Abstract
S. cerevisiae from different environments are subject to a wide range of selective pressures, whether intentional or by happenstance. Chemicals classified by their application, such as herbicides, fungicides and antibiotics, can affect non-target organisms. First marketed as RoundUp™, glyphosate is the most widely used herbicide. In plants, glyphosate inhibits EPSPS, of the shikimate pathway, which is present in many organisms but lacking in mammals. The shikimate pathway produces chorismate which is the precursor to all the aromatic amino acids, para-aminobenzoic acid, and Coenzyme Q10. Crops engineered to be resistant to glyphosate contain a homolog of EPSPS that is not bound by glyphosate. Here, we show that S. cerevisiae has a wide-range of glyphosate resistance. Sequence comparison between the target proteins, i.e., the plant EPSPS and the yeast orthologous protein Aro1, predicted that yeast would be resistant to glyphosate. However, the growth variation seen in the subset of yeast tested was not due to polymorphisms within Aro1, instead, it was caused by genetic variation in an ABC multiple drug transporter, Pdr5, and an amino acid permease, Dip5. Using genetic variation as a probe into glyphosate response, we uncovered mechanisms that contribute to the transportation of glyphosate in and out of the cell. Taking advantage of the natural genetic variation within yeast and measuring growth under different conditions that would change the use of the shikimate pathway, we uncovered a general transport mechanism of glyphosate into eukaryotic cells.
Introduction
RoundUp™ is a non-selective herbicide containing glyphosate and a variety of additives such as detergents. As a broad-spectrum herbicide, glyphosate, the active ingredient in RoundUp™, inhibits production of chorismate, the precursor for tryptophan, tyrosine, phenylalanine, Coenzyme Q10, and para-Aminobenzoic acid (PABA), and its supplementation circumvents this growth inhibition in plants [1]. Glyphosate inhibits 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) in plants, an enzyme of the shikimate pathway (Fig 1A). Sensitive alleles of EPSPS are directly bound by glyphosate [2, 3]. Changing amino acids in the glyphosate binding site of EPSPS or overexpression of EPSPS confers RoundUp™ resistance [4, 5]. The functional ortholog of EPSPS in yeast is Aro1 and it contains additional enzymatic functions (S1A Fig) that are encoded by separate proteins in plants and bacteria [6].
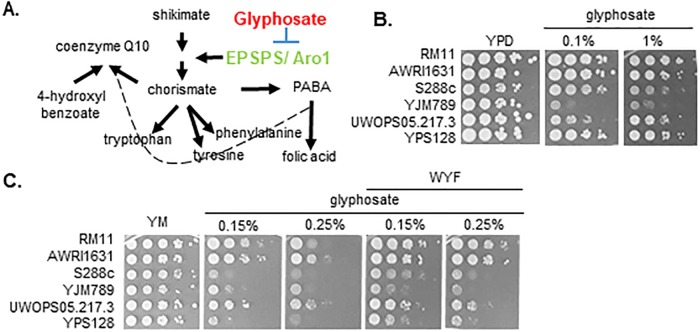
Fig 1. Genetic variation effects growth inhibition by glyphosate.
A. Shikimate pathway produces the precursor for phenylalanine, tyrosine, tryptophan, para-Aminobenzoic acid (PABA), folic acid and Coenzyme Q10. The canonical target of glyphosate is EPSPS in plants and Aro1 is the yeast homolog of EPSPS. PABA and 4-hydroxylbenzoate can be converted to Coenzyme Q10. B. Serial dilution of genetically diverse yeast on rich media (YPD) with dilutions of glyphosate as indicated (1% vol/ vol is equivalent to 78 mM). C. Serial dilution of genetically diverse yeast on minimal media with glyphosate. Aromatic amino acids, tryptophan (W), tyrosine (Y) and phenylalanine (F) were added to YM plates to make WYF.
Relatively low human acute toxicity and the broad range of susceptible plants has encouraged widespread use of glyphosate, and its effects on non-target organisms are becoming pervasive [7]. Humans do not have EPSPS and cannot make aromatic compounds, hence acquire these essential nutrients through their diet or microbiota [8, 9]. The few reports of acute glyphosate toxicity in humans are likely from the detergents that are part of the commercial formulations [10]. Spraying glyphosate on crops and other weeds also exposes nearby organisms to glyphosate, including insects, bacteria and fungi. Saccharomyces cerevisiae, a species with tremendous genetic diversity, occupies a wide range of niches, making it well suited for investigations of adaptation to new environmental stressors. There is more genetic diversity (SNPs/ Kb) between two different strains of yeast than among the entire human species [11–14]. There are 60,000 SNPs in the 12 Mb yeast genome [15] and 10 million SNPs in the 3,200 Mb human genome. [16]. Therefore, yeast is an ideal model organism to address acquisition of glyphosate resistance by examining variable alleles.
Two different mechanisms of glyphosate resistance have been uncovered in plants and bacteria. The first mechanism involves changing the ability of EPSPS to be bound by glyphosate, either by mutations that alter glyphosate binding efficacy (reviewed in [17]) or amplification of the EPSPS gene. The second mechanism involves increased levels of transport pathway elements such as ABC transporters, that move it to the vacuole and in turn neutralize the effect of glyphosate [18]. Pdr5 is an ABC transporter that is orthologous to the human Mdr1 and is often amplified in chemotherapeutic resistant cancers [19].
To determine if there were other underlying mechanisms of tolerance, in this study we characterized the yeast response to glyphosate. In different growth conditions, yeast exhibited a wide range of growth inhibition by glyphosate. The only known target of glyphosate is Aro1; however, polymorphisms in Aro1 were not responsible for the genetic variation of growth inhibition by glyphosate. Supplementing aromatic amino acids, which should bypass the shikimate pathway, improved growth in a dose-dependent manner but there was still variation in growth. To address the media-dependent genetic variation in glyphosate tolerance, Quantitative Trait Loci (QTL) analysis was carried out between two strains that demonstrated the greatest divergence in phenotypic response to glyphosate. In minimal media, the variation in glyphosate resistance was mapped to an amino acid permease, Dip5. While DIP5 deletion increased glyphosate resistance, expression of the resistant allele further improved growth of yeast on glyphosate which suggested that the resistant allele has additional functions. Dip5 function was decreased by the addition of aspartic acid, resulting in relieved growth inhibition of all yeast tested in response to glyphosate. However, the magnitude of increased DIP5 mRNA levels in yeast that were treated with glyphosate did not correlate with changes in growth across strains. Together this suggests that, regardless of the genetic variation in DIP5 among the various yeast strains, the Dip5 protein was similarly regulated by aspartic and glutamic acid. Dip5 can import glyphosate into the yeast cell and the resistant allele has additional functions that the sensitive allele lacks in response to glyphosate. In addition, variation in glyphosate resistance in rich media was mapped to an ABC pleiotropic drug transporter, Pdr5. Deletion of PDR5 resulted in the loss of glyphosate tolerance, while expression of the resistant allele conferred glyphosate resistance and expression of the sensitive allele did not. While it is likely that there are many proteins that regulate the response to and the transport of glyphosate, this study sought to identify the divergent genes between the two strains.
