome5,12,17,18, and H3K27ac encodes gene regulatory elements16. Reprogramming appeared specific, as we observed no consistent/recurrent changes in H3K27me3 or H3K36me3 across samples (Fig. 1d) and the reprogrammed modifications themselves were not dependent on proliferation rates or PDAC chemotherapy (Supplementary Fig. 2, Supplementary Table 1). Finally, western blots on cell lines isolated from matched primary tumor and distant metastatic subclones collected from a patient who presented with widespread distant metastases1,19 also showed reductions in H3K9me3 and H4K20me3 between primary tumor subclones that were inherited by the distant metastasis (Supplementary Fig. 1f), further suggesting that reprogramming initiated in primary tumor subclones. Thus, in vivo tissue and in vitro cell culture findings across the collective 30 patient samples strongly suggested that global epigenetic state was reprogrammed during the evolution of distant metastasis.
The epigenomic landscape of PDAC subclonal evolution
We next wished to map the locations of reprogrammed chromatin modifications across the PDAC genome. To this end, we comprehensively mapped the epigenetic landscape of PDAC evolution with chromatin immunoprecipitation followed by sequencing (ChIP-seq) for histone modifications with well-understood functions (heterochromatin: H3K9me2, H3K9me3, H3K27me3; euchromatin: H3K27ac, H3K36me3). To capture the evolutionary diversity of malignant progression, ChIP-seq was performed on sequence-verified cell lines isolated from a peritoneal metastasis (A38Per) matched to a lung metastasis (A38Lg) from the same patient1, and two primary tumor subclones (A13Pr1, A13Pr2) that were also matched to a lung metastasis (A13Lg) from the same patient1,19. We also performed RNA-seq in parallel to identify matched gene expression changes. Finally, we complemented these datasets with whole genome bisulfite sequencing (WGBS) across these cell lines and frozen tumor tissues that corresponded to a subset of the formalin-fixed tumor sections. In all, we generated 160 datasets with 12.7 x 109 uniquely aligned sequencing reads, including >15.0 x 106 (median: 32.3 x 106) uniquely aligned reads for each ChIP-seq experiment as recommended by ENCODE guidelines20. Experiments were performed as biological replicates, with strong correlation between replicates (Supplementary Table 2). To our knowledge, this represents the first comprehensive genome-wide analysis of epigenetic reprogramming during the evolutionary progression of a human cancer.
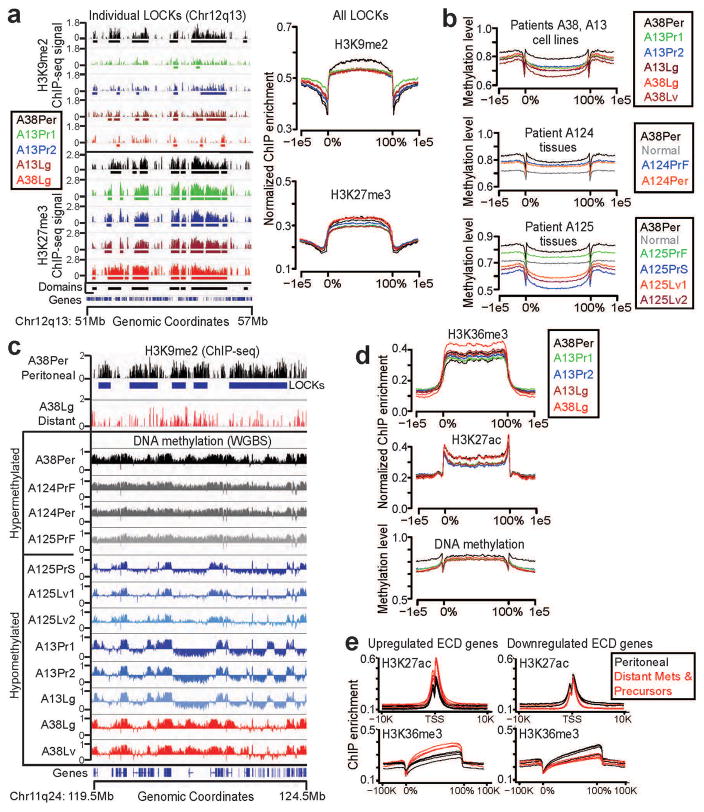
Because of the striking divergence in global epigenetic state between peritoneal and distant metastatic samples, we compared the peritoneal subclone against the distant metastases and their matched primary tumor subclones. This revealed a striking and unexpected degree of epigenomic reprogramming that was targeted to thousands of large chromatin domains. These included heterochromatin regions corresponding to LOCKs5 that occupied approximately half of the genome, gene-rich euchromatin domains (ECDs) that occupied a third of the genome, and a smaller subset of other very large LOCK domains. Domain characteristics are detailed in Supplementary Table 3, and comprehensive statistical tests of significance presented in Supplementary Table 4.
We first analyzed heterochromatin domains defined by broad enrichments of H3K9me2/3 and H3K27me3 (Fig. 2a, Supplementary Fig. 3a). Thousands of large (median lengths: 232Kb-311Kb), block-like heterochromatin domains were detected in each sample (detailed in the Supplementary Note), and these significantly overlapped with previously reported LOCK domains (avg: 76.7 +/− 16.9% overlap; p<0.01 by permutation testing). Domain calls were robust: varying threshold parameters across multiple sensitivity analyses produced nearly identical enrichments (Supplementary Fig. 3b) with overlapping genomic coordinates (Supplementary Table 4g). Similar to the immunostain and western blot data, we detected strong H3K9me2 enrichment across LOCKs in the peritoneal subclone, whereas these same regions displayed global reductions of H3K9me2 in the distant metastases and their matched primary tumor subclones (Fig. 2a, average: 591Mb/1,470Mb; X2 p-value<2.2e-16; Supplementary Table 4a). In contrast, high global levels of H3K27me3 were detected from these regions across all subclones (Fig. 2a), similar to western blot findings. We further detected patient-specific patterns of H3K9me3 reduction from reprogrammed LOCKs that also matched the western blot findings (Supplementary Note and Supplementary Fig. 4). Finally, we detected localized, reciprocal changes in H3K27ac and H3K9me2 over promoters coupled to similar reciprocal changes in H3K36me3 and H3K27me3 over gene bodies within LOCK genes that were differentially expressed (DE) between subclones (Supplementary Fig. 5), suggesting that DE genes resided within hybrid LOCK sub-regions with gene regulatory potential that resembled euchromatin islands (LOCK-EIs21).
Figure 2. Epigenomic reprogramming of chromatin domains during PDAC subclonal evolution.
(a) Representative (left panels) and total summarized (right panels) ChIP-seq experiments revealed loss of H3K9me2 from LOCKs between peritoneal (A38Per) and distant metastatic and primary tumor precursor subclones (others). H3K27me3 remained strong in all subclones. (b) WGBS data on cell lines (A38, A13, top panel) and frozen tissue samples (A124, A125 bottom panels) showed that samples from local regional spread and parental clones (A38Per, A124PrF, A124Per, A125PrF) possessed hypermethylated LOCKs. In contrast, distant metastatic subclones (A125Lv1/2, A13Lg, A38Lv, A38Lg) and their primary tumor subclones (A125PrS, A13Pr1, A13Pr2) showed hypomethylation of DNA across the same LOCK regions. (c) Examples of individual LOCKs displaying hyper- vs. hypomethylation across subclones, as described above. WGBS signals are plotted above (0.5–1.0) and below (0.0–0.5) a midpoint of 0.5 in IGV, which represents the background (basal) level of methylation across all samples. (d) Global levels of H3K36me3, H3K27ac, and DNA methylation within ECDs did not show any consistent differences between subclones. (e) In contrast, distant metastatic subclones and primary tumor subclones (red lines) displayed local reprogramming of H3K36me3 and H3K27ac specifically over DE genes within ECDs, compared to the same DE genic ECD regions from A38Per controls (black lines) compared against each subclone independently.