Results
There is a tremendous amount of phenotypic variation among yeast strains in response to different forms of stress. As rich media (YPD) contains all the required amino acids, it represses amino acid biosynthetic pathways and permits yeast to transport amino acids from the media into the cell. Six genetically diverse yeast isolated from different environments were grown on YPD supplemented with glyphosate (Fig 1B), RM11(a wine yard isolate), AWRI1631 (used for commercial wine making), S288c (a laboratory strain), YJM789 (a clinical isolate), UWOPS05.217.3 (isolated from bertam palm nectar), and YPS128 (isolated from the soil under an oak tree). These strains showed little change in growth on solid media in the presence of 0.1% volume/volume glyphosate (78 mM) on YPD. The growth of YJM789, a clinical isolate, was reduced by the addition of 0.1% glyphosate in YPD (Fig 1B). At levels ten times higher, the growth of YJM789 was nearly completely inhibited and the growth of S288c was slightly reduced. The commercial preparation of glyphosate contains additives such as detergents to increase the tissue penetration of glyphosate. To better reflect conditions that organisms would be exposed to, we chose to use the commercial preparation of glyphosate (RoundUp™) rather than pure glyphosate. In minimal media (YM), yeast were required to make all amino acids using ammonium sulfate as the sole nitrogen source. Under this growth condition, S288c, YJM789, and YPS128 were sensitive to 0.15% glyphosate and all the strains tested, grew more slowly on 0.25% glyphosate treatment (Fig 1C). AWRI1631, RM11, and UWOPS05.217.3, which are agricultural isolates, demonstrated higher glyphosate tolerance in YM. Hence, among the agricultural isolates RM11 was found to be the most resistant to glyphosate exposure, followed by AWRI1631 and UWOPS05.217.3. Whereas, YJM789 was the most sensitive followed by YPS128 and S288c. When aromatic amino acids (tryptophan (W), tyrosine (Y), and phenylalanine (F)) were added back to YM (WYF), growth inhibition of S288c and YPS128 were alleviated (Fig 1C) and the growth of YJM789 showed improvement at lower glyphosate levels.
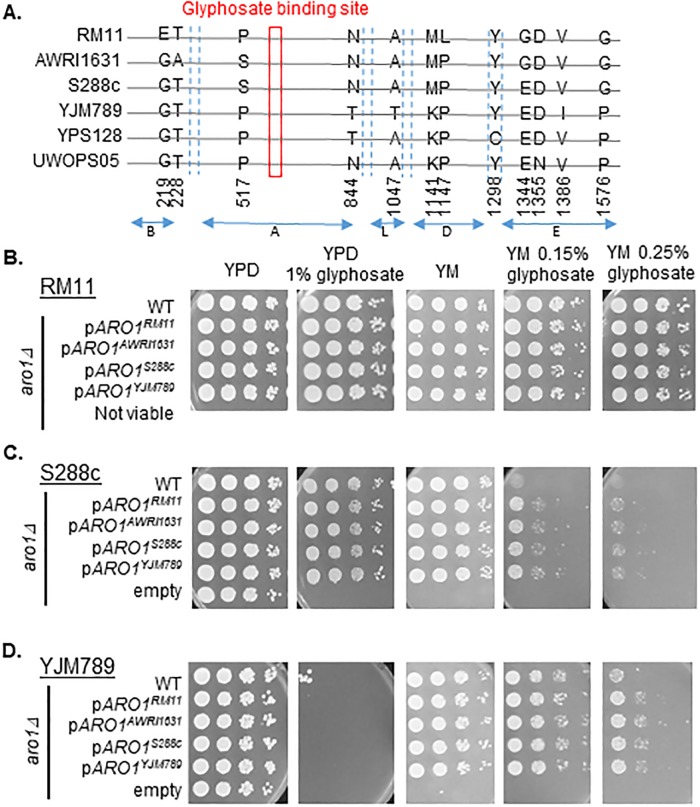
Numerous polymorphisms in the plant and bacterial homologs of Aro1 result in glyphosate resistance [17]. We predicted that yeast would be resistant to RoundUp™ by comparing mutations that have been mapped in AroA from E. coli and the yeast amino acid sequence of Aro1 [4, 20, 21] (S1B Fig). Yet, different strains of yeast showed differences in sensitivity to glyphosate that could be rescued with the supplementation of WYF. The polymorphisms in Aro1 across these six different yeast strains were outside the glyphosate binding domain (Fig 2A). To determine if polymorphisms in Aro1 contributed to the variation in glyphosate resistance, ARO1 was knocked out. The RM11heterozygous aro1Δ/ ARO1 strain was sporulated and the tetrads were dissected onto YPD. A lethal mutation segregated in a 2:0 pattern that was linked to NatR in two independent knockouts, i.e., the aro1Δ was inviable in RM11 (S1D Fig). ARO1 was successfully knockout in S288c and YJM789 haploid yeast and could grow if WYF was supplemented (Fig 2).
Fig 2. Contribution of the genetic variation within Aro1 to glyphosate resistance.
A. Protein alignment of Aro1 from genetically diverse yeast strains. B. Serial dilutions of haploid aro1Δ in RM11, C. S288c, and D. YJM789, expressing different alleles of plasmid encoded ARO1 were grown on YPD and YM with and without glyphosate. Rows labeled empty have pGS36 plasmid with no ARO1. Parental strains with the endogenous ARO1 expressed from the chromosome labeled WT carry an empty plasmid (pGS36).
To determine if ARO1 was an essential gene in RM11 or if RM11 contained an allele that was synthetically lethal in combination with an aro1 deletion, RM11 (wild-type) was crossed with S288c aro1Δ (viable mutant), sporulated and dissected onto YPD (S1E Fig). Fourteen tetrads were dissected and 34 spores were viable. Twelve viable aro1Δ segregants from the F1 hybrids ruled out the possibility that Aro1RM11 gained an essential function, but instead aro1Δ was synthetically lethal with an unknown allele present in the RM11 genome and not present in the S288c genome. The unknown allele was found to be unlinked to the size of the colony, the MAT, HO, and ARO1 loci. In conclusion, the Aro1 deletion in most strains was found to be viable, with an exception of RM11.
To assess the impact of the genetic variation of Aro1 in response to glyphosate, four different alleles of ARO1 were cloned under their endogenous promoter and terminator into plasmids and transformed into aro1Δ yeast. ARO1 from AWRI1631 and RM11 represent alleles that are present in glyphosate resistant yeast, while ARO1 from S288c and YJM789 are alleles from less tolerant strains. While ARO1 was essential in RM11, plasmids encoding alleles of ARO1 were transformed into the heterozygous diploid, and the haploid knockouts with the plasmid were recovered after sporulation (Fig 2B). S288c aro1Δ and YJM789 aro1Δ were viable on YPD and WYF but not on YM (Fig 2C and 2D). It can be concluded that all alleles of ARO1 could complement aro1Δ mutation in RM11, S288c and YJM789 yeast, because the aro1Δ yeast with plasmid expressed ARO1 grew on YM (Fig 2) to similar levels as the wild-type parents carrying an empty plasmid. When grown on YM with glyphosate, yeast cells showed no difference in growth irrespective of the ARO1 allele expressed. However, S288c and YJM789 with ARO1 expressed from the plasmid consistently grew slightly better than yeast expressing chromosomal ARO1. The mRNA levels of ARO1 from RM11, YJM789 and YJM789 aro1Δ were quantified. In YJM789, ARO1 expressed from both the chromosomal location and from a plasmid was approximately the same (S1C Fig). There was a slight decrease in the levels of ARO1 mRNA found when YJM789 was exposed to glyphosate. In YJM789, expression of ARO1RM11 was no different from ARO1YJM798. Yet, there was an increase in ARO1 mRNA levels in RM11 compared to YJM789 in untreated growth conditions.
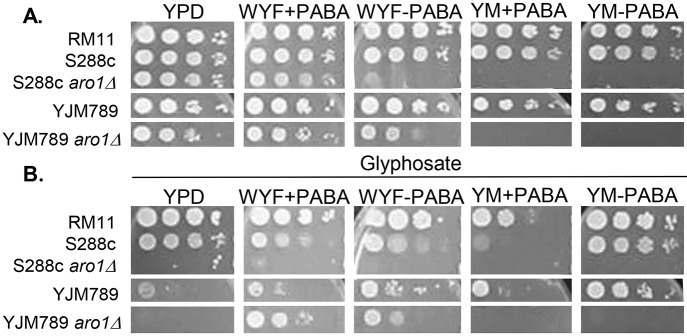
The primary target of glyphosate is inhibition of Aro1 in the chorismate pathway and the genetic variation resulting in growth inhibition in response to glyphosate, was not due to genetic differences in Aro1. Therefore, the genetic variation in response to glyphosate could be due to genetic variation in other components of the chorismate pathway or in an unrelated pathway. To explore the differences in the chorismate pathway between S288c and YJM789 yeast, the aro1 knockouts were tested on various media. Chorismate is the precursor for aromatic amino acids, PABA, and Coenzyme Q10 (CoQ10). para-Aminobenzoic acid (PABA) is essential for the production of folic acid derivatives and can be converted to Coenzyme Q10 precursor (Fig 1A). CoQ10 is involved in respiration and is not essential for yeast viability. The commercial preparation of YM contains PABA and folate [22] and is noted here as YM+PABA. S288c aro1Δ could not grow on WYF-PABA (YM without PABA, supplemented with WYF), while the wild-type parent could grow. In YJM789 aro1Δ, growth was only slightly reduced on WYF-PABA as compared to its wild-type parent; this difference in growth between S288c aro1Δ and YJM789 aro1Δ suggested the existence of an alternative pathway present in YJM789 that is absent in S288c. YJM789 was able to convert WYF to PABA and influence cell growth. RM11 required ARO1 for viability and therefore the knockout could not be tested (Fig 3A). Additionally, PABA-free, and folate-free media was tested, where no change in growth was seen in comparison to PABA-free media, and thus this avenue was not further examined.
Fig 3.
A. Different responses of aro1Δ yeast on media supplemented with different aromatic metabolites. Wild-type RM11 was compared to S288c (GSY147) and YJM789 with and without ARO1, three days in minimal media supplemented with aromatic amino acids (+WYF) or para-aminobenzoic acid (+PABA) or without these metabolites (-WYF or -PABA). B. Different responses of aro1Δ yeast on glyphosate. Wild-type RM11 was compared to S288c (GSY147) and YJM789 with and without ARO1 on 0.15% glyphosate three days in minimal media supplemented with +WYF or +PABA or without these metabolites (-WYF or -PABA).
To determine if supplementation of yeast with downstream metabolites synthesized from chorismate could bypass the growth inhibition in the presence of glyphosate, yeast cells were grown on media supplemented with WYF in either the presence or absence of PABA. To assess whether PABA had a role in glyphosate growth inhibition, yeast cells were also tested on PABA-free media, supplemented with WYF. S288c aro1Δ were more sensitive than the wild-type parent to glyphosate on WYF media (Fig 3B), while YJM789 aro1Δ was more resistant to glyphosate than the wild-type parent. Only growth of RM11 on YM with glyphosate showed a slight decrease, while no change was detected when RM11 was grown in other conditions.
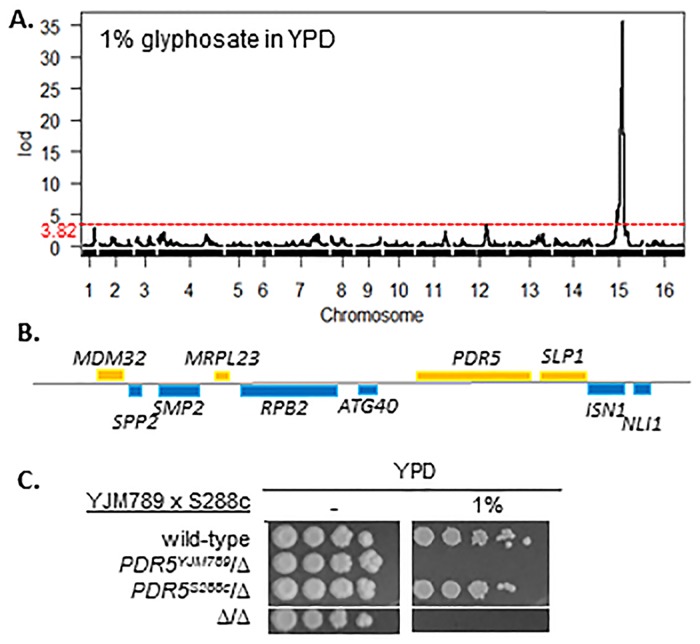
While genetic variation leads to differences in glyphosate resistance, the phenotypic response to glyphosate was not due to variation in ARO1. We chose to map the genes associated with glyphosate resistance in S288c and YJM789 because of the variation in their growth in YPD, WYF and YM with glyphosate. We tested the growth of 125 recombinant haploid segregants from a hybrid of S288c and YJM789 [23]. For S288c in YPD with 1% glyphosate, one locus of interest was identified on chromosome 15 with a LOD score of 35.5 (Fig 4A). PDR5, an ABC transporter that confers resistance to a wide-range of structurally diverse chemicals [24, 25] was located within this region (Fig 4B). PDR5 has previously been implicated in plant response to glyphosate [18]. The role of each allele of PDR5 was tested in YJM789 and S288c diploid hybrids. PDR5 was knocked out from each parent and mated with the other parent. The resulting hemizygous strains are identical except for the PDR5 allele that the hybrid inherited from one parent (Fig 4C). The PDR5YJM789/Δ hybrid was sensitive to glyphosate on YPD, while the PDR5S288c/Δ hybrid demonstrated the same level of tolerance as that of the wild-type hybrid. The sensitivity of the PDR5YJM789/Δ hybrid was similar to that of the homozygous knockout mutant. Pdr5 is a highly polymorphic protein, where the hyper-variability of its genetic sequence allows for greater specificity and tolerance to a wide array of chemicals per allele [26]. Between YJM789 and S288c, there is a 5% difference in amino acid sequence. Among twelve available sequences that were analyzed, no obvious polymorphism was found in common among the yeast that were sensitive to glyphosate on YPD (S2 Fig).
Fig 4. Genetic linkage analysis of glyphosate sensitivity in glyphosate on rich media.