Because LOCKs correspond to a subset of block-like regions that are DNA hypomethylated in pancreatic and other human cancers6,7, we also asked whether DNA methylation changes were targeted to LOCKs during PDAC subclonal evolution. For this analysis, we performed WGBS on all of the cell lines with ChIP-seq data reported above. We also selected seven frozen tissue samples for in vivo WGBS that were matched to the same formalin-fixed tissues with IHC data presented in Fig. 1a–b. Normal pancreas was included with frozen tissue samples as an internal control. Similar to ChIP-seq for H3K9me2, these experiments revealed significant reductions in LOCK-wide DNA methylation across cell lines isolated from distant metastases relative to peritoneal carcinomatosis (Fig. 2b–c, Supplementary Table 5). Analysis of the same LOCK regions from the frozen tissue samples also revealed relatively high DNA methylation in LOCKs from patient A124 (peritoneal spread) and the founder clone from patient A125, while the primary tumor and distant metastatic subclone descendants displayed striking loss of DNA methylation (Fig. 2b–c, Supplementary Table 6). We also detected localized DNA hypomethylation from down-regulated DE genes in the hybrid LOCK-EI sub-regions, while up-regulated genes remained hypermethylated with sharp dips at the 5′-ends of genes, similar to H3K9me2 (Supplementary Fig. 5). Thus, DNA methylation was globally and locally reprogrammed across LOCKs from primary tumor and distant metastatic subclones, similar to histone modifications. Based on the collective immunostain, western blot, ChIP-seq, and WGBS data (summarized in Supplementary Table 1), we conclude that a substantial fraction of global reprogramming events was targeted to heterochromatin domains (LOCKs) during the evolution of distant metastasis.
We next analyzed reprogramming within ECDs, which were defined by enrichments for global euchromatin modifications H3K27ac and H3K36me3 with depletion of heterochromatin modifications (Supplementary Fig. 6a). Similar to heterochromatin, we detected thousands of large block-like ECDs in each sample (Supplementary Note), and ECD calls were also robust by sensitivity analyses (Supplementary Fig. 6b). Globally, all subclones displayed broad H3K36me3 signals over gene bodies that were flanked by sharp peaks of H3K27ac and dips in DNA methylation at gene regulatory elements, consistent with actively transcribed euchromatin (Fig. 2d). However, mapping DE genes from RNA-seq data to ECDs identified clear patterns of local reprogramming within chromatin encoding these genes (Fig. 2e). Genes up-regulated from ECDs acquired increased levels of both H3K36me3 and H3K27ac, which could reflect a permissive chromatin state or hyperactive transcription. In contrast, down-regulated genes displayed greatly reduced H3K36me3 with only modest reductions in H2K27ac, which could reflect an inactive yet poised chromatin state or direct transcriptional repression. Thus, reprogramming in ECDs was largely localized to chromatin encoding DE genes.
Finally, we detected patient-specific reprogramming targeted to a unique subset of very large LOCK domains (median lengths: 730Kb–1,340Kb, Supplementary Fig. 7). Although these regions were situated within DNA hypomethylated blocks similar to other LOCKs6,7, they differed in several other respects. As detailed in the Supplementary Note, these domains were substantially larger than other LOCKs, they were strongly enriched with H3K9me322 yet depleted of H3K9me2/H3K27me3, and reprogramming was characterized by striking loss of H3K9me3 coupled to increased H3K9me2 and DNA methylation. Although the functional significance of these findings is uncertain, they strongly correlated with patterns of genome instability (outlined below).
Reprogrammed chromatin domains specify malignant heterogeneity
Subclonal evolution may generate significant phenotypic heterogeneity within an individual patient23. We therefore wished to investigate in-depth whether reprogrammed chromatin domains might encode heterogeneous malignant properties between PDAC subclones from the same patient. To this end, we selected matched subclones from the same patient (A38Per and A38Lg), performed gene ontology (GO) analyses on reprogrammed LOCK and ECD genes that were differentially expressed between the subclones (Supplementary Tables 7–11), and tested whether GO results matched actual phenotypic differences with experimental assays.
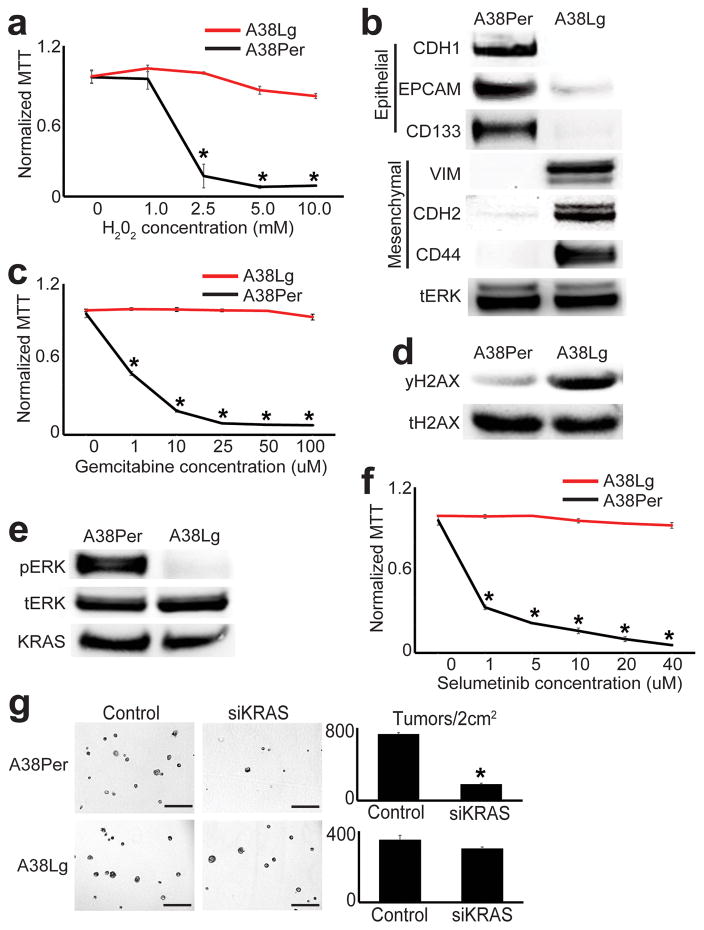
This revealed that reprogrammed LOCKs and ECDs encoded substantial malignant divergence. First, a large number of DE genes with oxidoreductase activity were up-regulated from LOCKs in A38Lg (Supplementary Table 8). This subclone was accordingly highly resistant to H202-mediated oxidative stress (Fig. 3a), and possessed higher oxidoreductase activity and NADPH levels than A38Per (Supplementary Fig. 8a–b). Second, genes encoding differentiation state (epithelial vs. mesenchymal) were reciprocally expressed from A38Per and A38Lg LOCKs (Supplementary Table 9, Fig. 3b). Consistent with this, A38Per maintained well-differentiated (epithelial) morphology while A38Lg was poorly differentiated (EMT-like) across multiple in vitro culture conditions (Supplementary Fig. 8c), and EMT emerged in the primary tumor subclone that seeded the A38Lg metastasis in vivo (Supplementary Fig. 8d). Immune-related genes were also differentially expressed from reprogrammed LOCKs (Supplementary Table 9), which could hold implications for PDAC immunotherapy. Third, genes involved in DNA repair were up-regulated from ECDs in A38Lg, including genes crucial for maintenance of genome integrity (e.g. Fanconi anemia complex, non-homologous end joining, Supplementary Table 10). This subclone was accordingly highly resistant to PDAC chemotherapy (Fig. 3c), and western blots showed hyper-phosphorylation of the DNA repair target histone H2AX S139 (γH2AX, Fig. 3d). Fourth, genes involved in oncogenic signal transduction were down-regulated in ECDs from A38Lg, especially KRAS/ERK-related genes (Supplementary Table 11). Indeed, A38Lg showed loss of phosphorylated ERK (Fig. 3e), resistance to ERK inhibition (Fig. 3f), and minimal response to knockdown of oncogenic KRAS in 3D tumor forming assays (Fig. 3g, Supplementary Fig. 8e–f), despite possessing identical KRASG12V mutations as A38Per1. Finally, mapping previously reported rearrangements from this patient10 to chromatin domains revealed that rearrangements were preferentially targeted to ECDs and the small subset of large LOCK domains, whereas other LOCKs were strongly depleted (Supplementary Fig. 9).