A. Genetic linkage of sensitivity of YJM789 to 1% glyphosate in YPD. LOD score (y-axis) was mapped across the yeast genome (x-axis) with chromosomes numbered left to right. The LOD significant levels (alpha = 0.05) was 3.82 and was marked by a red dashed line. B. The genomic loci under the peak located on chromosome 15 contains 10 genes. Genes encoded on the top strand are in yellow and genes encoded on the bottom strand are blue. C. Serial dilution of S288c (GSY147) x YJM789 hybrids in which the entire PDR5 coding region in either parent was deleted (Δ), crossed, and the resulting hemizygotes were grown on rich media with and without 1% of glyphosate.
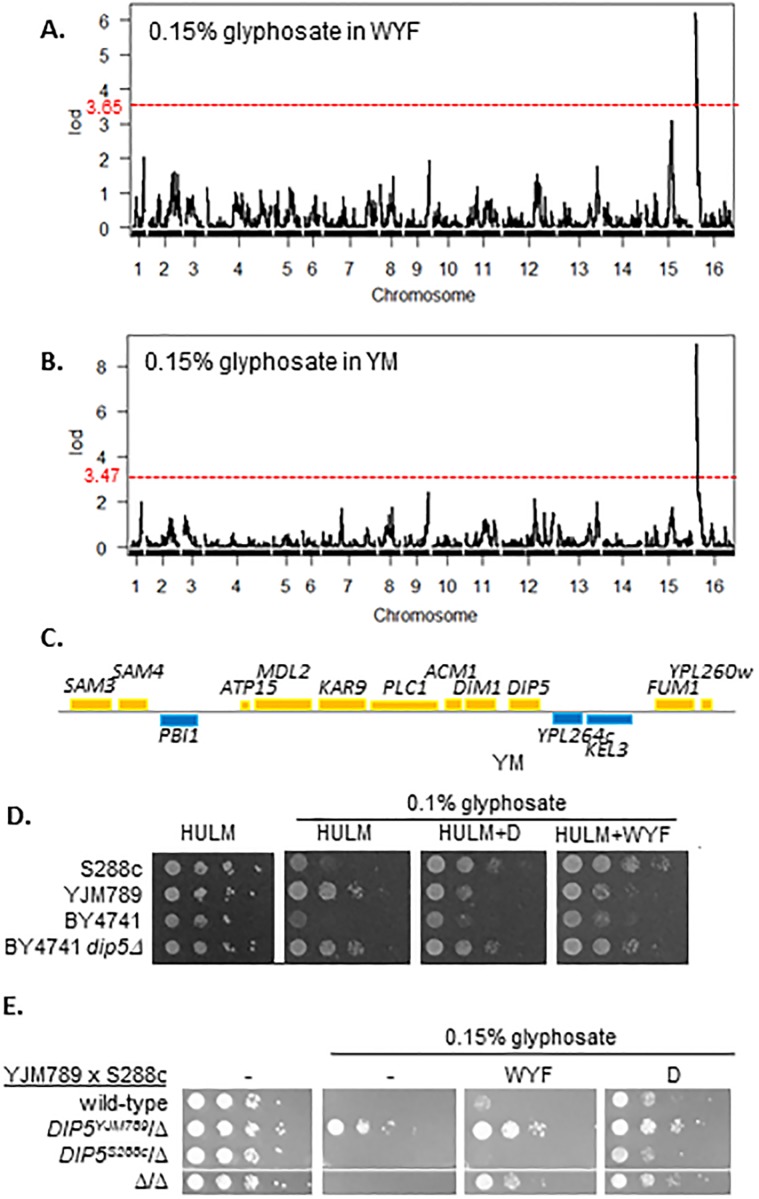
Among the S288c and YJM789 segregants grown on WYF, an additional peak on chromosome 16 was found to be linked to glyphosate response (Fig 5A). The aforementioned peak on chromosome 15 associated with PDR5 in YPD was observed, however the peak fell below the level of significance (LOD < 3.65) in YM with WYF. Additionally, the same peak on chromosome 16 was also identified when yeast were grown on YM without WYF supplementation (Fig 5B). Fourteen candidate genes were in the region under this peak (Fig 5C) and were evaluated using the yeast knockout collection, where the likely candidate was DIP5 [27]. The knockout collection was constructed in an S288c related strain, BY4741 that has multiple amino acid auxotrophies and several other differences from GSY147 (another S288c related strain) [28]. To circumvent the amino acid auxotrophies and achieve normal growth, BY4741 was supplemented with histidine, uracil, methionine, and leucine (HULM). BY4741 is highly sensitive to glyphosate and to accommodate for this, the level of glyphosate was reduced to 0.1%. This reduction allowed for a more pronounced rescue of BY4741 dip5Δ.
Fig 5. Genetic Linkage analysis of glyphosate sensitivity in glyphosate on minimal media with and without aromatic amino acids.
A. Genetic linkage of sensitivity of YJM789 to 0.15% glyphosate in yeast minimal media supplemented with aromatic amino acids (WYF). LOD score (y-axis) was mapped across the yeast genome (x-axis). The LOD significant levels (alpha = 0.05) was 3.65 and was marked by a red dashed line. B. Genetic linkage of sensitivity of YJM789 to 0.15% glyphosate in yeast minimal media (YM). LOD score (y-axis) was mapped across the yeast genome (x-axis). The LOD significant levels (alpha = 0.05) was 3.47 and was marked by a red dashed line. C. The genomic loci under the peak located on chromosome 16 contains 14 genes. Genes encoded on the top strand are in yellow and genes encoded on the bottom strand are blue. D. Serial dilutions of S288c (GSY147), YJM789 and BY4741 with DIP5 knocked out grown on YM (HULM), WYF, aspartic acid (D) with glyphosate at the concentrations indicated. Histidine, uracil, leucine and methionine (HULM) were supplemented for growth of BY4741. E. Serial dilutions of S288c (GSY147) x YJM789 hybrids. The entire DIP5 coding region in either parent was deleted (Δ), crossed, and the resulting hemizygotes were grown on solid media YM, WYF, aspartic acid (D) with glyphosate at the indicated concentrations and supplemented.
Dip5 is a high affinity permease for aspartic and glutamic acid [29] and when these amino acids are in excess, the localization of the protein at the plasma membrane is reduced [30]. To assess if down-regulation of Dip5 at the plasma membrane could rescue glyphosate-induced growth inhibition, cells were grown in glyphosate media supplemented with aspartic acid. Aspartic acid (D) rescue was similar to that of WYF rescue and was specific. Whereas, the addition of other amino acids (HULM) did not rescue glyphosate sensitive yeast (Fig 5D). Aspartic acid is not a product of the shikimate pathway. Hence, the increased glyphosate tolerance of yeast when supplemented with aspartic acid suggests that glyphosate was imported into yeast via Dip5, where the increased tolerance is achieved through aspartic acid-mediated downregulation of Dip5. The role of each allele of DIP5 was tested in reciprocal hemizygous YJM789 and S288c diploid hybrids. DIP5 was knocked out from each parent, which was then mated with the other parent. The resulting hemizygous strains are identical except for the allele of DIP5 the hybrid inherited from one parent (Fig 5E). The wildtype hybrid, the homozygous mutant, and DIP5S288c/Δ hybrid were sensitive to glyphosate on minimal media, while the DIP5YJM789/Δ hybrid was resistant. The addition of WYF, supplements amino acids from the shikimate pathway. With 0.15% glyphosate, only the homozygous mutant diploid hybrid could be rescued by the addition of WYF. However, the addition of aspartic acid recused all strains.
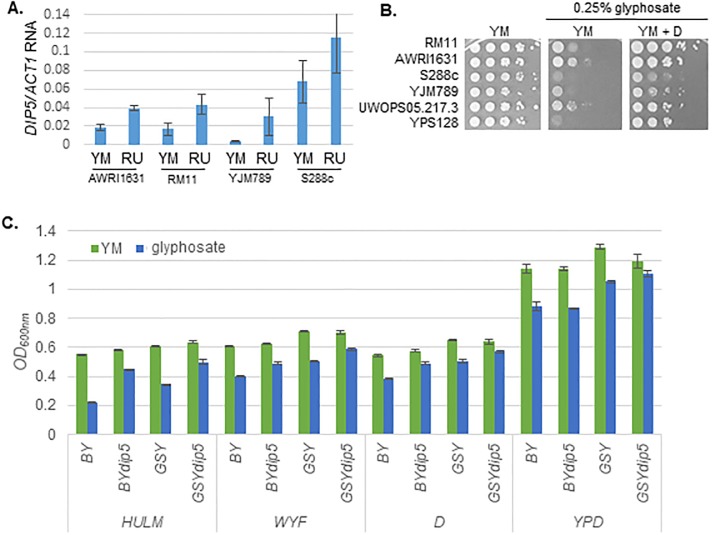
As in the case of Pdr5, we could not identify a single polymorphism that was associated with all the resistant strains. However, as the presence of the DIP5S288c allele decreased glyphosate resistance while presence of the DIP5YJM789 conferred tolerance, the responsible polymorphism could be contributing to a hypomorphy-induced resistance—such as a mutation in the promoter sequence resulting in decreased expression of DIP5. This hypomorphy could be suspected to be responsible for the resistance of the YJM789 DIP5 allele, as DIP5 is expressed in levels greater than two-fold higher in S288c yeast as compared to YJM789 in YM [31]. In contrast, the expression of DIP5 increased in glyphosate treatment in YJM789 and to a lesser extent in AWRI1631 and RM11, but not S288c (Fig 6A). Growth inhibition of the other strains of yeast on exposure to glyphosate was rescued by addition of aspartic acid (Fig 6B). Additionally, the homozygous dip5Δ mutant, did have improved growth with the addition of WYF compared to the DIP5S288c hemizygous diploid. To quantitate the ability of different amino acids to regulate the function of Dip5, quantitative growth assays were performed with two haploid S288c strains (BY4741 and GSY147). In the absence of glyphosate, deletion of DIP5 did not change the growth of either strain, except GSY147 dip5Δ grew slightly slower than GSY147 in YPD (Fig 6C). Both dip5Δ strains grew better then wild-type controls when glyphosate was added (Fig 6C). The addition of aspartic acid downregulates Dip5 at the plasma membrane [30]. The improved growth of the dip5 mutants on addition of aspartic acid suggests that import of glyphosate by Dip5 is a general mechanism, however there are other aspartic acid regulated modes of transport into the cell.
Fig 6. Regulation of DIP5 by glyphosate and aspartic acid.
A. RNA expression levels of DIP5 mRNA in AWRI1631, RM11, YJM789 and S288c grown in YM with and without 0.25% glyphosate (RU). Q RT-PCR of DIP5 mRNA levels are normalized to ACT1 mRNA. B. Serial dilution of genetically diverse yeast on minimal media with glyphosate supplemented with aspartic acid. C. Different responses of dip5Δ yeast in liquid media supplemented with different aromatic metabolites and amino acids on exposure to glyphosate. BY4741 and GSY147 were grown in the presence of glyphosate (0.1% in HULM, WYF and D and 1% in YPD) and the optical density was measured in log phase (10 hr).
Because aro1Δ mutants are sensitive to many drugs [32], we transformed alleles of PDR5 into both wild-type and aro1Δ mutants to determine if expression of PDR5S288c could bypass sensitivity of aro1Δ in the presence of glyphosate. On YPD, both S288c and YJM789 aro1Δ mutants did not grow in the presence of glyphosate. Expression of each allele of PDR5 in YJM789 aro1Δ decreased growth in WYF media and completely abolished growth when supplemented with glyphosate (S4 Fig). Surprisingly, the YJM789 aro1Δ mutant grew on WYF with glyphosate but not on YPD with glyphosate, where growth was abolished with the ectopic expression of any allele of PDR5.
Discussion
When grown in minimal media, the canonical target of glyphosate, the shikimate pathway, was affected and could be rescued by supplementation of downstream metabolites in most yeast. Utilizing an existing recombinant haploid yeast collection, two different loci linked to genetic differences in glyphosate response in S288c and YJM789 yeast were identified. Expression of PDR5S288c in YJM789 rescued sensitivity to glyphosate in rich media, while deletion in S288c conferred sensitivity. Pdr5 is the yeast ortholog of Mdr1, a multiple drug transporter that is highly polymorphic in yeast [15]. The second gene linked to glyphosate response was DIP5, which encoded an aspartic acid and glutamic acid permease [29]. The identification of these two genes furthers our understanding of the mechanisms by which glyphosate is transported in and out of the cell.
S288c is a domesticated strain of S. cerevisiae and has been the subject of extensive phenotypic, molecular and genetic analysis. Conducting studies on one genetic background limits the perspective. For example, aro1 mutants displayed differences in viability in RM11, S288c, and YJM789 strains and their growth on nutrient limiting media. Unlike S288c, ARO1 was essential for growth of RM11 on rich media but that was not from polymorphisms or a novel function in Aro1RM11 but from a synthetic lethal interaction with an unknown allele in the RM11 genome. YJM789 aro1 mutants grew poorly on glyphosate and WYF alone, and were rescued by expressing an extra copy of PDR5 but only in WYF, when the function of Aro1 to produce chorismate was thought to be bypassed. Despite pdr5 mutants having multiple drug sensitivities in S288c, aro1 mutants show no increase in growth when expressing one extra copy of PDR5. RNA analysis of a slightly different glyphosate formulation found genes regulating membrane stress in response to glyphosate [33]. The sensitivity to the commercial preparation of glyphosate could only be in part a response due to other additives for two reasons. The yeast growth profiles were not the same in YPD and WYF and expression of Pdr5 alleles in WYF only showed a small rescue. Previous studies did not directly identify the target of glyphosate in rich media, but protein coding polymorphisms in Pdr5S288c and Pdr5AWRI1631 would affect export of glyphosate.
The best characterized target of glyphosate is 5-enolpyruvylshikimate-3-phosphate synthase in the shikimate pathway, and indeed all strains showed improved growth with WYF remediation. In addition, S288c and YJM789 aro1Δ cells grew slower on WYF with glyphosate than wild-type parental strains. This may be an indirect effect, as S288c aro1Δ yeast have been found to be sensitive to multiple drugs [32]. An alternative explanation is that there are other targets of glyphosate that become affected when the primary target of glyphosate is deleted. Nevertheless, the increased toxicity of glyphosate when downstream metabolites of the shikimate pathway were provided, could be an indication of the presence of non-canonical glyphosate targets. Even sensitivity to glyphosate in rich media was unexpected. The shikimate pathway is down-regulated by the presence of aromatic amino acids, and the expression of Aro1 decreased in YPD compared to YM in S288c [34]. No change in Dip5 protein levels was detected in S288c by previous studies, while Pdr5 protein levels decreased in YM compared to YPD [34]. In rich media, nutrient transport pathways are unregulated and biosynthetic pathways are down regulated. While in minimal media, the opposite effect is observed in pathway regulation and expression. Also, there may be additional glyphosate-sensitive targets and future work will address their identification. Reports have found that glyphosate chelates calcium, manganese, iron, and magnesium [35] and glyphosate-sensitive soybean have lower levels of these minerals [36]. The shikimate pathway was originally identified years after the invention of glyphosate, by identifying increased levels of shikimate in glyphosate treated plants [2], similar methods may be applied to identify other affected pathways.