Figure 3. Reprogrammed chromatin domains encode divergent malignant properties.
(a) A38Lg was remarkably resistant to H202 treatments compared to A38Per. MTT signals reflect cell viability normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.03 by two tailed t-tests). (b) Western blots for proteins involved in epithelial and EMT differentiation were differentially expressed between A38Per and A38Lg, as predicted by GO analyses of reprogrammed DE genes from LOCKs. (c) A38Lg was highly resistant to gemcitabine compared to A38Per, as predicted by GO analyses of reprogrammed DE genes from ECDs. MTT signals reflect cell viability normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). (d) A38Lg possessed elevated levels of γH2AX by western blot, consistent with activation of DNA repair pathways. (e) Western blots showed that A38Lg lost hyper-phosphorylated ERK and (f) was resistant to ERK targeted therapy, compared to A38Per. MTT signals reflect cell viability, normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.03 by two tailed t-tests). (g) A38Lg also lost sensitivity to KRAS knockdown by matrigel 3D tumor forming assays, compared to A38Per (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
Thus, reprogrammed chromatin domains collectively specified malignant gene expression programs, divergent phenotypic properties, and patterns of genome instability that emerged during subclonal evolution in patient A38. We note that a SMARCA2 point mutation of unclear significance was present in A38Lg (CID, unpublished observations), raising the possibility that some epigenetic and/or phenotypic changes could have occurred downstream of this mutation. Although the nature and extent of such findings will certainly vary among patients, they imply that PDAC is capable of acquiring substantial epigenetic and malignant diversity during subclonal evolution, even in the same cancer from the same patient.
Anabolic glucose metabolism controls epigenetic state and tumorigenicity
We next asked whether a recurrent, metastasis-intrinsic pathway might have been selected for during subclonal evolution to exert upstream control over global epigenetic state and tumorigenic potential. Several recent studies have linked metabolism to global levels of histone modifications24–29. Because PDAC distant metastases are largely tropic for organs (liver, lung2,3) that provide a rich supply of glucose, we asked whether these subclones might have evolved a dependence on specific aspects of glucose metabolism.
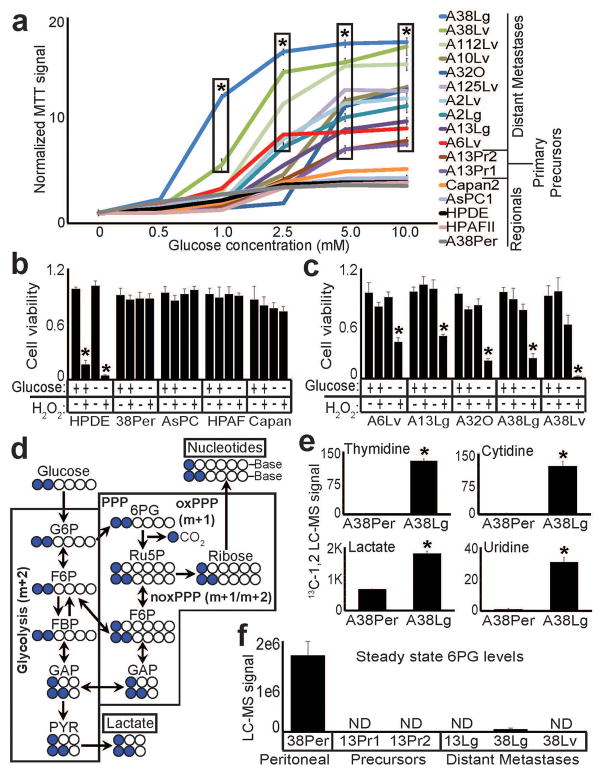
Altered glucose metabolism (Warburg effect) is a well-known property of neoplastic and highly proliferative cells30,31. Although most of our metastatic subclones actually displayed modest proliferative rates in culture (e.g. Supplementary Fig. 2) and in vivo1, we nonetheless asked whether distant metastases might have acquired further adaptations in glucose metabolism. Surprisingly, relative to immortalized pancreatic ductal epithelial (HPDE) cells and regional PDAC samples, glucose strongly stimulated metabolic (oxidoreductase) activity across distant metastatic subclones (Fig. 4a), and glucose was accordingly required for these subclones to withstand oxidative stress (Fig 4b–c). Distant metastases also hyper-consumed glucose, as we detected elevated glucose uptake and lactate secretion in distant metastases and their precursors (Supplementary Fig. 10a). To determine if excess glucose uptake was specifically incorporated into downstream metabolic pathways, we selected matched peritoneal and distant metastatic subclones from the same patient, incubated them with 13C[1–2]-labeled glucose, and measured glucose incorporation into metabolic products with liquid chromatography followed by high resolution mass spectrometry (LC-HRMS). These experiments revealed elevated incorporation of both 13C1- and 13C1,2-labeled glucose into lactate and nucleotides in the distant metastasis (Fig. 4d–e), consistent with enhanced glucose entry into both glycolysis and the pentose phosphate pathway (PPP).
Figure 4. Hyperactive glucose metabolism and 6PG depletion in distant metastatic subclones.
(a) MTT assays performed on equal numbers of viable, growth-arrested cells from the indicated subclones showed elevated signal (oxidoreductase activity) across distant metastatic subclones, compared to HPDE and regional PDACs (n=4 technical replicates, error bars: s.d.m., *p<10−5 by two tailed t-tests). (b) Normalized cell counts for the indicated samples incubated with (+) or without (−) 10mM glucose and treated with 1mM H202 as indicated (+,−) showed that HPDE cells were sensitive to H202 under either glucose condition (as expected), whereas regional PDAC samples were resistant to H202 irrespective of glucose availability (n=3 technical replicates, error bars: s.d.m). (c) In contrast, distant metastatic subclones were sensitive to H202 when glucose was not present in the media (n=3 technical replicates, error bars: s.d.m, *p<0.001 by two tailed t-tests). (d) Simplified schematic of 13C-(1,2)-labeled glucose flow through glycolysis and the PPP. Glucose that enters the oxidative branch of the PPP has one labeled carbon cleaved during conversion of 6PG to Ru5P (m+1), whereas glucose that travels through glycolysis retains both labeled carbons (m+2). Note that cross-talk allows glucose with either labeling pattern to re-enter the other pathway and incorporate. (e) LC-MS for nucleotides and lactate showed that these downstream metabolites acquired greatly elevated 13C-1,2 labels from glucose in A38Lg (n=3 biological replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). (f) Steady state LC-HRMS measurements for 6PG showed either complete (ND: not detected) or near complete loss of metabolite across distant metastases and their precursors.