Dip5 is located at the plasma membrane, and it transports aspartic acid and glutamic acid into the yeast cell. When there is excessive aspartic acid, Dip5 is targeted for endocytosis via arrestins and through ubiquitination it is targeted for degradation [30, 37]. Deletion of DIP5 and expression of DIP5YJM789 further increased glyphosate tolerance of the cell. In yeast cells that expressed only the DIP5YJM789 allele, glyphosate resistance increased in YM, compared to yeast containing the DIP5S288c allele which did not show the same. We proposed that Dip5 at the plasma membrane is at least one of the proteins involved in transporting glyphosate into the cell. This process is regulated by phosphorylations that promote ubiquitination [30], but there were no polymorphisms at any of these known residues (S3 Fig). Within the first 74 nucleotides there were three SNPs. From global transcriptomics [31] and mRNA expression it has been determined that the level of DIP5 mRNA is two-fold lower in YJM789. As S288c (GSY147) dip5Δ rescue was less pronounced than BY4741 dip5Δ, it can be concluded that Dip5 may be differently regulated between these strains. In addition, DIP5S288c may be regulated differently than DIP5YJM789 because there was no rescue in DIP5S288c with the addition of aspartic acid to levels of glyphosate tested here. Yeast expressing DIP5YJM789 was not the same as the knockout, which suggests that there is an additional function of DIP5YJM789 compared to DIP5S288c. The lower levels of Dip5 in YJM789 at the membrane which will internalize glyphosate may be downregulated faster than Dip5S288c. The addition of glyphosate increased the amount of DIP5 mRNAs in all the strains tested. The addition of aspartic acid rescued all strains including dip5 mutants suggesting that there are other transporters of glyphosate.
In this study, we have uncovered one path of glyphosate import and one path of glyphosate export, and identified the differences within these transporters in the various strains. The polymorphisms in DIP5 and PDR5 determine the entry and pumping out of glyphosate from the cell, respectively. We propose that polymorphisms, and differences in the Dip5 protein levels, change the amount of glyphosate transported into the cell. Dip5YJM789 transports less glyphosate than Dip5S288c and both alleles are down-regulated by the addition of aspartic acid through ubiquitination (Ub) and endocytosis. Once inside the yeast, glyphosate inhibits Aro1 and possibly other non-canonical targets. Either glyphosate or metabolized products are then transported out of the yeast by Pdr5 with the S288c allele being more active than the YJM789 allele. The allele present, in turn has a correlation with growth inhibition on exposure to glyphosate. Additional studies may reveal targets of glyphosate outside the shikimate pathway that could be classified as non-canonical targets. This study focused on differences of two yeast strains that varied in their glyphosate transport pathways. With the widespread use of glyphosate, encroachment of developments into pristine areas, and the efforts to control weeds and invasive species in state parks, glyphosate resistance is likely to continue its spread in the wild.
Materials & methods
Yeast strains and plasmids
Previously published strains and their derivatives are in S1 Table. ARO1, DIP5 and PDR5 were knocked out using homologous recombination with the dominant drug resistance NatR or KanR as previously described [38] and listed in S1 Table. In YJM789, DIP5 was replaced with dip5::KanR using BY4741 dip5 as the template and the following primers 5’ AAA GTA CCA CAT ATC TAA CG 3’ and 5’ GTG ATA CCT GTA CAC TAT GGT TCC 3’. Cloning of ARO1 alleles was done by PCR amplified from genomic DNA using primers as follows 5’ARO1 5’ATG ACC ATG ATT ACG CCA AGC TTG CAT GCC TGC AGG TCG AGC CAA TCT CAC AGA TTT AAT ATA G3’, 3’ARO1 5’TAT ATT GAT CAC CGA TAT ATG GAC TTC CAC ACC AAC TAG TAA TTC TTC AGT GAA TAA ACG GGC C3’, 5’PDR5 5’GAT TAC GCC AAG CTT GCA TGC CTG CAG GTC GAC TCT AGA CTA ATC CAA TTC AGT TGT CTC3’, and 3’PDR5 5’ATC ACC GAT ATA TGG ACT TCC ACA CCA ACT AGT TTC GGA CAG ATA ATG ATA TAA TAT ATC3’. All cloned genes were accompanied by their surrounding intergenic sequences up to the neighboring upstream and downstream genes. Genes were cloned via homologous recombination into the XbaI and SpeI sites of pGS36 plasmid with hygromycin resistance [39]. Plasmids were kept under selection with hygromycin after LiAc chemical transformations [40]. ARO1 was knocked out with a PCR cassette containing NatR in YJM789K5a, GSY147, and RM11 MATa/MATα and plated onto YPD containing 1μg/ml nourseothricin. RM11 MATα, ho::KanR was crossed to GSY147 MATa, aro1:NatR and selected on YM with nourseothricin and G418. RM11 aro1Δ/ ARO1 diploid carrying plasmids expressing ARO1 were sporulated and dissected onto YPD with hygromycin to select for the plasmid. There was genetic variation in tolerance of strains to hygromycin in YM. RM11 strains grown in YM required twice as much hygromycin (250 μg/ml) as required in YPD to maintain plasmids under hygromycin selection. Hemizygous hybrid yeast strains were constructed by transforming the wild-type parent with pGS36 and mating it with the mutant parent. Hemizygous yeast were selected with hygromycin and nourseothricin or G418 depending on the dominant selectable markers. The respective hemizygous genotypes and their markers are listed in S1 Table. The haploid recombinant segregant collection between YJM789 and S288c (S96) was previously generated [23].
Media and chemicals
All yeast strains were grown in nutrient rich media (YPD) or minimal media (YM) which includes 2% dextrose, 6.7g/L yeast nitrogen base and 20g/L agar in solid media. WYF plates contained 20 μg/ml tryptophan, 30 μg/ ml tyrosine and 50 μg/ ml phenylalanine added to YM plates while D plates were supplemented with 100 μg/ml aspartic acid. Plates lacking para-Aminobenzoic acid (PABA) were made from yeast nitrogen base lacking PABA (Sunrise®). Credit® 41 Extra contained 41% glyphosate and surfactants. The haploid recombinant segregant collection contained lys2 and lys5 alleles segregating in the cross and therefore, lysine was added to media for these strains. YM and WYF media were also supplemented with histidine, uracil, leucine, and methionine for BY4741 strains. Yeast were grown overnight to saturation and then diluted to 0.1 OD units (approximately 1X107 cells) and serially diluted 10-fold. Dilutions were then stamped on to plates. Growth was scored on solid media after 2–3 days of growth, relative to a control without glyphosate to account for how many of the spots grew. Quantitative growth assays were carried out in a TECAN automatic plate reader as previously described [41].
RNA quantitation
Total RNA was isolated by hot phenol extraction [42] and precipitated from cultures grown to mid-log phase and treated for 90 minutes with 0.25% glyphosate. RNA treated with DNAse I was then converted to cDNA using Invitrogen SuperScript® III First-Strand kit according to the manufacturer’s directions. cDNA from biological duplicates was amplified in triplicate using specific primers in SSO FAST on a BioRad real-time PCR system. ARO1 primers used were upstream 5’ACCGACTGGTTAGGTATCCG3’ and downstream 5’CCTAAACTGTGCAAGGCGTA 3’. 25S rRNA and genomic DNA were used to normalize samples with the following primers upstream 5’GACTACTTGCGTGCCTTGTTG3’ and downstream 5’CCGTTCCCTTGGCTGTG3’. PDR5 primers used were 5’GTT GGC TGT TGG TGT TGC TA3’ and downstream 5’AAC TAC AGG TGT CAG TGG CA3’.
DIP5 primers used were upstream 5’CTG CTG CTT TGG TCA TTC AA3’ and downstream 5’ TGG TTA GGA CCT CCA CCA AG3’.
Mapping genetic linkage
Quantitative trait loci (QTL) mapping for S96 x YJM789 haploid segregants in three different conditions was carried out as previously described [43]. The statistical threshold for each trait was calculated independently with 1000 permutations using R package qtl with EM method. Genes and coordinates under the peak of association for each condition was referenced using Saccharomyces Genome Database [44]. QTL scores are listed in S2–S4 Tables.
Supporting information
A. Schematic of enzyme functions in Aro1 with bacterial proteins AroB (3-dehydroquinate synthase amino acids 1–392), AroA (EPSPS amino acids 404–861), AroL (shikimate kinase amino acids 887–1060), AroD (3-dehydroquinase amino acids 1061–1295), and AroE (shikimate dehydrogenase amino acids 1306–1599). B. Alignment of the ESPS glyphosate binding site across different species. In red are residues that when mutated confer resistance to glyphosate in E. coli. C. RNA expression levels of ARO1 mRNA from RM11, YJM789 and YJM789 aro1Δ carrying different alleles ARO1 grown in YM with and without 0.25% glyphosate. Q RT-PCR mRNA of ARO1 levels are normalized to 25S rRNA. D. Tetrad dissections of RM11 heterozygous knockout of ARO1 compared to wild-type RM11 diploid on YPD. Tetrads were numbered and haploid segregant germinating from a single spore are lettered. Plates were incubated at 30°C for two days. E. Tetrad dissections of RM11 wildtype and S288c aro1Δ hybrids (F1) were incubated for five days before being photographed. Haploid segregants from F1 yeast with aro1Δ were circled.
(TIF)
Branch length was determined by UPGMA in ClustalW. Relative growth of yeast on glyphosate was normalized to growth with no treatment.
(TIF)
Branch length was determined by UPGMA in ClustalW. Relative growth of yeast on glyphosate was normalized to growth with no treatment.
(TIF)
ARO1 was knocked out in S288c and YJM789. PDR5 was cloned and expressed from its native promoter from a plasmid. Yeast were grown on YPD (rich media) with 1% glyphosate, YM (minimal media) with 0.25% glyphosate and WYF (yeast minimal media supplemented with aromatic amino acids) with 0.25% glyphosate.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Ashok Bidwai for critical reading of the manuscript. Thanks to Luke Evans for suggesting the use of RoundUp™. Barbara Dunn and Gavin Sherlock provided the yeast used here and pGS36. Angela Lee generously shared the entire BY4741 yeast knockout collection. Zhenglong Gu and Xiaoxian Guo shared the YJM789 pdr5 knockout. West Virginia University PSCoR and West Virginia University Senate Grant provided initial funding. ACB was funded by WVU Summer Undergraduate Research Experience. ZRL was funded by NSF REU-Biological Responses to the Environment from Genes to the Ecosystem DBI 1156627. This work was funded by NSF-MCB 1614573.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
West Virginia University PSCoR and West Virginia University Senate Grant provided initial funding. ACB was funded by WVU Summer Undergraduate Research Experience. ZRL was funded by NSF REU-Biological Responses to the Environment from Genes to the Ecosystem DBI 1156627. This work was funded by NSF-MCB 1614573. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haderlie LC, Widholm JM, Slife FW. (1972) Effect of glyphosate on carrot and tobacco cells. Plant Pathology 60(1): 40–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrhein N, Deus B, Gehrke P, Steinrücken HC. (1980) The site of the inhibition of the shikimate pathway by glyphosate. 66(5): 830–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JNS, et al. (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proceedings of the National Academy of Sciences 98(4): 1376–1380. doi: 10.1073/pnas.98.4.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Healy-Fried ML, Funke T, Priestman MA, Han H, Schönbrunn E. (2007) Structural basis of glyphosate tolerance resulting from mutations of Pro101 in escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. Journal of Biological Chemistry 282(45): 32949–32955. doi: 10.1074/jbc.M705624200 [DOI] [PubMed] [Google Scholar]
- 5.Rogers SG, Brand LA, Holder SB, Sharps ES, Brackin MJ. (1983) Amplification of the aroA gene from escherichia coli results in tolerance to the herbicide glyphosate. Applied and Environmental Microbiology 46(1): 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan K, Edwards RM, Coggins JR. (1987) The pentafunctional arom enzyme of saccharomyces cerevisiae is a mosaic of monofunctional domains. The Biochemical Journal 246(2): 375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanney JB, Hutchison LJ. (2010) The effects of glyphosate on the in vitro linear growth of selected microfungi from a boreal forest soil. Canadian Journal of Microbiology 56(2): 138–44. doi: 10.1139/w09-122 [DOI] [PubMed] [Google Scholar]
- 8.Reeds PJ. (2000) Dispensable and indispensable amino acids for humans. J Nutr 130(7): 1835S–40S. [DOI] [PubMed] [Google Scholar]
- 9.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312(5778): 1355–1359. 312/5778/1355. doi: 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradberry SM, Proudfoot AT, Vale JA. (2004) Glyphosate poisoning. Toxicological Reviews 23(3): 159–67. [DOI] [PubMed] [Google Scholar]
- 11.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458(7236): 337–41. doi: 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brem RB, Kruglyak L. (2005) The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proceedings of the National Academy of Sciences of the United States of America 102(5): 1572–7. doi: 10.1073/pnas.0408709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315(5813): 848–53. doi: 10.1126/science.1136678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keinan A, Clark AG. (2012) Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336(6082): 740–3. doi: 10.1126/science.1217283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, et al. (2007) Genome sequencing and comparative analysis of saccharomyces cerevisiae strain YJM789. Proceedings of the National Academy of Sciences of the United States of America 104(31): 12825–30. doi: 10.1073/pnas.0701291104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. (2004) Finishing the euchromatic sequence of the human genome. Nature 431(7011): 931–45. doi: 10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- 17.Pollegioni L, Schonbrunn E, Siehl D. (2011) Molecular basis of glyphosate resistance-different approaches through protein engineering. The FEBS Journal 278(16): 2753–66. doi: 10.1111/j.1742-4658.2011.08214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge X, d'Avignon DA, Ackerman JJ, Sammons RD. (2010) Rapid vacuolar sequestration: The horseweed glyphosate resistance mechanism. Pest Manag Sci 66(4): 345–348. doi: 10.1002/ps.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman MM, Fojo T, Bates SE. (2002) Multidrug resistance in cancer: Role of ATP–dependent transporters. Nature Reviews Cancer 2(1): 48–58. doi: 10.1038/nrc706 [DOI] [PubMed] [Google Scholar]