We next asked whether distant metastases might have evolved a dependence on specific enzymatic steps in either of these glucose-driven pathways, which we hypothesized would manifest as severe depletion of metabolite substrate secondary to hyper-consumption. To test this, we surveyed glycolytic and PPP metabolite profiles across a diverse panel of samples including HPDE cells, peritoneal and distant metastases, and primary tumor precursors. Analysis of all detected glycolytic and pentose phosphate metabolites (Supplementary Fig. 10b) revealed a striking, recurrent depletion of 6-phosphogluconic acid (6PG) across distant metastases and their precursors (Fig. 4f). 6PG is the substrate for 6-phosphogluconate dehydrogenase (PGD), an enzyme involved in anabolic glucose metabolism that operates within the oxidative branch of the PPP.
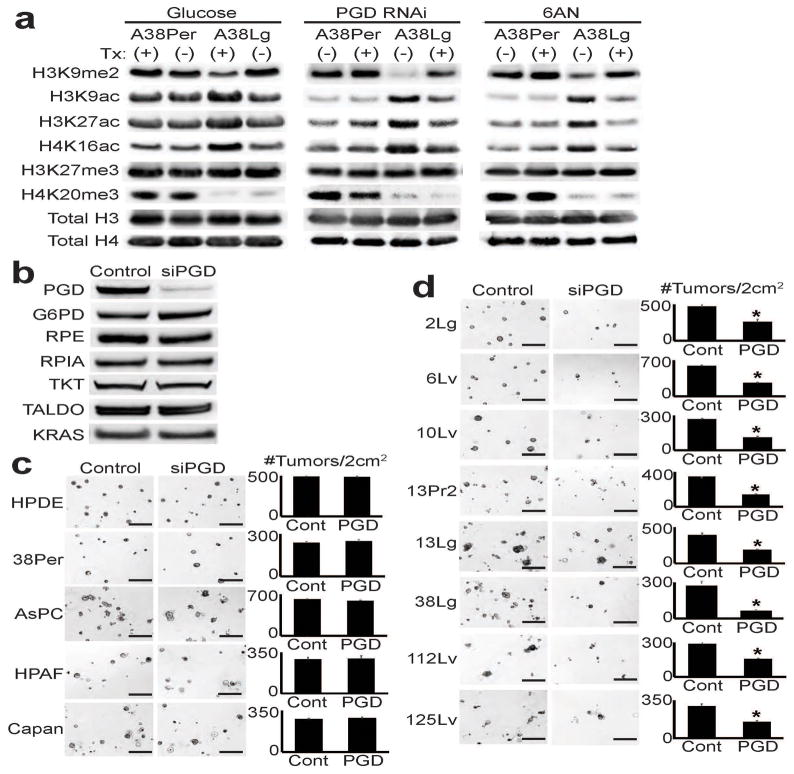
Glucose may enter the PPP via the oxidative (oxPPP) or the non-oxidative (noxPPP) branch of the pathway, which are thought to be uncoupled22. Although some studies have suggested that PGD is an important oncogene32, it is KRAS-mediated noxPPP activation that drives primary tumor growth in mouse models of PDAC22. Because KRAS and other driver mutations are acquired early in PDAC progression and shared by all subclones that evolve thereafter1, we hypothesized that PGD dependence might have been selected for during the evolution of distant metastasis. Glucose deprivation, RNAi against PGD, and 6-aminonicotinamide (6AN, a nicotinamide antimetabolite33 prodrug reported to preferentially inhibit PGD34) had no effect on global chromatin modifications in the peritoneal subclone, while all treatments reversed the reprogrammed chromatin state of the distant metastasis from the same patient (Fig. 5a). PGD loss-of-function appeared specific, as PGD knockdown did not alter expression of KRAS or other PPP components (Fig. 5b).
Figure 5. PGD-dependence in distant metastatic subclones.
(a) Western blots against indicated histone modifications performed on peritoneal (A38Per) and distant metastatic (A38Lg) subclones from the same patient showed that global levels of reprogrammed H3K9me2 and acetylation in A38Lg were reversed by removal of glucose from the media (left panel), PGD RNAi (middle panel), and 6AN treatments (right panel). (b) Western blots on A38Lg indicated that PGD knockdown by RNAi did not perturb expression of other PPP components or KRAS. (c) PGD RNAi did not affect the ability of HPDE cells or regional PDAC samples to form tumors in 3D matrigel assays (representative photomicrographs shown with quantified numbers of tumors/well, n=4 technical replicates, error bars: s.d.m.). (d) In contrast, PGD RNAi significantly reduced tumor formation across distant metastatic subclones (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
We next asked whether PGD knockdown might affect intrinsic tumor forming capacity across a larger panel of subclones. Despite their aggressive behavior in patients, distant metastatic subclones were unable to effectively form metastatic tumors in immunodeficient mice, and PGD RNAi was not toxic in routine 2D cultures (data not shown). To bypass these limitations, we treated cells with RNAi and used 3D matrigel tumor-forming assays35 to measure the effects of PGD knockdown on intrinsic tumor-forming capacity. PGD RNAi had minimal effect on the ability of HPDE cells to form spheres or regional PDACs to form tumors (Fig. 5c). In contrast, PGD RNAi universally interfered with the ability of distant metastatic subclones to form tumors (Fig. 5d). These findings suggested that PGD might represent a therapeutic target with selectivity for distant metastases. Because 6AN could represent a lead compound for targeted therapies, we stringently tested it for activity against distant metastases.
6AN treatments slowed rates of glucose consumption and lactate secretion with no effect on glutamine consumption or glutamate secretion in distant metastatic and precursor subclones (Supplementary Fig. 11a), and 6AN reversed the previously detected high incorporation of glucose into lactate and nucleotides (Supplementary Fig. 11b). Furthermore, steady state levels of glucose and metabolites directly upstream of the PGD reaction were dramatically elevated in response to 6AN with corresponding reductions in downstream metabolites (Supplementary Fig. 11c), consistent with strong PGD inhibition34.
We next examined our RNA-seq datasets to explore whether the above metabolic changes might be coupled to malignant gene expression programs (Supplementary Table 7h–i). Remarkably, over half (1032/1968, 52%, X2 p-value<2.2e-16) of 6AN down-regulated genes from A38Lg corresponded to genes that were over-expressed in this subclone (compared to A38Per). A large fraction of 6AN up-regulated genes also matched DE genes that were repressed (915/2192, 42%, X2 p-value<2.2e-16). Even more striking, nearly one-third (255/891, 29%, X2 p-value<2.2e-16) of recurrently over-expressed genes across distant metastatic subclones were down-regulated by 6AN. The magnitude of 6AN expression changes was partial when compared to A38Per transcript levels as reflected by modest correlation coefficients (r=0.38, Supplementary Fig. 11d). This suggested that 6AN might quantitatively target key DE genes important to maintain malignancy, rather than reversing differentiation state. Indeed, GO analyses revealed that 6AN specifically targeted recurrently over-expressed genes with malignant functions, including those involved mitotic cell cycle progression, protein acetylation, chromosome stability, DNA repair, and biosynthetic capacity (Supplementary Tables 12–14).
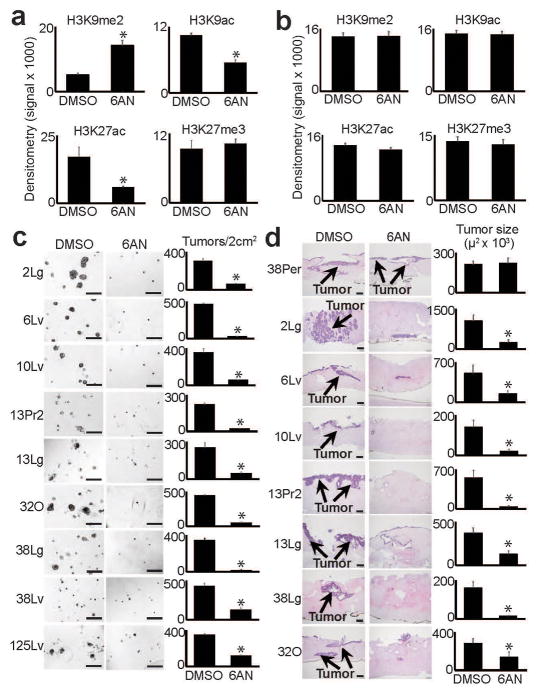
The gene expression changes prompted us to test the effects of 6AN on epigenetic state. 6AN treatments quantitatively reversed several reprogrammed chromatin modifications across distant metastatic subclones with minimal effect on normal cells or regional PDACs (Supplementary Fig. 12a–b; summarized in Fig. 6a–b), and this effect persisted upon removal of 6AN from the media (Supplementary Fig. 12c). Because these changes mirrored aspects of LOCK reprogramming, we examined the chromatin state of LOCK DE genes regulated by 6AN. This revealed that these DE genes were located within the reprogrammed hybrid LOCK-EI regions (Supplementary Fig. 13a). ChIP-seq experiments on control and 6AN-treated A38Lg cells further showed that the quantitative increase of global H3K9me2 was targeted to LOCK regions that were reprogrammed in A38Lg vs. A38Per (Supplementary Fig. 13b), while reduced H3K27ac was specifically targeted to genes repressed from LOCKs (Supplementary Fig. 13c). Similar to gene expression, this also reflected a partial (quantitative) reversal (r=0.49, Supplementary Fig. 13d). Levels of H3K27me3 remained stable across all regions in response to 6AN (Supplementary Fig. 13b–e), similar to western blot findings. Collectively, these experiments demonstrated that 6AN selectively and quantitatively targeted chromatin changes that emerged during the evolution of distant metastasis.
Figure 6. Reversal of reprogrammed chromatin and tumorigenic potential by 6AN.
(a) Densitometry summary of western blots shown in Supplementary Fig. 12a (n=8 biological replicates, error bars: s.e.m., *p≤0.01 by two tailed t-tests). 6AN selectively reversed reprogrammed H3K9me2 and H3K9/27ac. (b) Densitometry summary of western blots shown in Supplementary Fig. 12b (n=6 biological replicates, error bars: s.e.m.). 6AN had minimal effects on histone modifications across normal (HPDE, fibroblast) or regional PDAC samples. (c) 6AN ablated tumor formation in 3D matrigel assays (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm. (d) 6AN also blocked the ability of distant metastatic subclones to form tumors when injected into 3D organotypic stromal cultures (n=3 technical replicates, error bars: s.d.m., *p<0.05 by two tailed t-tests). Scale bars: 200μm.
Because 6AN appeared to target malignant epigenetic programs, we hypothesized that it might also selectively interfere with tumorigenic capabilities in distant metastatic subclones. Strikingly, 6AN selectively and strongly blocked tumor formation in distant metastatic and primary tumor precursor subclones but not regional PDACs across multiple 3D tumorigenic experimental platforms, including suspension tumorsphere assays36 (Supplementary Fig. 14), matrigel tumor forming assays35 (Fig. 6b), and injection of PDAC cells into organotypic stroma that recapitulates aspects of in vivo patient tumors37,38 (Fig. 6c). Thus, like PGD knockdown, chemical inhibition of PGD by 6AN selectively blocked the tumorigenic potential of distant metastases.
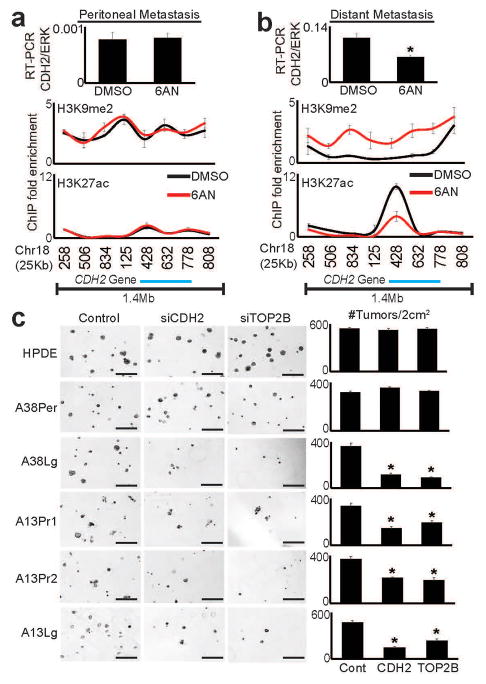
The above findings suggested that tumorigenicity might be mediated in part through metabolic-epigenetic control of malignant gene expression. To more directly test this possibility, we selected two cancer gene candidates (Supplementary Note) that were recurrently over-expressed and selectively down-regulated by 6AN: N-cadherin (CDH2) and topoisomerase 2β (TOP2B). ChIP-qPCR performed on 6AN and control treated cells from matched peritoneal and distant metastatic subclones from the same patient showed minimal changes of H3K9me2 and H3K27ac across these gene loci in the peritoneal subclone (Fig. 7a and Supplementary Fig. 15a). In contrast, 6AN induced broad enrichment of H3K9me2 across both loci in the distant metastasis with concordant reductions in H3K27ac over the CDH2 gene itself (Fig. 7b and Supplementary Fig. 15b). Loss of expression was important functionally: knockdown of CDH2 and TOP2B with RNAi partially blocked 3D tumor formation in subclones that over-expressed these genes, with minimal effect on HPDE cells or the peritoneal subclone (Fig. 7c). These findings further supported the hypothesis that tumorigenic capacity was selectively maintained through metabolic-epigenetic control of malignant gene expression programs.
Figure 7. CDH2 and TOP2B are epigenetic targets of 6AN that selectively regulate tumorigenic potential.