- 20.Eschenburg S, Healy ML, Priestman MA, Lushington GH, Schönbrunn E. (2002) How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from escherichia coli. Planta 216(1): 129–135. doi: 10.1007/s00425-002-0908-0 [DOI] [PubMed] [Google Scholar]
- 21.Funke T, Han H, Healy-Fried ML, Fischer M, Schonbrunn E. (2006) Molecular basis for the herbicide resistance of roundup ready crops. Proceedings of the National Academy of Sciences of the United States of America 103(35): 13010–5. doi: 10.1073/pnas.0603638103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marbois B, Xie LX, Choi S, Hirano K, Hyman K, Clarke CF. (2010) Para-aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in saccharomyces cerevisiae. Journal of Biological Chemistry 285(36): 27827–27838. doi: 10.1074/jbc.M110.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, et al. (2002) Dissecting the architecture of a quantitative trait locus in yeast. Nature 416(6878): 326–30. doi: 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- 24.Leppert G, McDevitt R, Falco SC, Van Dyk TK, Ficke MB, Golin J. (1990) Cloning by gene amplification of two loci conferring multiple drug resistance in saccharomyces. Genetics 125(1): 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. (1994) PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. Journal of Biological Chemistry 296: 2206–2214. [PubMed] [Google Scholar]
- 26.Guan W, Jiang H, Guo X, Mancera E, Xu L, Xu L. (2010) Antagonistic changes in sensitivity to antifungal drugs by mutations of an important ABC transporter gene in a fungal pathogen. PloS One 5(6): e11309 doi: 10.1371/journal.pone.0011309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, et al. (2002) Functional profiling of the saccharomyces cerevisiae genome. Nature 418(6896): 387–91. doi: 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 28.Winston F, Dollard C, Ricupero-Hovasse SL. (1995) Construction of a set of convenient saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11(1): 53–5. doi: 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- 29.Regenberg B, Holmberg S, Olsen LD, Kielland-Brandt MC. (1998) Dip5p mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate in saccharomyces cerevisiae. Curr Genet 33(3): 171–177. [DOI] [PubMed] [Google Scholar]
- 30.Hatakeyama R, Kamiya M, Takahara T, Maeda T. (2010) Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol 30(24): 5598–5607. doi: 10.1128/MCB.00464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. (2010) Genetic analysis of variation in transcription factor binding in yeast. Nature 464(7292): 1187–91. doi: 10.1038/nature08934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, et al. (2008) The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science 320(5874): 362–365. doi: 10.1126/science.1150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sirisattha S, Momose Y, Kitagawa E, Iwahashi H. (2004) Genomic profile of roundup treatment of yeast using DNA microarray analysis. Environ Sci 11(6): 313–23. [PubMed] [Google Scholar]
- 34.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441(7095): 840–846. doi: 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- 35.Madsen HL, Christensen H, Gottlieb-Petersen C. (1978) Stability constants of copper (II), zinc, manganese (II), calcium, and magnesium complexes of N-(phosphonomethyl) glycine (glyphosate). Acta Chem Scand 32: 79–83. [Google Scholar]
- 36.Cakmak I, Yazici A, Tutus Y, Ozturk L. (2009) Glyphosate reduced seed and leaf concentrations of calcium, manganese, magnesium, and iron in non-glyphosate resistant soybean. Eur J Agron 31(3): 114–119. [Google Scholar]
- 37.O'Donnell AF, Huang L, Thorner J, Cyert MS. (2013) A calcineurin-dependent switch controls the trafficking function of alpha-arrestin Aly1/Art6. J Biol Chem 288(33): 24063–24080. doi: 10.1074/jbc.M113.478511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein AL, McCusker JH. (1999) Three new dominant drug resistance cassettes for gene disruption in saccharomyces cerevisiae. Yeast 15(14): 1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 39.Wenger JW, Schwartz K, Sherlock G. (2010) Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from saccharomyces cerevisiae. PLoS Genetics 6(5): e1000942 doi: 10.1371/journal.pgen.1000942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gietz RD, Schiestl RH. (2007) Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2(1): 1–4. doi: 10.1038/nprot.2007.17 [DOI] [PubMed] [Google Scholar]
- 41.Rong-Mullins X, Winans MJ, Lee JB, Lonergan ZR, Pilolli VA, Weatherly LM, et al. (2017) Proteomic and genetic analysis of S. cerevisiae response to soluble copper leads to improvement of antimicrobial function of cellulosic copper nanoparticles. Metallomics 9(9) 1304–1315. doi: 10.1039/c7mt00147a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collart MA, Oliviero S. (2001) Preparation of yeast RNA. Current Protocols in Molecular Biology. John Wiley & Sons, Inc. doi: 10.1002/0471142727.mb1312s23 [DOI] [PubMed] [Google Scholar]
- 43.Gallagher JE, Zheng W, Rong X, Miranda N, Lin Z, Dunn B, et al. (2014) Divergence in a master variator generates distinct phenotypes and transcriptional responses. Genes & Development 28(4): 409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christie KR, Hong EL, Cherry JM. (2009) Functional annotations for the saccharomyces cerevisiae genome: The knowns and the known unknowns. Trends in Microbiology 17(7): 286–94. doi: 10.1016/j.tim.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Schematic of enzyme functions in Aro1 with bacterial proteins AroB (3-dehydroquinate synthase amino acids 1–392), AroA (EPSPS amino acids 404–861), AroL (shikimate kinase amino acids 887–1060), AroD (3-dehydroquinase amino acids 1061–1295), and AroE (shikimate dehydrogenase amino acids 1306–1599). B. Alignment of the ESPS glyphosate binding site across different species. In red are residues that when mutated confer resistance to glyphosate in E. coli. C. RNA expression levels of ARO1 mRNA from RM11, YJM789 and YJM789 aro1Δ carrying different alleles ARO1 grown in YM with and without 0.25% glyphosate. Q RT-PCR mRNA of ARO1 levels are normalized to 25S rRNA. D. Tetrad dissections of RM11 heterozygous knockout of ARO1 compared to wild-type RM11 diploid on YPD. Tetrads were numbered and haploid segregant germinating from a single spore are lettered. Plates were incubated at 30°C for two days. E. Tetrad dissections of RM11 wildtype and S288c aro1Δ hybrids (F1) were incubated for five days before being photographed. Haploid segregants from F1 yeast with aro1Δ were circled.
(TIF)
Branch length was determined by UPGMA in ClustalW. Relative growth of yeast on glyphosate was normalized to growth with no treatment.
(TIF)
Branch length was determined by UPGMA in ClustalW. Relative growth of yeast on glyphosate was normalized to growth with no treatment.
(TIF)
ARO1 was knocked out in S288c and YJM789. PDR5 was cloned and expressed from its native promoter from a plasmid. Yeast were grown on YPD (rich media) with 1% glyphosate, YM (minimal media) with 0.25% glyphosate and WYF (yeast minimal media supplemented with aromatic amino acids) with 0.25% glyphosate.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.