(a) Real-time RT-qPCR (top panel) and ChIP-qPCR for H3K9me2 and H3K27ac showed that 6AN had minimal effect on CDH2 mRNA expression or epigenetic state (H3K9me2/H3K27ac) over the CDH2 locus in the peritoneal subclone. Numbers along the x-axis of ChIP panels correspond to coordinates of the ChIP-PCR primer positions across the 1.4Mb CDH2 locus (Chr18q 27Mb:primer position). (b) In contrast, RT-PCR (top panel) showed that the distant metastatic subclone (A38Lg) from the same patient over-expressed CDH2 relative to A38Per (compare y-axis values), and that expression was repressed by 6AN (n=4 technical PCR replicates from two biological replicate experiments, error bars: s.d.m., *p=0.002 by two tailed t-tests). ChIP assays further showed that 6AN induced enrichment of H3K9me2 across the 1.4Mb CDH2 locus with corresponding reductions of H3K27ac directly over the genic region (n=2 biological replicates, error bars: s.e.m.). (c) RNAi against both CDH2 and TOP2B (Supplementary Fig. 15c–d) selectively blocked 3D tumor formation in distant metastatic and precursor subclones that over-expressed these genes by RNA-seq, with no effect on A38Per or HPDE cells (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
DISCUSSION
The first major result of this study was widespread epigenetic reprogramming during the evolution of distant metastasis in the absence of metastasis-specific driver mutations. This manifested as global reprogramming of histone H3K9 and DNA methylation within large heterochromatin domains (LOCKs) as well as regional changes in gene regulatory modifications. Second, these changes specified several divergent malignant properties that emerged during subclonal evolution. Third, it was the PGD step of the oxPPP that controlled reprogrammed chromatin and tumorigenicity in distant metastatic subclones. Collectively, these results suggest that a non-genetic form of natural selection may have emerged during the evolution of PDAC progression, and raise several important mechanistic questions for further study (see Supplementary Note).
Our findings in conjunction with deep sequencing studies on many of the same samples reported here1,11 suggest a model whereby driver mutations arise early to initiate PDAC tumorigenesis, followed by a period of subclonal evolution that generates heterogeneous metabolic, epigenetic, and malignant properties. Like driver mutations, those properties that confer increased fitness to cells that acquire them may be selected for and clonally expanded during invasive tumor growth and metastatic spread. The strong oxPPP dependence we observe in distant metastatic subclones could reflect such selection: distant metastatic sites provide ample glucose to fuel the pathway, pathway products enhance biosynthesis and reduce reactive oxygen species, and the pathway itself is coupled to epigenetic programs that promote tumorigenesis. As such, reversal of malignant epigenetic programs by targeting the oxPPP could represent an effective therapeutic strategy for metastatic PDAC, one of the most lethal of all human malignancies.
ONLINE METHODS
Rapid autopsy samples and cell culture conditions
Tissue samples and cell lines were previously collected from PDAC patients by rapid autopsy, sequenced-validated, and monitored for mycoplasma as previously described1–3. Low passage (2–17) rapid autopsy cell lines were cultured at 37°C in DMEM with 10% fetal bovine serum (FBS, Gibco). For serum starvation experiments, cells were washed twice with PBS and incubated in DMEM without FBS.
For MTT assays, 15,000 cells/well were plated into 96 well plates in quadruplicate, treated 12 hours later, and assayed after 24hr (glucose responses) or 6 days with one media change at day 3 (chemotherapy) with CellTiter96 (Promega). For glucose response assays, nutrient-deplete DMEM (no glucose, glutamine, pyruvate, or serum) was used with addition of glucose as indicated. For glucose-dependent oxidative stress analysis, cells plated in triplicate (in 24 well plates) and grown to 80% confluence followed by incubation in nutrient-deplete DMEM containing 10% dialyzed FBS with or without 10mM glucose and 1mM H202 for 24 hours. Cells were then washed with PBS, trypsinized, and viable cells counted with a hemocytometer. For 6AN treatments in 2D, cells were grown to 70–80% confluency and treated for 3 days with 250uM 6AN or DMSO negative control. We note that similar results have been obtained in 2D and 3D experiments using much lower concentrations of 6AN (e.g. 10uM to 25uM, data not shown). The higher concentrations were used primarily to obviate any need for replenishment during long-term experiments and to ensure that negative controls (regional PDACs) were not sensitive to 6AN even at the higher concentrations due to off-target or nonspecific effects.
RNAi experiments were performed with siRNA transfections (Oligofectamine, Life Technologies) according to manufacturer protocols. Cells were incubated with negative control siRNA (Sigma, SIC001) and pre-designed siRNA oligonucleotides against indicated genes in parallel (Sigma, PGD: SASI_Hs02_00334150, CDH2: SASI_Hs01_00153995, TOP2B: SASI_Hs02_00311874). siRNAs against mutant KRASG12V (CUACGCCAACAGCUCCAAC) were custom designed. Cells were incubated with siRNAs for 4 days after transfection, harvested, and used for downstream experiments.
3D tumor forming assays
3D matrigel assays were adapted from ref.4. Briefly, 2D cultures were trypsinized into single cells, 4,000–8,000cells/mL were suspended and thoroughly mixed in ice-cold DMEM containing 5% matrigel (BD systems) and 2% FBS (+/− DMSO/6AN as needed), 500ul plated in quadruplicate into 24 well ultra-low attachment plates, and incubated for at least 7–21 days to allow tumor growth (depending on the cell line). Well-formed tumors were then counted and representative photographs taken with an EVOS instrument. 3D suspension tumorsphere assays were performed with 20,000 starting cells/well in ultra-low attachment 6 well plates as described5, and tumors counted/photographed after at least 7 days of tumor growth.
Organotypic tumor forming assays were adapted from refs6,7. Briefly, 6 well permeable transwell plates (Costar 3414) were overlayed with 1mL type 1 rat tail collagen (Fisher) containing 10X DMEM (acellular layer). Human dermal fibroblasts (ATCC) were suspended (12x106 cells/mL) in a mixture of ice cold 10X DMEM, 10% FBS, 52.5% collagen, and 17.5% matrigel (cellular layer), thoroughly mixed, and 2mL/well plated over the acellular layer. The mixture was allowed to partially solidify for approximately 15 minutes at 37°C, followed by triplicate injection of 1x106 PDAC cells (suspended in 20ul DMEM) into the partially solidified cellular layer, which holds the cell suspension in place to allow subsequent tumor formation. Cells were incubated for 24 hours in fibroblast growth media above and below the inserts to initiate contraction of the discs. Fresh media with DMSO or 6AN was then added and replenished every 2–3 days for 6 additional days, followed by addition of DMEM with DMSO or 6AN underneath the inserts (no media on the top) for an additional 7 days. Discs were harvested, fixed overnight in 10% formalin, inked for orientation, thinly sectioned, and submitted to the Vanderbilt core histology lab for paraffin embedding and H&E staining. Tumor slides were photographed and measured with an Olympus BX53 microscope using CellSens Standard software.
Protein, nucleic acid, and IHC experimental conditions
Histones were acid extracted as described8 and western blots performed on 3.5ug histones, which were checked by Ponceau stains prior to western blot to ensure equal loading. Visualized histones were excised from the membrane and incubated overnight with primary antibody in TBS-T and 5% BSA at 4°C. Antibodies are listed in Supplementary Table 15. Blots were imaged with a Syngene G:box instrument. Densitometry on western blot images was performed with ImageJ software. RNA was extracted with Trizol reagent (Life Technologies) and isopropanol precipitated. Genomic DNA for WGBS was purified with MasterPure DNA extraction reagents (Epicenter). ChIP assays were performed as previously described for fixed cells with sonication8 and DNA concentrations determined by Qbit high sensitivity assay (Invitrogen). For ChIP-qPCR experiments, equal amounts of paired input/IP DNA were amplified by real-time PCR (Roche LightCycler96) and fold enrichments calculated8. Immunohistochemistry, H&E staining, and immunofluorescence on formalin-fixed, paraffin-embedded (FFPE) rapid autopsy tissue microarray sections (TMAs) were performed according to standard procedures. Antibodies used for western blot, IHC, and ChIP are listed in Supplementary Table 15. Primer sequences are listed in Supplementary Table 16.
YSI metabolite analysis
Metabolite consumption (glucose and glutamine) and production (lactate and glutamate) were measured using a YSI 7100 Bioanalyzer. Indicated cell lines were plated at day −1 in a 6 well plate. At day 0 cells were counted (3 wells) or cultured in either regular medium or medium supplemented with the indicated compound. Tissue culture supernatants (1mL, n=3, each condition) were harvested 72 hours after cell plating. Tissue culture conditions were optimized to ensure nutrient availability and exponential cell growth. Metabolite consumption/ production data were normalized to cell number area under the curve. The area under the curve (AUC) was calculated as N(T)d/ln2(1-2-T/d), where N(T) is the final cell count, d is doubling time, and T is time of experiment. Doubling time was calculated as d = (T)[log(2)/log(Q2/Q1)], where Q1 is starting cell number and Q2 is final cell number, as determined by manual counting using a hemocytometer.
LC-HRMS Metabolite Profiling
LC-HRMS samples were prepared and analyzed as described in detail9. For glucose tracing experiments, cells were plated into 6 well plates in triplicate, grown in DMEM with 10% FBS until 70–80% confluent, washed 2X with nutrient deplete DMEM, and incubated in nutrient deplete DMEM containing 10mM 13C-1,2 labeled glucose (Cayman) and 10% dialyzed FBS (Invitrogen) for an additional 36 hours. Additional replicates were also included and counted at the end of the experiment for cell number normalizations. Metabolism was quenched by quickly removing media and adding 1mL pre-chilled (−80°C) LC-MS grade 80% methanol (Sigma), incubated at −80°C for at least 20 minutes, followed by scraping into the methanol and pelleting of metabolites by centrifugation. For drug treatments, cells were incubated in standard DMEM +/− DMSO/6AN for 36hours, followed by incubation in labeled glucose media +/− DMSO/6AN for an additional 36 hours, quenched, and pelleted as above. Pellets were reconstituted in equal volumes of 1:1 LC-MS grade acetonitrile:methanol and water and 5ul were injected to the LC-QE-MS for analysis. For steady state measurements cells were incubated in growth media (DMEM with 10% FBS for PDACs, keratinocyte serum-free media for HPDE) until they reached 80–90% confluence, followed by 48 hours in DMEM without serum (for PDACs, since the standard growth media for comparison HPDE cells also did not contain serum). Metabolism was then quenched with methanol and metabolites pelleted as above. Pellets were reconstituted into a volume normalized to protein content (15uL of 1:1 acetonitrile:methanol and 15uL of water was used per 1mg protein) and analyzed by LC-QE-MS. Raw data collected from the LC-QE-MS was processed on Sieve 2.0 (Thermo Scientific) using a targeted frame-seed that included glycolytic/PPP metabolites as required for the analysis. The output file including detected m/z and relative intensity in different samples is obtained after data processing, and replicates of selected metabolites from each sample were graphed and presented as shown in the figures.
Preparation of sequencing libraries
Libraries were prepared from 2–10 ng of IP ChIP DNA and 100ng of input DNA and sequenced on Illumina HiSeq. Briefly, samples were checked for quality and concentration from 150–250bp on a bioanalyzer. DNA was end-repaired using Klenow polymerase in 58ul of reaction buffer. For IP DNA, Klenow was diluted 1:5. Samples were incubated at 20°C for 30 minutes and subsequently purified on QIAquick PCR purification columns. A-tails were then added to the DNA with Klenow and dATP in NEB buffer 2 at 37°C for 30 minutes and cleaned with Qiagen MiniElute PCR purification columns. Sequencing adapters were then ligated onto the DNA for 15 minutes at room temperature followed by cleaning with MiniElute columns. Samples were then run on 2% agarose gels and DNA from 216bp-366bp (DNA plus adapters) were cut from the gel and purified with Qiagen Gel extraction kits. Concentrations were then checked on a bioanalyzer and 8ng were PCR amplified with Phusion polymerase (Fisher) for 15 cycles (10sec 98°C, 30sec 65°C, 30sec 72°C) followed by 5 minutes at 72°C. Samples were then cleaned with Ampure kits (Illumina) and washed with 80% ethanol. DNA samples were resuspended at the end of the cleanup into 17.5ul buffer EB (Qiagen) and subjected to next generation sequencing on Illumina HiSeq platform according to manufacturer’s instructions. For SOLID sequencing, ChIP DNA was prepared and samples were processed according to manufacturer’s protocols in the Johns Hopkins CRBII core facility.
BS-Seq data processing
100 bp paired-end HiSeq2000 sequencing reads were aligned by BSmooth bisulfite alignment pipeline (version 0.7.110) as previously described in detail10,11. Briefly, reads were aligned by Bowtie2 (version 2.0.1) against human genome (hg19) as well as the lambda phage genome. After alignment, methylation measurements for each CpG were extracted from aligned reads. We filtered out measurements with mapping quality <20 or nucleotide base quality on cytosine position <10 and we also removed measurements from the 5′ most 10 nucleotides of both mates. Then, bsseq package in BSmooth was used to identify small and large differentially methylated regions (DMRs). Only CpGs with at least coverage of 3 in all samples were included in our analysis. For small DMRs, smooth window of 20 CpGs or 1kb was used, and t-statistic cutoff of −4.6, 4.6 and methylation difference greater than 20% were used for identifying small DMRs. While for large DMRs, smooth window of 200 CpGs or 10,000 bps was used, and t-statistic cutoff of −2, 2, methylation difference greater than 10% and length of DMRs > 5 kb were used for identifying large DMRs.
RNA-Seq data processing
100 bp paired-end HiSeq2000 sequencing reads were aligned against human genome (hg19) by OSA (version 2.0.112) with default parameters. After alignment, only uniquely aligned reads were kept for further analysis. Gene annotation information was downloaded from ENSEMBL (www.ensembl.org, release 66). Reads count for each gene of all samples were estimated using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) and then were used to identify differentially expressed (DE) genes using DESeq package13. Genes with FDR <0.01 and fold-change >1.5 were considered DE genes.
Chip-seq data processing
For 46 bp paired-end Illumina HiSeq2000 sequencing data, reads were aligned against human genome (hg19) using BWA with default parameters14. After alignment, duplicate reads were removed and only uniquely aligned reads were kept for further analysis. For 48 bp single-end Solid sequencing data, reads were aligned using Bowtie15 with default parameters and only uniquely aligned reads were kept for further analysis. IP sequencing reads were normalized to their corresponding inputs, and all input libraries were checked to ensure that no contaminating mycoplasma sequencing reads were detected. For narrow histone modification peaks (H3K4me3 and H3K27ac), MACS2 were used for peak calling with default parameters16. For broad histone modification enrichments (H3K36me3, H3K27me3, and H3K9me2/3), peak calling was performed using RSEG which is based on hidden Markov model (HMM) and specifically designed for identifying broad histone peaks17.
Identifying large chromatin domains
We define LOCK domains for heterochromatin modifications (H3K9me2/H3K27me3) based on the peak calling results from RSEG. Briefly, peaks shorter than 5 kb were first removed to prevent regions with many nearby, short peaks being called as LOCKs. Then, neighboring peaks with distance less than 20 kb were merged to into one domain. Merged regions greater than 100Kb identified in both biological replicates were called LOCKs. We noticed another unique subset of heterochromatin domains (“large LOCKs”) that were invariably larger than 500 kb, strongly enriched with H3K9me3, depleted of H3K9me2 and H3K27me3, and flanked by strong peaks of H3K27me3 at the boundaries. Because of this, we defined these LOCKs by H3K9me3 regions with length greater than 500 kb and less than 50% of their length overlapped with H3K27me3. Finally, the large regions (>50 kb) between heterochromatin domains that contained at least one gene with corresponding euchromatic H3K4me3/H3K27ac regulatory peaks were defined as ECDs. Because we found that H3K27ac alone was sufficient for these calls, H3K4me3 was also used for the initial test dataset with A38Per/Lg, but not required in subsequent datasets (A13Pr1/2, A13Lg).
Defining different gene groups
Genes were classified as belonging within euchromatin (> 50% of genic region located in ECDs) or heterochromatin (> 50% of genic region located in those heterochromatin domains). A handful of other genes that did not fit these criteria and were classified as “other”.
Quantifying and enrichment plotting of ChIP-seq and RNA-seq
To plot each histone modification on defined large chromatin domains and their flanking regions, we divided flanking sequences of chromatin domains into bins with fixed length (in bp) and domains themselves into bins with fixed percentage of each domain length. ChIP enrichment was measured and normalized as described previously18. In brief, the number of reads per kilobase of bin per million reads sequenced was calculated for each ChIP and its input control (denoted as RPKMChIP and RPKMinput). ChIP enrichment is measured as ΔRPKM=RPKMChIP – RPKMinput and ChIP enrichment regions should have ΔRPKM>0. Then all ΔRPKM were normalized to a scale between 0 and 1 and the average normalized ChIP enrichment signals across all large chromatin domains were plotted for each histone mark. RNA-Seq data was also normalized by the number of reads per kilobase of bin per million reads sequenced and plotted similarly.
Statistical tests of significance
Tests for statistical significance (two-tailed students t-test) were performed on data collected from technical replicate (performed in parallel at the same time) or biological replicate (performed at different times) experiments as indicated in the figure legends using excel software for western blot densitometry, MTT assays, and tumor measurements. Permutation tests for overlapping genomic intervals were performed in the R package using GenometriCorr19. For comparisons between groups, Chi-square and Wilcoxon rank sum tests were performed using R software (version 3.3.1).
Data availability
All ChIP-seq, WGBS, and RNA-seq datasets are available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxyjkaqsvfalheh&acc=GSE63126.
Supplementary Material
Acknowledgments
We thank J. Zacharias for technical assistance with sample processing, and members of the Johns Hopkins DNA sequencing core facility for ChIP-seq on SOLID formats. This work was supported by NIH grant CA38548 (APF), NIH grants CA140599 and CA179991 (CID), the AACR Pancreatic Cancer Action Network Pathway to Leadership grant (OGM), Vanderbilt GI SPORE (OGM), Vanderbilt-Ingram Cancer Center (OGM), and NIH grant CA180682 (AMM).
Footnotes
Accession codes. All ChIP-seq, WGBS, and RNA-seq data are accessible through the Gene Expression Omnibus (GSE63126): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxyjkaqsvfalheh&acc=GSE63126.
URLs. Gene annotation information was downloaded from ENSEMBL (www.ensembl.org, release 66). Genic sequencing read counts were estimated using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). GO analyses were performed using DAVID functional annotation software (https://david.ncifcrf.gov/summary.jsp)
AUTHOR CONTRIBUTIONS
OGM, CID, and APF conceived the work and wrote the manuscript. APF oversaw epigenomic sequencing, performed at JHU. CID selected patient samples, performed pathologic review, analyzed immunostain data, and oversaw whole genome sequencing studies, performed at MSKCC. OGM designed and performed or oversaw all experiments and data analysis, performed at Vanderbilt. XL and HW performed the computational bioinformatics and statistical analyses. OGM and APF guided the bioinformatics analyses. TS performed immunohistochemical and immunofluorescence experiments. RT prepared sequencing libraries and performed sequencing runs, assisted by THS. SM, MW, and JWL performed that LC-HRMS measurements of metabolites. OGM and AEW analyzed and plotted LC-HRMS data. AC and KEW performed and analyzed YSI glucose/lactate measurements. SN and KS maintained cell culture, performed NADPH assays, and a subset of western blots. YZ performed animal injection experiments. AMM performed whole genome sequencing. OGM prepared the figures. All authors approved of the final version.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Embuscado EE, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–54. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iacobuzio-Donahue CA, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–13. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timp W, et al. Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome Med. 2014;6:61. doi: 10.1186/s13073-014-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–99. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NG-A42854R3. Nature Genetics [Google Scholar]
- 12.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WH, et al. Human pancreatic adenocarcinoma: in vitro and in vivo morphology of a new tumor line established from ascites. In Vitro. 1982;18:24–34. doi: 10.1007/BF02796382. [DOI] [PubMed] [Google Scholar]
- 14.Metzgar RS, et al. Antigens of human pancreatic adenocarcinoma cells defined by murine monoclonal antibodies. Cancer Res. 1982;42:601–8. [PubMed] [Google Scholar]
- 15.Kyriazis AA, Kyriazis AP, Sternberg CN, Sloane NH, Loveless JD. Morphological, biological, biochemical, and karyotypic characteristics of human pancreatic ductal adenocarcinoma Capan-2 in tissue culture and the nude mouse. Cancer Res. 1986;46:5810–5. [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–8. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 18.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 19.Duesberg P, et al. Origin of metastases: subspecies of cancers generated by intrinsic karyotypic variations. Cell Cycle. 2012;11:1151–66. doi: 10.4161/cc.11.6.19580. [DOI] [PubMed] [Google Scholar]
- 20.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–31. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen B, et al. Euchromatin islands in large heterochromatin domains are enriched for CTCF binding and differentially DNA-methylated regions. BMC Genomics. 2012;13:566. doi: 10.1186/1471-2164-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon GC, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–58. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klco JM, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–92. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–19. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mentch SJ, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22:861–73. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cluntun AA, et al. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015;3:10. doi: 10.1186/s40170-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–6. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41:211–8. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan C, et al. Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol Cell. 2014;55:552–65. doi: 10.1016/j.molcel.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson WJ, McColl JD. 6-Aminonicotinamide--a Potent Nicotinamide Antagonist. Science. 1955;122:834. doi: 10.1126/science.122.3174.834. [DOI] [PubMed] [Google Scholar]
- 34.Kohler E, Barrach H, Neubert D. Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett. 1970;6:225–228. doi: 10.1016/0014-5793(70)80063-1. [DOI] [PubMed] [Google Scholar]
- 35.Cheung WK, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–38. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227.e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med. 2010;16:1450–5. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andl CD, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–30. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 1.Embuscado EE, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–54. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iacobuzio-Donahue CA, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–13. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung WK, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–38. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227.e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Andl CD, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–30. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 7.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat Med. 2010;16:1450–5. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem. 2014;86:2175–84. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Ge H, Newman M, Liu K. OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics. 2012;28:1933–4. doi: 10.1093/bioinformatics/bts294. [DOI] [PubMed] [Google Scholar]
- 13.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Q, Smith AD. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011;27:870–1. doi: 10.1093/bioinformatics/btr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–91. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favorov A, et al. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol. 2012;8:e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ChIP-seq, WGBS, and RNA-seq datasets are available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sxyjkaqsvfalheh&acc=GSE63126.